Summary
Mutations in leucine-rich repeat kinase 2 (LRRK2) are a common cause of familial and sporadic Parkinson's disease (PD). Elevated LRRK2 kinase activity and neurodegeneration are linked, but the phosphosubstrate that connects LRRK2 kinase activity to neurodegeneration is not known. Here, we show that ribosomal protein s15 is a key pathogenic LRRK2 substrate in Drosophila and human neuron PD models. Phospho-deficient s15 carrying a threonine 136 to alanine substitution rescues dopamine neuron degeneration and age-related locomotor deficits in G2019S LRRK2 transgenic Drosophila and substantially reduces G2019S LRRK2-mediated neurite loss and cell death in human dopamine and cortical neurons. Remarkably, pathogenic LRRK2 stimulates both cap-dependent and cap-independent mRNA translation, and induces a bulk increase in protein synthesis in Drosophila, which can be prevented by phospho-deficient T136A s15. These results reveal a novel mechanism of PD pathogenesis linked to elevated LRRK2 kinase activity and aberrant protein synthesis in vivo.
Introduction
Following the identification of pathogenic LRRK2 mutations that lead to PD in 2004 (Paisan-Ruiz et al., 2004; Zimprich et al., 2004), LRRK2 mutations are now known to be the most common genetic cause of PD, accounting for up to 40% of familial cases in certain populations (Martin et al., 2011). LRRK2 mutations result in clinical and pathological features that closely resemble the more common sporadic PD, suggesting that understanding LRRK2-linked disease mechanisms may be a gateway to understanding sporadic PD. Multiple lines of evidence indicate that LRRK2 kinase activity is key to PD development, particularly the findings that (i) the common G2019S mutation bestows increased kinase activity towards generic kinase substrates (Anand et al., 2009; Covy and Giasson, 2009; Greggio et al., 2006; Luzon-Toro et al., 2007; Smith et al., 2006; West et al., 2007) and (ii) LRRK2 toxicity is kinase-dependent (Deng et al., 2011; Greggio et al., 2006; Lee et al., 2010; MacLeod et al., 2006; Smith et al., 2006). To understand the link between LRRK2 kinase activity and PD development, it is imperative to identify authentic LRRK2 substrates that are linked to neurodegeneration in PD (Tsika and Moore, 2012).
LRRK2 is a large multidomain protein with several protein-protein interaction domains and a catalytic core containing GTPase, COR (C-terminal of ROC) and kinase domains where most PD-linked mutations are found. It is likely that its physiologic and pathophysiologic functions are mediated through protein-protein interactions and/or phosphorylation of LRRK2 substrates (Cookson, 2010). A number of candidate LRRK2 substrates have been identified. LRRK2 kinase activity is part of an endophilin A phosphorylation cycle that promotes efficient synaptic vesicle endocytosis at the neuromuscular junction (Matta et al., 2012). Phosphorylation of ezrin/radixin/moesin by LRRK2 promotes the rearrangement of the actin cytoskeleton in neuronal morphogenesis (Parisiadou et al., 2009). LRRK2 phosphorylation of eukaryotic initiation factor 4E (eIF4E)-binding protein (4E-BP) has been suggested to couple increased LRRK2 kinase activity to aberrant translation of a small number of mRNAs through 4E-BP-dependent perturbation of microRNA activity (Gehrke et al., 2010; Imai et al., 2008). However, while this study raises the possibility that LRRK2 may affect translation at some level, a mechanistic link involving phospho-4E-BP is lacking as there was no direct demonstration that LRRK2 phosphorylation of 4E-BP mediates altered translation of these mRNAs. Moreover, recent studies suggest that 4E-BP may not be an important in vivo LRRK2 substrate (Kumar et al., 2010; Trancikova et al., 2012) raising additional doubt on the role of 4E-BP as a kinase substrate important in LRRK2 toxicity. Thus, how elevated LRRK2 kinase activity is coupled to aberrant mRNA translation and neurodegeneration in PD remains to be clarified.
In order to understand the connection between LRRK2 kinase activity and neurotoxicity, candidate LRRK2 substrates were identified through LRRK2 tandem affinity purification and in vitro kinase screening of LRRK2-interacting phosphoproteins. We find that ribosomal proteins are major LRRK2 interactors and LRRK2 kinase targets, and that LRRK2 is markedly enriched in the ribosomal subcellular fraction. Blocking phosphorylation of the small ribosomal subunit protein s15 rescues LRRK2 neurotoxicity in human dopamine neurons and Drosophila PD models. We demonstrate that pathogenic LRRK2 induces an increase in bulk protein synthesis in flies, which is blocked by phospho-deficient s15. Moreover, the global protein synthesis inhibitor anisomycin rescues the locomotor deficits and dopamine neuron loss in aged G2019S LRRK2 transgenic Drosophila. Our findings identify s15 as a key pathogenic LRRK2 substrate that links elevated LRRK2 kinase activity to Parkinson's disease pathogenesis via altered translation.
Results
Identifying candidate LRRK2 kinase substrates
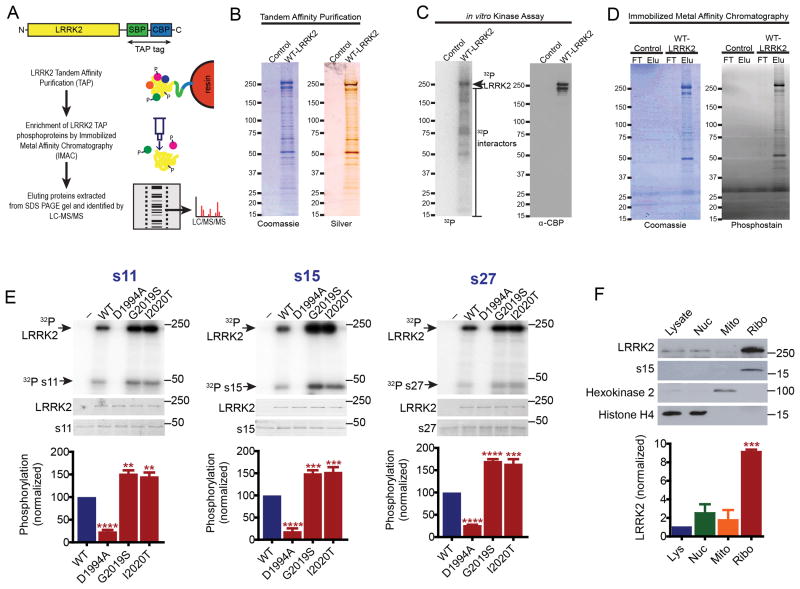
LRRK2-interacting phosphoproteins were identified by tandem affinity purification (TAP) of LRRK2 from HEK293 cells. A LRRK2 TAP tag was generated by fusing a streptavidin binding peptide and a calmodulin binding peptide to the C-terminus of LRRK2. TAP-tagged LRRK2 complex retains kinase activity (Fig. 1A-C). LRRK2-interacting phosphoproteins were enriched by immobilized metal affinity chromatography (IMAC) (Fig. 1D) and then separated via SDS-PAGE. The gel was divided into multiple evensized pieces and protein bands were identified by mass spectrometry. To control for non-specific binding, proteins identified by TAP of two unrelated proteins, Botch and GADD45ß were subtracted from the mass spectrometry data set for LRRK2, leaving 161 interacting proteins (Table S1). A Cytoscape network analysis reveals that the major LRRK2 interacting proteins identified by this approach are functionally categorized into four major groups; structural constituents of the ribosome, proteins involved in transporter activity, nucleotide binding and cation binding (Fig. S1A). To determine which of these interacting proteins are direct LRRK2 substrates, GST-fusion proteins were generated by Gateway cloning and screened via an in vitro LRRK2 kinase assay to assess phosphorylation in the presence of wild type LRRK2, kinase-dead LRRK2 (D1994A), and two disease-causing variants with kinase domain mutations, G2019S LRRK2 and I2020T LRRK2 (Fig. S1B). Approximately 60% of all GST-fusion proteins were amenable to purification (Table S1). 11 proteins are phosphorylated by wild type, G2019S and I2020T LRRK2, but not by kinase-dead D1994A LRRK2. 10 candidate substrates are ribosomal proteins and the other protein is lactate dehydrogenase B (LDHB) (Figs. 1E and S1C). Consistent with our finding that LRRK2 interacts with and phosphorylates ribosomal proteins, endogenous LRRK2 is highly enriched in the ribosomal subcellular fraction suggesting that LRRK2 might play an important role in ribosomal function and mRNA translation (Fig. 1F). Similar levels of enrichment are observed for overexpressed wild type, D1994A and G2019S LRRK2 (data not shown), suggesting that the physical association of LRRK2 with ribosomes is not kinase-dependent. To further determine the scope of LRRK2 actions at the ribosome, all known ribosomal proteins belonging to the human 40S and 60S subunits amenable to purification were subjected to in vitro LRRK2 kinase assays (Table S2). A total of 19 ribosomal proteins are phosphorylated by LRRK2 (Table S2). Three substrates belonging to the 40S ribosomal subunit, s11, s15 and s27, exhibit significantly increased phosphorylation by the pathogenic mutants G2019S and I2020T LRRK2 (Fig. 1E). We reasoned that substrates exhibiting elevated phosphorylation with pathogenic LRRK2 variants might be involved in LRRK2 toxicity. To identify phosphorylation sites on these 3 ribosomal proteins, tandem mass spectrometry analysis was performed following LRRK2 phosphorylation in vitro (Fig. S2A-B). LRRK2 phosphorylates threonine 136 of s15 and threonines 28, 46 and 54 of s11 (Fig. S2A-B). We are unable to detect phosphorylation of s27 by LRRK2 via mass spectrometry (data not shown).
Figure 1. Identification of candidate LRRK2 substrates.
(A), cartoon of LRRK2 TAP construct with C-terminal streptavidin binding peptide (SBP) and calmodulin binding peptide (CBP) tags and scheme for identifying LRRK2-interacting phosphoproteins. (B), LRRK2 TAP co-purified proteins visualized by coomassie and silver stained polyacrylamide gels. Control is empty vector. (C), phosphorimaging of LRRK2 complex purified by TAP demonstrating that it was kinase active. (D), IMAC enrichment of phosphoproteins from TAP eluate. FT is flow-through, Elu is eluate. (E), LRRK2 kinase substrates exhibiting significantly increased phosphorylation (γ-32P-ATP incorporation) by G2019S and I2020T LRRK2 compared to WT LRRK2 (individual oneway ANOVAs for effect of LRRK2 variant, p<0.0001, Bonferroni's post-test, ** p<0.01, *** p<0.001, **** p<0.0001, n=3-7). Coomassie brilliant blue-stained gels are shown. (F), Endogenous LRRK2 levels in HEK293 cell whole lysates, nuclear (Nuc), mitochondrial (Mito) or ribosomal (Ribo) subcellular fractions. Endogenous LRRK2 is enriched in ribosomal fractions (ANOVA followed by Bonferroni's post-test, *** p < 0.001, n = 3). Data are mean ± SEM. See also Figure S1 and Table S1 and Table S2.
s15 is a pathogenic LRRK2 substrate in human dopamine neurons
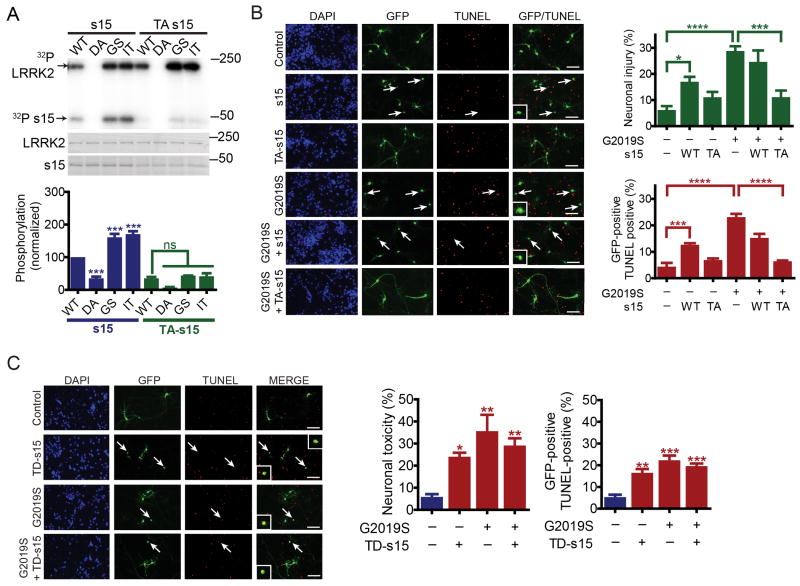
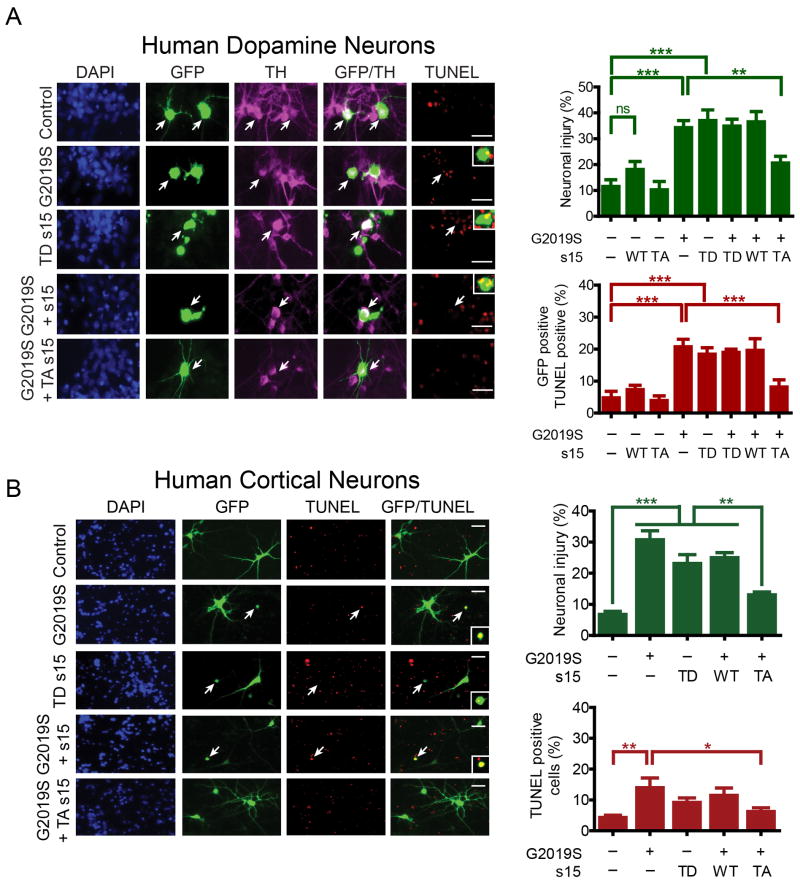
A substitution of threonine 136 to alanine in s15 significantly reduces wild type, G2019S and I2020T LRRK2 phosphorylation of s15 (Fig. 2A). A triple mutation of threonines 28, 46 and 54 to alanines in s11 eliminates phosphorylation of s11 by wild type, G2019S and I2020T LRRK2 (Fig. S2C). To ascertain whether s15 or s11 phosphorylation are required for LRRK2 toxicity, the effects of T136A s15 and triple mutant (T28A, T46A, T54A) s11 were examined. G2019S LRRK2 has been repeatedly shown to cause kinase-dependent neuronal toxicity characterized by neurite loss and cell death in rodent and human neurons (Greggio et al., 2006; Lee et al., 2010; Ramsden et al., 2011; Smith et al., 2006; West et al., 2007). Phospho-deficient T136A s15 is markedly protective against G2019S LRRK2 toxicity (neurite loss and cell death) whereas triple mutant s11 failed to influence LRRK2 toxicity in rat cortical neurons (Figs. 2B and S2C). Overexpression of s15 is modestly toxic to rat cortical neurons but overexpression of T136A s15, s11 or triple mutant s11 are not (Fig. 2B and S2C). T136A s15 is additionally protective against the kinase domain mutant I2020T LRRK2, but not the pathogenic ROC domain mutant R1441C LRRK2 (Fig. S2D). Coexpression of wild type s15 did not substantially influence toxicity for any of the LRRK2 variants tested. To further explore the role of s15 phosphorylation in LRRK2 toxicity, the effect of phosphomimetic T136D s15 was examined. T136D s15 alone is sufficient to induce neuronal toxicity and when expressed together with G2019S LRRK2, did not exacerbate LRRK2 toxicity (Fig. 2C) consistent with G2019S LRRK2 toxicity being mediated via s15 phosphorylation. To extend our investigation and probe the role of s15 in human neurotoxicity caused by G2019S LRRK2, human midbrain dopamine neurons and cortical neurons were derived from human embryonic stem cells and characterized (Figs. S3A and B). G2019S LRRK2 causes neurotoxicity in human midbrain dopamine neurons (Fig. 3A) and in human cortical neurons (Fig. 3B), where it results in significantly elevated endogenous phospho-T136 s15 levels (Fig. S3C), observed using a validated phospho-specific antibody to phospho-T136 (Fig. S3D-F). LRRK2 toxicity in both human dopamine and cortical neurons is significantly attenuated by T136A s15 but not wild type s15. Phospho-mimetic T136D s15 is toxic to both types of neurons (Figs. 3A and B). Partial knock-down (~50%) of s15 in human cortical neurons partially rescues neuronal toxicity caused by G2019S LRRK2, similar to that observed for T136A s15 (Fig. S3G), while expression of G2019S LRRK2 is not affected (Fig. S3G). Finally, increasing wild-type LRRK2 overexpression leads to a modest increase in s15 phosphorylation and neuronal toxicity, but not to the extent observed via G2019S LRRK2 (Fig. S3H and I). Taken together, these data reveal that s15 is a pathogenic LRRK2 substrate for LRRK2 mutations within the kinase domain of LRRK2 and that LRRK2 and s15 act in the same cell death pathway.
Figure 2. Phospho-deficient s15 protects against LRRK2 toxicity.
(A), LRRK2 in vitro kinase assay. T136A (TA) s15 reduces phosphorylation by wild type (WT), D1994A (DA), G2019S (GS) and I2020T (IT) LRRK2 (two-way ANOVA, p<0.0001 for effect of T136A mutation, n=3), and prevents the increase in s15 phosphorylation via G2019S and I2020T LRRK2 (Bonferroni's post-test, *** p <0.001; ns, not significant). (B), T136A (TA) s15 blocks G2019S LRRK2 toxicity (neurite shortening and cell death) in rat cortical neurons (individual ANOVAs, Bonferroni's post-tests, * p<0.05, *** p<0.001, **** p<0.0001, n =5). Arrows indicate neurons lacking neurites, a subset of which are TUNEL-positive (see inset magnifications) as indicated. The vast majority of GFP-positive neurons (~90%) co-labeled for LRRK2 and s15 when co-transfected (not shown). (C), phospho-mimetic T136D (TD) s15 mimics G2019S LRRK2 toxicity (individual ANOVAs, Bonferroni post-test, * p<0.05, ** p<0.01, *** p<0.001, n =3). Scale bar, 25 µM. Data are mean ± SEM. See also Figure S2.
Figure 3. Phospho-deficient s15 protects against G2019S LRRK2 toxicity in human dopamine and cortical neurons.
G2019S LRRK2 toxicity (neurite shortening and cell death) in human midbrain dopamine neurons (A) (white arrows) and human cortical neurons (B) is phenocopied by phospho-mimetic T136D (TD) s15 and rescued by phospho-mutant T136A (TA) s15 but not wild type s15. Individual ANOVAs, Bonferroni's post-test, * p<0.05, ** p<0.01, *** p<0.001, n = 4). Scale bar, 25 µM. Data are mean ± SEM. See also Figure S3.
s15 is an authentic LRRK2 substrate in vivo
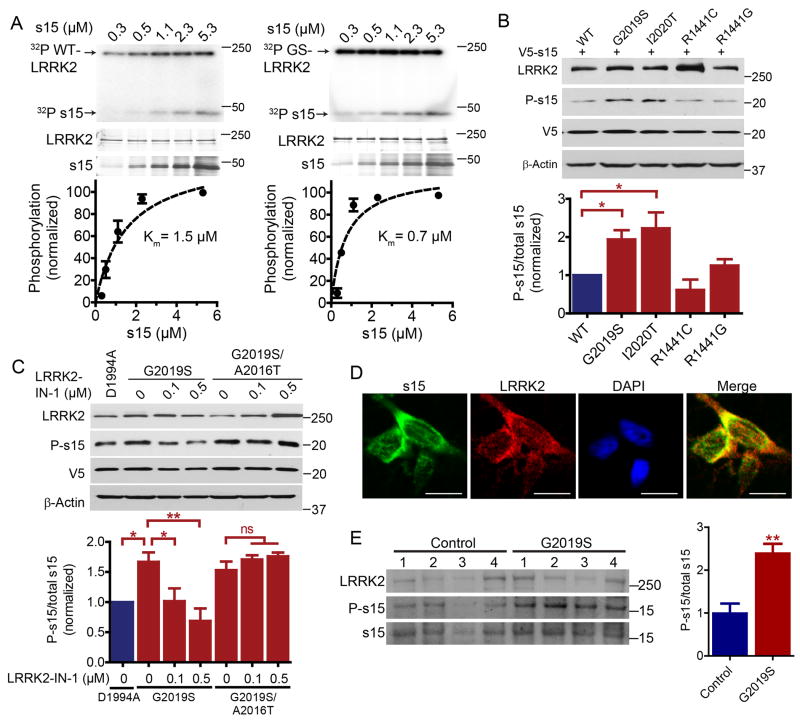
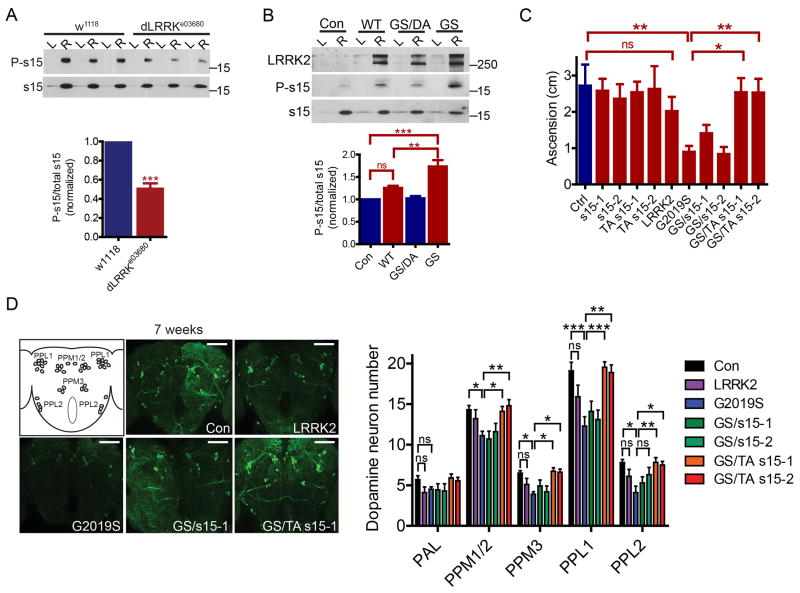
Since phosphorylation of s15 by LRRK2 was found to be required for LRRK2 neurotoxicity, the interaction and phosphorylation of s15 by LRRK2 was further investigated. Recombinant s15 is phosphorylated by wild type LRRK2 with a Km of 1.5 µM and by G2019S LRRK2 with a Km of 0.7 µM, suggesting a stronger substrate affinity between s15 and G2019S LRRK2 compared to that with wild type LRRK2 (Fig. 4A). G2019S and I2020T LRRK2 both increase phospho-s15 levels in cells whereas overexpression of the ROC domain mutants R1441C or R1441G do not (Fig. 4B). These phosphorylation results are consistent with the ability of the T136A s15 mutant to rescue G2019S and I2020T LRRK2 neurotoxicity but not R1441C neurotoxicity (see Figs. 2 and S2). LRRK2-IN-1 and CZC-25146, two potent LRRK2 kinase inhibitors with non-overlapping off-target kinase inhibition (Deng et al., 2011; Ramsden et al., 2011) block G2019S LRRK2 phosphorylation of s15 both in kinase assays in vitro and in cell culture (Figs. 4C, S4A and B). The G2019S/A2016T LRRK2 variant is resistant to LRRK2-IN-1 kinase inhibition, and phosphorylation of s15 by this LRRK2 variant is unaffected by LRRK2-IN-1 treatment (Fig. 4C), further supporting that reduced s15 phosphorylation is caused specifically by LRRK2 kinase inhibition. Endogenous LRRK2 and s15 both exhibit punctate immunostaining with partial perinuclear co-localization in human cortical neurons (Figs. 4D and S4C), which is also found for overexpressed G2019S LRRK2 and s15 (Fig. S4C). Endogenous s15 and LRRK2 co-immunoprecipitate and s15 interacts with the WD40 protein interaction domain of LRRK2 (Fig. S4D). Post-mortem cortex from human G2019S carriers exhibit increased s15 phosphorylation in ribosomal fractions (Fig. 4E). LRRK2 expression was not significantly different in G2019S carrier ribosomal fractions or whole lysates compared to control patient brain samples (Figs. 4E and S4F). Finally, ribosomal fractions from dLRRK (Drosophila LRRK2 homolog) null fly heads exhibit significantly reduced s15 phosphorylation (Fig. 5A) whereas phospho-s15 is significantly increased in head ribosomal fractions from G2019S transgenic flies but not kinase-dead G2019S/D1994A LRRK2 flies (Fig. 5B). As in human cells, endogenous s15 and dLRRK exhibit partial colocalization around the nucleus of Drosophila S2 cells (Fig. S4G). These data collectively demonstrate that s15 is an authentic and direct in vivo LRRK2 kinase substrate.
Figure 4. Phosphorylation of s15 by pathogenic LRRK2 variants and block by LRRK2-IN-1.
(A), Kinetics of LRRK2 or G2019S LRRK2 initial enzyme velocity at various s15 concentrations for derivation of the Michaelis-Menten constant, Km. Data are mean ± SEM, n =3. Silver stained gels are shown. (B), Phospho-s15 levels following co-transfection of HEK293 cells with V5-s15 and LRRK2. Densitometry revealed a significant effect of G2019S and I2020T LRRK2 variants on phospho-s15 levels (ANOVA followed by Bonferroni post-test, * p<0.05, n =5). (C), LRRK2-IN-1 blocked V5-s15 phosphorylation by G2019S LRRK2 but not a drug-resistant variant, G2019S/A2016T LRRK2 in co-transfected HEK293 cells (ANOVA, Bonferroni's posttest, * p<0.05, ** p<0.01, n =3). (D), endogenous LRRK2 and s15 in human cortical neurons exhibit punctate immunostaining, predominantly in the perinuclear region where they colocalize. Scale bars, 10 µM. (E), phospho-s15 is increased in ribosomal fractions from G2019S LRRK2 human post-mortem brains (Student's t-test, ** p<0.01). Whole lysates were run separately (see Fig. S4) due to gel space constraints. Data are mean ± SEM. See also Figure S4.
Figure 5. Rescue of dopamine neuron degeneration and locomotor dysfunction in aged G2019S LRRK2 Drosophila by phospho-mutant s15.
(A), Phospho-s15 is reduced in dLRRKe03680 (dLRRK null) homozygotes compared to isogenic w1118 controls (Student's t-test, p<0.001, n = 4 groups of 100 fly heads/genotype). L, whole lysates; R, ribosomal fractions (B), G2019S LRRK2 transgenic flies (Ddc-Gal4; UAS-G2019S-LRRK2) exhibit increased phospho-s15 (ANOVA, Bonferroni's post-test, ** p<0.01; *** p <0.001; ns, not significant, n = 4 groups of 100 fly heads/genotype). L, whole lysates; R, ribosomal fractions. LRRK2 exhibits an additional high molecular weight band (likely SDS-insoluble LRRK2). (C), Aged flies expressing G2019S LRRK2 exhibit a significant locomotor deficit in negative geotaxis assays, which is rescued by T136A (TA) s15 expression (ANOVA followed by Bonferroni's post-test, * p<0.05, ** p<0.01, n =30-40 flies). Genotypes are Ddc-Gal4/+; +/+ (Control), UAS-s15/+; +/+ (s15), UAS-T136A s15/+; +/+ (TA s15), Ddc-Gal4/+; UAS-LRRK2/+ (LRRK2), Ddc-Gal4/+; UAS-G2019S-LRRK2/+ (G2019S), Ddc-Gal4/UAS-s15; UAS-G2019S-LRRK2/+ (GS/s15) and Ddc-Gal4/UAS-T136A s15; UAS-G2019S-LRRK2/+ (GS/TA s15). (D), dopamine neuron loss in aged G2019S LRRK2 flies is rescued by T136A s15 co-expression. Confocal projection images through the posterior fly brain show five major dopamine neuron clusters (PPM1, PPM2, PPM3, PPL1, PPL2). Scale bar, 60 µM. For quantitation, individual ANOVAs followed by Bonferroni's post-test, * p<0.05, ** p<0.01 *** p<0.001, n =10 fly brains per genotype. Genotypes are as in (C). Data are mean ± SEM. See also Figure S5.
Phospho-deficient s15 blocks neurodegeneration in G2019S LRRK2 transgenic Drosophila
To determine whether s15 phosphorylation underlies LRRK2-dependent neurodegeneration and PD-related phenotypes in vivo, we used the robust and efficient Drosophila LRRK2 transgenic model in which flies expressing human G2019S LRRK2 rapidly exhibit aging-related dopaminergic neurodegeneration and locomotor deficits linked to increased LRRK2 kinase activity (Liu et al., 2008). Dopamine neuron degeneration and locomotor dysfunction observed in aged G2019S LRRK2 transgenic Drosophila are completely rescued by co-expression of T136A s15 whereas wild type s15 is not protective (Figs. 5C and D and S5A). Neither s15 nor T136A s15 co-expression affects G2019S LRRK2 levels (Fig S5B). There were no effects of LRRK2, s15 or T136A s15 overexpression observed in young flies in these assays, consistent with previous reports for LRRK2 transgenic flies at this age (Liu et al., 2008) (Figs. S5C and D). Also, neither s15 nor T136A s15 expression alone significantly affects negative geotaxis or dopamine neuron survival in aged flies, although there was a slight reduction in climbing ability with s15 overexpression (Figs. S5E and F). s15 is an integral ribosomal protein and knocking-down s15 by RNAi is lethal in Drosophila, via whole-body or targeted neuronal expression (data not shown).
LRRK2 stimulates cap-dependent and cap-independent translation
The role of s15 in LRRK2 toxicity raises the possibility that pathogenic LRRK2 might modulate the activity of ribosomes to impact protein synthesis in an s15-dependent manner. Although little is known about the role of s15 in translation, it is located on the surface of the 40S ribosomal subunit and its C-terminal tail (where T136 lies) is highly conserved among eukaryotes and may extend into the ribosomal decoding site during mRNA translation (Khairulina et al., 2010; Pisarev et al., 2006). mRNA translation can occur by both cap-dependent and cap-independent mechanisms of ribosome binding. To simultaneously probe the effects of LRRK2 on cap-dependent and cap-independent translation, a bicistronic reporter composed of a FLAG readout to monitor cap-dependent translation followed by an IRES site with a c-Myc readout to monitor cap-independent translation from the same transcript was utilized (Fig. 6A). G2019S LRRK2 dose-dependently increases both cap-dependent and cap-independent reporter translation in a kinase-dependent manner (Fig. 6B), while reporter mRNA levels are not significantly affected by LRRK2 expression (Fig. S6A). Wild type s15 stimulates both cap-dependent and cap-independent reporter translation in a dose-dependent manner (Fig. 6C). Importantly, these effects are augmented by the phosphomimetic T136D mutation and subtly decreased by T136A s15, without significant changes in reporter transcript levels (Fig. 6C and S6A). To determine whether s15 phosphorylation mediates the effects of LRRK2 on translation, cap-dependent and cap-independent reporter translation induced by G2019S LRRK2 was monitored in the presence of wild type s15 or T136A s15. Wild type s15 modestly enhances the stimulation of translation by G2019S LRRK2, whereas T136A s15 blocked stimulation of both cap-dependent and cap-independent translation by G2019S LRRK2 (Fig. 6D). Reporter mRNA levels are not significantly different between the groups (Fig. 6D). Partial knock-down of s15 similarly attenuates cap-dependent and cap-independent reporter translation (Fig. S6B) further supporting the link between s15 and LRRK2 effects on translation. LRRK2 was previously reported to phosphorylate human 4E-BP1 and it was suggested that phospho-4E-BP1 affects the expression of certain genes by repressing miRNA activity (Gehrke et al., 2010). As expected, knock-down of dicer led to reduced miRNA levels (data not shown) but failed to stimulate bicistronic reporter expression (Fig. S6C) suggesting that G2019S LRRK2 does not promote reporter translation by impairing the miRNA pathway. Similarly, overexpressing hAgo2, which was proposed to be functionally impeded by phospho-4E-BP binding, did not attenuate cap-dependent or cap-independent reporter expression (Fig. S6D). Since phosphorylation of 4E-BP blocks its ability to bind to and repress eIF4E, it is possible that elevated levels of eIF4E could stimulate translation upon an increase in phospo-4E-BP (Imai et al., 2008). However, while 4E-BP overexpression was previously reported to rescue loss of dopamine neurons observed in flies expressing pathogenic dLRRK variants (Imai et al., 2008), it did not reduce G2019S LRRK2-mediated reporter expression in cells (Fig. S6D). Moreover, we did not detect an increase in phospho-4E-BP1 levels following G2019S LRRK2 expression in cells (Fig. S6E) or G2019S LRRK2 transgenic fly heads (Fig. S6F), consistent with other subsequent studies (Kumar et al., 2010; Trancikova et al., 2012) that failed to find an increase in 4E-BP1 phosphorylation in mammalian cells expressing pathogenic LRRK2 variants or LRRK2 transgenic mouse brain. Additionally, an increase in 4E-BP phosphorylation would not account for an increase in cap-independent translation since neither eIF4E nor 4E-BP1 are known to be involved in this mode of translation. Finally, partial loss of eIF4E expression did not block neurodegenerative phenotypes (age-related loss of dopamine neurons and locomotor deficits) observed in aged G2019S transgenic flies (Figs. S6G-I). Collectively, these results suggest that miRNA pathway impairment or elevated free eIF4E levels via 4E-BP1 phosphorylation does not play a major role in LRRK2-induced translation.
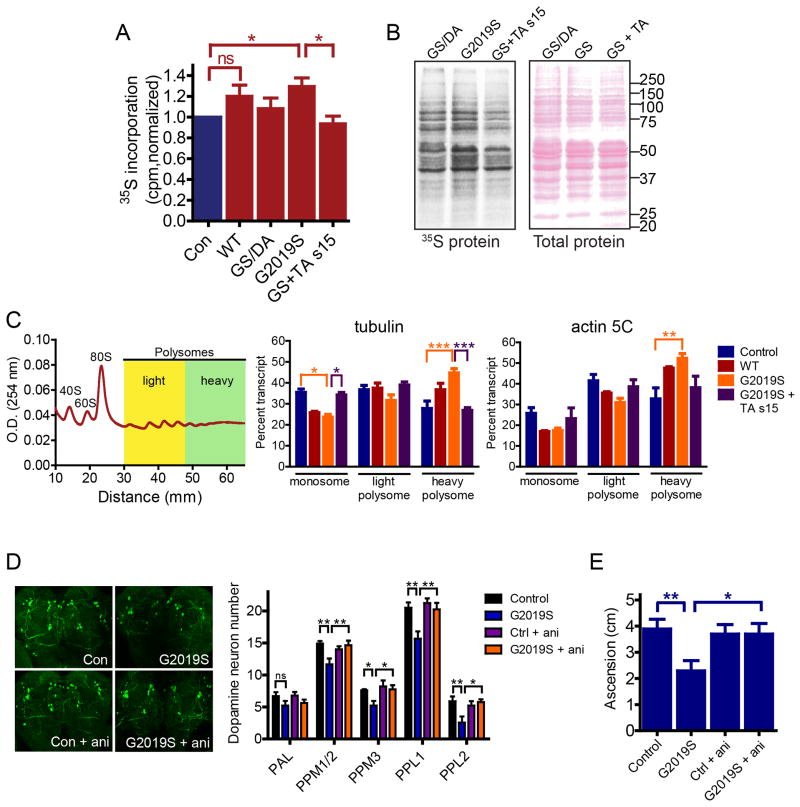
Figure 6. G2019S LRRK2 stimulates cap-dependent and cap-independent translation.
(A), bicistronic reporter assay for assessing effects of LRRK2 on cap-dependent and cap-independent translation. (B), G2019S LRRK2 stimulates cap-dependent (FLAG-GFP) and cap-independent (c-Myc-GFP) bicistronic reporter translation in human neuroblastoma (SH-SY5Y) cells (individual two-way ANOVAs, effect of LRRK2 dose, p ≤ 0.0075, and genotype, p < 0.002, n =3), in a kinase-dependent manner vs. G2019S/D1994A (GS/DA) (Bonferroni's post-tests, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, n =3). (C), V5-s15 stimulates cap-dependent and cap-independent reporter translation which is reduced by T136A and potentiated by T136D mutations (individual two-way ANOVAs, effect of s15 variant, p ≤ 0.0073, effect of dose, p ≤ 0.0099, n=3). (D), G2019S LRRK2-stimulated cap-dependent (c.d.) and cap-independent (c.i.) reporter translation is attenuated by V5-T136A s15 (individual ANOVAs, p ≤ 0.0014, Bonferroni's post-test, * p <0.05, ** p<0.01, n =8) while bicistronic reporter mRNA was not significantly different (ANOVA, ns, n=8). (E), in human cortical neurons, G2019S LRRK2-stimulated reporter expression, toxicity and cell death are rescued by T136A s15 co-expression (individual ANOVA, Bonferroni's post-test, * p <0.05, *** p <0.001, n =4). Scale bar, 25 µM. Data are mean ± SEM. See also Figure S6.
Reporter expression in human ES cell-derived cortical neurons is stimulated in G2019S LRRK2-expressing neurons and this is blocked by T136A s15 coexpression (Fig. 6E). If the stimulatory effect of G2019S LRRK2 on translation underlies its neuronal toxicity, then it might be expected that reporter-positive neurons would exhibit disproportionately high levels of toxicity. Indices of translation-linked neuronal injury and translation-linked cell death in reporter-positive neurons indicate that cell injury and death are coupled to increases in translation (Fig. 6E). G2019S LRRK2 stimulates reporter translation and increases both toxicity indices by almost 3-fold compared to kinase-dead G2019S/D1994A LRRK2 in human cortical neurons (Fig. 6E). This effect is blocked by T136A s15 coexpression. Comparison of reporter-positive neurons with neighboring reporter-negative neurons reveals 5-fold higher levels of neurite toxicity in reporter-positive neurons, and a similar difference in levels of cell death under all conditions, further suggesting a link between translation stimulation and LRRK2 toxicity (Fig. S6J). Taken together, these results suggest that phosphorylation of s15 on T136 by LRRK2 mediates enhanced cap-dependent and cap-independent reporter translation and that a stimulatory effect of LRRK2 on mRNA translation contributes to LRRK2 toxicity.
G2019S LRRK2 increases bulk translation in Drosophila
From our in vitro reporter assays, we hypothesized that pathogenic LRRK2 may cause a bulk shift in translation that, as shown in other neurodevelopmental and neurodegenerative diseases, can impair neuronal function. Consistent with our in vitro observations, 35S-Methionine/Cysteine pulse labeling of newly synthesized protein was significantly increased in G2019S LRRK2 transgenic Drosophila heads (Fig. 7A), suggesting an increase in protein synthesis rates by LRRK2. In accordance with its ability to block LRRK2 toxicity, T136A s15 co-expression abolishes this increase (Fig. 7A). SDS-PAGE analysis of lysates indicates a bulk increase in many proteins across a large range of molecular weights by G2019S LRRK2 (Fig. 7B). Assessment of housekeeping gene mRNAs undergoing translation in monosome and polysome fractions from fly heads reveals that G2019S LRRK2 expression leads to an enrichment of mRNAs in heavy polysome fractions, an effect which is attenuated by T136A s15 (Fig 7C). This increase in density of ribosomes associated with mRNA indicates that LRRK2 stimulates mRNA translation and that an increased rate of protein synthesis underlies the augmented protein levels observed via 35S-methionine/cysteine labeling. Increased ribosomal density typically indicates an increase in translation initiation, but could theoretically signal a slower rate of ribosomal elongation or release upon termination. Reduced elongation/termination rates are not consistent with an increase in protein production, however, and G2019S LRRK2 did not affect rates of ribosomal runoff following treatment of cells with harringtonine, a drug which prevents translocation of ribosomes engaged in initiation but not elongation (Fig S7A-D). Hence, these data are consistent with an effect of LRRK2 on initiation, the rate-limiting step in translation. To independently test whether a bulk increase in protein synthesis underlies G2019S LRRK2 toxicity, flies were treated with the global protein synthesis inhibitor anisomycin and chronic low-dose anisomycin treatment throughout adulthood rescued locomotor deficits and dopamine neuron loss in aged G2019S LRRK2 transgenic Drosophila, whereas no effects of the drug were seen in control flies (Fig. 7D and E). Anisomycin treatment did not affect LRRK2 expression levels (Fig. S7E) or s15 phosphorylation (Fig. S7F) but did reduce the increase in translation observed in G2019S LRRK2 transgenic flies (Fig. S7G). The rescue effect of anisomycin supports our conclusion that elevated bulk protein synthesis underlies G2019S LRRK2 toxicity.
Figure 7. Elevated protein synthesis in G2019S LRRK2 transgenic flies is blocked by T136A s15.
(A), de novo protein synthesis, measured by 35S-met/cys incorporation is significantly increased in protein precipitates from G2019S LRRK2 transgenic fly heads, which is blocked by T136A s15 co-expression (* p <0.05, n = 5 groups of 50 heads/genotype). Genotypes are Da-Gal4 alone (Control), Da-Gal4; UAS-LRRK2 (WT), Da-Gal4/ UAS-G2019S/D1994A LRRK2 (GS/DA), Da-Gal4; UAS-G2019S LRRK2 (G2019S), Da-Gal4/UAS-T136A s15; UAS-G2019S LRRK2 (GS+TA s15) (B), autoradiography from lysates reveals a widespread increase in 35S-met/cys-labeled protein abundance. Ponceau staining for total protein. (C), fractions collected from fly head polysome profiles and used for RT-PCR of translating mRNA indicates a G2019S LRRK2-mediated shift in tubulin and actin 5C to heavy polysome fractions, prevented by T136A s15 (two-way ANOVA, Bonferroni's post-test, * p <0.05, ** p <0.01, *** p <0.001, n =3 groups of 100 fly heads/genotype). (D), Confocal z-stack projection images and quantitation of dopamine neurons in Drosophila brains. Anisomycin (10 μM) treatment to food throughout adulthood rescued dopamine neuron loss in aged G2019S transgenic flies (individual ANOVAs, Bonferroni's post-test, * p<0.05, ** p<0.01, n =8-10 fly heads per genotype). (E), anisomycin treatment prevented age-related locomotor deficits in G2019S flies (ANOVA, Bonferroni's post-test, * p<0.05, ** p<0.01, n =25 flies per genotype). Genotypes for (D) and (E) are as in (A). Data are mean ± SEM. See also Figure S7.
Discussion
The major finding from this study is the identification, via a screen for LRRK2 kinase substrates, of s15 as a novel in vivo LRRK2 substrate that underlies PD-related phenotypes in Drosophila and directly links LRRK2 toxicity to altered mRNA translation. Screening for LRRK2-interacting proteins revealed numerous ribosomal proteins and led to the discovery that LRRK2 is enriched in the ribosomal subcellular fraction. Further screening of all ribosomal proteins as candidate LRRK2 substrates revealed that s15 is a strong LRRK2 substrate, which although not identified in the original screen, interacts directly with LRRK2 in co-immunoprecipitation studies. In vitro phosphorylation assays indicate that wild type LRRK2 phosphorylates a number of ribosomal proteins, of which s15 phosphorylation transduces LRRK2-related toxicity and regulation of cap-dependent and cap-independent translation. s15 fulfills several criteria as a substrate for LRRK2 including enhanced phosphorylation in LRRK2 transgenic Drosophila brain and human brain expressing the common G2019S mutation, and in HEK293 cells expressing G2019S LRRK2, where it is blocked by two specific and unrelated LRRK2 kinase inhibitors. Moreover, s15 phosphorylation is significantly reduced in flies null for the Drosophila LRRK2 homolog, dLRRK. s15 phosphorylation is central to G2019S LRRK2 toxicity since T136A s15 blocks G2019S LRRK2 toxicity in human dopamine neurons and additionally blocks neurodegeneration, locomotor deficits and elevated protein synthesis observed in a Drosophila in vivo PD model. While wild type s15 co-expression appears to provide a minor protective effect in rat cortical neuron cultures overexpressing G2019S LRRK2 (Fig 2B), it was not protective in other neuronal culture experiments or in G2019S LRRK2 transgenic flies, in contrast to T136A s15 co-expression, which produces a clear and significant protective effect in all neuronal culture experiments and Drosophila neurodegenerative phenotypes examined. It is possible that the small protective effect seen in rat cortical neurons may be related to a minor competitive effect of extra cytosolic s15 that might reduce phosphorylation of s15 by LRRK2 directly at the ribosome in this context. Informatively, wild type LRRK2 overexpression in Drosophila causes a slight increase in s15 phosphorylation and, as observed in previous studies (Liu et al., 2008), leads to a modest effect on locomotion and dopamine neuron viability, which are more pronounced, with G2019S LRRK2 expression. This supports a well-established link between LRRK2 kinase activity and neurotoxicity and suggests that wild type LRRK2 overexpression could in theory result in pathology if expressed at high enough levels. Consistent with this, elevated wild-type LRRK2 overexpression in neuronal cultures results in slightly increased s15 phosphorylation and neuronal toxicity, but not to the extent observed with G2019S LRRK2 expression, suggesting, as previously reported, that additional mechanisms may differentially regulate the kinase activities of wild type and mutant forms of LRRK2 in vivo, effectively dampening the kinase activity of wild type LRRK2 (Sen et al., 2009; Webber et al., 2011). While we observe an increase in P-s15 and rescue effect of T136A s15 for the kinase domain mutants G2019S and I2020T LRRK2, R1441C LRRK2 does not show similar phenotypes, suggesting that alternative pathogenic mechanisms leading to PD may result from this mutation, possibly involving other pathogenic substrates or perhaps even kinase-independent pathways. Future detailed studies are necessary to understand the consequences of ROC domain mutations on LRRK2 function and neurotoxicity.
Studies in Drosophila models of LRRK2 toxicity suggest that LRRK2 mutants with increased kinase activity can stimulate 4E-BP1 phosphorylation and expression of transcription factors e2f1 and dp through unclear mechanisms possibly involving elevated levels of free eIF4E or perturbed miRNA function (Gehrke et al., 2010; Imai et al., 2008; Tain et al., 2009). Evidence showing that 4E-BP is a direct LRRK2 substrate or that 4E-BP has a role in miRNA function is lacking, however, and importantly there has been no demonstration that LRRK2-mediated phosphorylation of 4E-BP directly mediates altered translation of the few mRNAs mentioned. Subsequent studies have failed to detect an increase in P-4E-BP in cells or mouse brain following increased LRRK2 kinase activity (Kumar et al., 2010; Trancikova et al., 2012). Our data from in vitro reporter assays and Drosophila suggest that G2019S LRRK2 actually promotes an increase in bulk translation, and provide a mechanism by which G2019S LRRK2 directly stimulates cap-dependent and cap-independent translation at the ribosomal level through phosphorylation of s15. Our results also indicate that pathogenic LRRK2 does not cause a systematic effect on either mode of translation via 4E-BP phosphorylation or disruption of the miRNA pathway. Thus, the observation that overexpression of 4E-BP or reduction in dLRRK levels can be protective in Drosophila models of PINK1 and Parkin pathology may be accounted for by a potential for modulated 4E-BP expression to dampen aberrant protein production as opposed to direct interference with LRRK2 substrate phosphorylation-mediated signaling (Tain et al., 2009). Consistent with this notion are our observations that the global protein synthesis inhibitor anisomycin rescues the locomotor deficits and dopamine neuron loss in aged G2019S LRRK2 transgenic Drosophila. Further studies investigating a possible role of altered translation following other genetic mutations linked to PD are warranted by this finding.
Defects in translational regulation underlie a number of inherited diseases. These diseases exhibit a high degree of heterogeneity despite the fact that protein synthesis is a fundamental process of all cells (Scheper et al., 2007). In the nervous system, excessive protein synthesis has long been implicated in the development of Fragile X syndrome (reviewed in (Bhakar et al., 2012)). More recently, systematically altered translation has been identified as a fundamental mechanism driving prion disease neurodegeneration (Moreno et al., 2012) and perturbed RNA metabolism through mutations in RNA binding proteins such as TAR DNA binding protein-43 (TDP-43) and fused in sarcoma/translocated in sarcoma (FUS/TLS) has been implicated in the pathogenesis of amyotrophic lateral sclerosis and frontotemporal dementia (Lagier-Tourenne et al., 2012; Polymenidou et al., 2011; Tollervey et al., 2011). Thus, loss of translational control is emerging as an important mediator of diverse neurologic diseases. A molecular understanding of how aberrant protein synthesis leads to neurodegeneration in these diseases will be an important future priority.
In summary, these findings support a role for dysregulated translation in the pathogenesis of PD. Consistent with this notion is the recent observation that mutations in the translation initiation factor eIF4G1 cause autosomal dominant PD (Chartier-Harlin et al., 2011; Nuytemans et al., 2013). Whether mutations in LRRK2 and eIF4G1 converge on common or overlapping pathogenic outputs are under detailed investigation. As mentioned above, elevated or impaired translation has been identified as a causative factor in numerous neurological diseases and emphasizes the importance of protein homeostasis in neuronal function. Thus, further understanding the role of translation in the degenerative process of PD will provide new targets for disease modification in PD.
Experimental Procedures
LRRK2 tandem affinity purification
LRRK2-interacting phosphoproteins were identified using tandem affinity purification of TAP tagged LRRK2 from HEK293 cells, IMAC phosphoprotein enrichment and mass spectrometry in series. Full description of the method is provided in the Supplemental Information.
LRRK2 in vitro kinase assays
Recombinant LRRK2 (aa 970-2527) and recombinant GST-tagged substrate candidates were incubated in kinase assay buffer containing γ-P32-ATP for phosphorimaging-based visualization of protein phosphorylation following separation of the reaction mixture by SDS-PAGE. See Supplemental Information for full details.
LRRK2 in vitro toxicity assays
Toxicity (loss of neurites and appearance of TUNEL-positive nuclei) in rodent and human neurons overexpressing LRRK2, substrate and GFP for neurite tracing in a 10:10:1 ratio was assessed using previously described methods 48 h following plasmid transfection (Lee et al., 2010; Li et al., 2010; Ramsden et al., 2011; Xiong et al., 2010). The vast majority of GFP positive neurons (~90%) also co-labeled for overexpressed LRRK2 and substrate indicating that the effects of LRRK2 and s15/s11 overexpression could be assessed in these neurons, as previously described (Lee et al., 2010). See Supplemental Information for more details.
Generation of s15 and T136A s15 transgenic lines
Human s15 and T136A s15 constructs were generated by subcloning full-length cDNA into pUAST between EcoR1 and Xba1 restriction sites. After sequence verification, constructs were microinjected into w1118 fly embryos. Transgenic UAS-s15 and UAS-T136A s15 expression were confirmed by western blot on fly heads following expression driven via daughterless-Gal4 (Fig. S5A). Details on additional fly lines used are provided in Supplemental Information.
Phospho-s15 assessment in cells and Drosophila
A polyclonal antibody to phospho-Thr 136 was generated and validated as described in Supplemental Information. In cells, phosphorylation of overexpressed s15 was assessed from whole lysates extracted using 1% NP-40 lysis buffer and separated by SDS-PAGE for immunoblotting. In homogenized heads of LRRK2 transgenic or dLRRK-null flies, phosphorylation of endogenous s15 was assessed in ribosomal fractions that were isolated essentially as described before (Belin et al., 2010) with the addition of phosphatase inhibitors to the extraction buffer.
Drosophila dopamine neuron immunohistochemistry and locomotor function
Dopamine neuron immunohistochemistry and negative geotaxis assessment of locomotor function were performed as described in Supplemental Information.
De novo protein synthesis and mRNA polysome profiling in Drosophila
For protein synthesis measurement, 35S-methionine/cysteine (100 µCi/ml) incorporation into flies fed with labeled food for 24h was assessed by liquid scintillation counting and expressed relative to total protein amount. Polysomes were isolated from fly heads via sucrose gradient centrifugation and fractionated for RNA extraction, cDNA synthesis and real-time PCR measurement of genes relative to a spike-in luciferase synthetic RNA added prior to RNA extraction as previously described (Trancikova et al., 2012). See Supplemental Information for full details.
Statistical analyses
All statistical analyses, including Student's t-tests, ANOVA and associated Bonferroni's post-tests were performed using GraphPad Prism software.
Supplementary Material
Highlights.
Ribosomal proteins are major LRRK2 interactors and kinase substrates
G2019S LRRK2 toxicity is mediated through phosphorylation of ribosomal protein s15
Phosphorylation of s15 links LRRK2-neurodegeneration to increased mRNA translation
LRRK2-neurodegeneration is blocked through translation inhibition
Acknowledgments
The authors acknowledge the Adrienne Helis Malvin Medical Research Foundation and the Diana Helis Henry Medical Research Foundation and their direct engagement in the continuous active conduct of medical research in conjunction with The Johns Hopkins Hospital and The Johns Hopkins University School of Medicine and the Foundation's Parkinson's Disease Programs. The authors acknowledge the Johns Hopkins Stem Cell Core Facility for providing a subset of the human neurons used in this study. Funding for a portion of the research described in this article was provided by Merck KGAA. Under a licensing agreement between Merck KGAA and The Johns Hopkins University, Dr. Dawson and the University shared fees received by the University on licensing of some of the reagents described in this article. Dr. Dawson also was a paid consultant to Merck KGAA. The terms of this arrangement are being managed by The Johns Hopkins University in accordance with its conflict of interest policies. Mayo Clinic and Dr. Wszolek have a financial interest in a technology (LRRK2 gene) referenced in this manuscript. This technology has been licensed to a commercial entity and Mayo Clinic and Dr. Wszolek receive royalties from that license. Dr. Wszolek's annual royalties are less than $200 per year. This work was also supported by NIH/NINDS P50NS038377, the JPB Foundation and the MDSCRF 2007-MSCRFI-0420-00, 2009-MSCRFII-0125-00, MDSCRF 2013-MSCRFII-0105-00 to T.M.D. and V.L.D, NIH/NINDS P50 NS072187 to D.D. and Z.K.W. and a New York Stem Cell Foundation-Druckenmiller Fellowship to I.M. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Footnotes
Author Contributions: I.M., V.L.D. and T.M.D. formulated the hypothesis, initiated and organized the study and wrote the manuscript. I.M., J.K., B.D.L., H.K., M.S.K., J.Z., H.J., A.P., T.M.D. and V.L.D. designed experiments and analyzed data. I.M., J.K. J.S., B.D.L., H.K., J-C.X., M.S.K., J.Z., M.K., S.A. and H.J. performed experiments.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anand VS, Reichling LJ, Lipinski K, Stochaj W, Duan W, Kelleher K, Pungaliya P, Brown EL, Reinhart PH, Somberg R, et al. Investigation of leucine-rich repeat kinase 2 : enzymological properties and novel assays. FEBS J. 2009;276:466–478. doi: 10.1111/j.1742-4658.2008.06789.x. [DOI] [PubMed] [Google Scholar]
- Belin S, Hacot S, Daudignon L, Therizols G, Pourpe S, Mertani HC, Rosa-Calatrava M, Diaz JJ. Purification of ribosomes from human cell lines. Curr Protoc Cell Biol. 2010;Chapter 3:40. doi: 10.1002/0471143030.cb0340s49. Unit 3. [DOI] [PubMed] [Google Scholar]
- Bhakar AL, Dolen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Annu Rev Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Dachsel JC, Vilarino-Guell C, Lincoln SJ, Lepretre F, Hulihan MM, Kachergus J, Milnerwood AJ, Tapia L, Song MS, et al. Translation initiator EIF4G1 mutations in familial Parkinson disease. Am J Hum Genet. 2011;89:398–406. doi: 10.1016/j.ajhg.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson's disease. Nat Rev Neurosci. 2010;11:791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covy JP, Giasson BI. Identification of compounds that inhibit the kinase activity of leucine-rich repeat kinase 2. Biochem Biophys Res Commun. 2009;378:473–477. doi: 10.1016/j.bbrc.2008.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, Lee JD, Patricelli MP, Nomanbhoy TK, Alessi DR, et al. Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nat Chem Biol. 2011;7:203–205. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, Lee JD, Patricelli MP, Nomanbhoy TK, Alessi DR, et al. Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nature chemical biology. 2011;7:203–205. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairulina J, Graifer D, Bulygin K, Ven'yaminova A, Frolova L, Karpova G. Eukaryote-specific motif of ribosomal protein S15 neighbors A site codon during elongation and termination of translation. Biochimie. 2010;92:820–825. doi: 10.1016/j.biochi.2010.02.031. [DOI] [PubMed] [Google Scholar]
- Kumar A, Greggio E, Beilina A, Kaganovich A, Chan D, Taymans JM, Wolozin B, Cookson MR. The Parkinson's disease associated LRRK2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation. PLoS One. 2010;5:e8730. doi: 10.1371/journal.pone.0008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BD, Shin JH, VanKampen J, Petrucelli L, West AB, Ko HS, Lee YI, Maguire-Zeiss KA, Bowers WJ, Federoff HJ, et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nat Med. 2010;16:998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Moore DJ, Xiong Y, Dawson TM, Dawson VL. Reevaluation of phosphorylation sites in the Parkinson disease-associated leucine-rich repeat kinase 2. J Biol Chem. 2010;285:29569–29576. doi: 10.1074/jbc.M110.127639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang X, Yu Y, Li X, Wang T, Jiang H, Ren Q, Jiao Y, Sawa A, Moran T, et al. A Drosophila model for LRRK2-linked parkinsonism. Proc Natl Acad Sci U S A. 2008;105:2693–2698. doi: 10.1073/pnas.0708452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzon-Toro B, Rubio de la Torre E, Delgado A, Perez-Tur J, Hilfiker S. Mechanistic insight into the dominant mode of the Parkinson's disease-associated G2019S LRRK2 mutation. Hum Mol Genet. 2007;16:2031–2039. doi: 10.1093/hmg/ddm151. [DOI] [PubMed] [Google Scholar]
- MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Martin I, Dawson VL, Dawson TM. Recent advances in the genetics of Parkinson's disease. Annu Rev Genomics Hum Genet. 2011;12:301–325. doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta S, Van Kolen K, da Cunha R, van den Bogaart G, Mandemakers W, Miskiewicz K, De Bock PJ, Morais VA, Vilain S, Haddad D, et al. LRRK2 controls an EndoA phosphorylation cycle in synaptic endocytosis. Neuron. 2012;75:1008–1021. doi: 10.1016/j.neuron.2012.08.022. [DOI] [PubMed] [Google Scholar]
- Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, Halliday M, Morgan J, Dinsdale D, Ortori CA, et al. Sustained translational repression by eIF2alpha-P mediates prion neurodegeneration. Nature. 2012;485:507–511. doi: 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuytemans K, Bademci G, Inchausti V, Dressen A, Kinnamon DD, Mehta A, Wang L, Zuchner S, Beecham GW, Martin ER, et al. Whole exome sequencing of rare variants in EIF4G1 and VPS35 in Parkinson disease. Neurology. 2013;80:982–989. doi: 10.1212/WNL.0b013e31828727d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Parisiadou L, Xie C, Cho HJ, Lin X, Gu XL, Long CX, Lobbestael E, Baekelandt V, Taymans JM, Sun L, et al. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci. 2009;29:13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. Specific functional interactions of nucleotides at key -3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden N, Perrin J, Ren Z, Lee BD, Zinn N, Dawson VL, Tam D, Bova M, Lang M, Drewes G, et al. Chemoproteomics-based design of potent LRRK2-selective lead compounds that attenuate Parkinson's disease-related toxicity in human neurons. ACS Chem Biol. 2011;6:1021–1028. doi: 10.1021/cb2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper GC, van der Knaap MS, Proud CG. Translation matters: protein synthesis defects in inherited disease. Nat Rev Genet. 2007;8:711–723. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- Sen S, Webber PJ, West AB. Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. J Biol Chem. 2009;284:36346–36356. doi: 10.1074/jbc.M109.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nature neuroscience. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci. 2009;12:1129–1135. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trancikova A, Mamais A, Webber PJ, Stafa K, Tsika E, Glauser L, West AB, Bandopadhyay R, Moore DJ. Phosphorylation of 4E-BP1 in the mammalian brain is not altered by LRRK2 expression or pathogenic mutations. PLoS One. 2012;7:e47784. doi: 10.1371/journal.pone.0047784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsika E, Moore DJ. Mechanisms of LRRK2-mediated neurodegeneration. Curr Neurol Neurosci Rep. 2012;12:251–260. doi: 10.1007/s11910-012-0265-8. [DOI] [PubMed] [Google Scholar]
- Webber PJ, Smith AD, Sen S, Renfrow MB, Mobley JA, West AB. Autophosphorylation in the leucine-rich repeat kinase 2 (LRRK2) GTPase domain modifies kinase and GTP-binding activities. J Mol Biol. 2011;412:94–110. doi: 10.1016/j.jmb.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, et al. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Coombes CE, Kilaru A, Li X, Gitler AD, Bowers WJ, Dawson VL, Dawson TM, Moore DJ. GTPase activity plays a key role in the pathobiology of LRRK2. PLoS Genet. 2010;6:e1000902. doi: 10.1371/journal.pgen.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.