Abstract
Problem
Inflammatory biomarkers are associated with preeclampsia (PE) and poor fetal growth; however, genetic epidemiologic studies have been limited by reduced gene coverage and the exclusion of African American mothers.
Method of study
Cases and controls (N = 1646) from a pregnancy cohort were genotyped for 503 tagSNPs in 40 genes related to inflammation. Gene-set analyses were stratified by race and were followed by a single SNP analysis within significant gene sets.
Results
Gene-level associations were found for IL6 and KLRD1 for term small for gestational age (SGA) among African Americans. LTA/TNF and TBX21 were associated with PE among European Americans. The strongest association was for PE among European Americans for an upstream regulator of TNF with RR = 1.8 (95% CI 1.1–2.7).
Conclusion
Although previous studies have suggested null associations, increased tagging and stratification by genetic ancestry suggests important associations between IL6 and term SGA for African Americans, and a TNF regulator and PE among European Americans (N = 149).
Keywords: African Americans, genetic association studies, preeclampsia, pregnancy, small for gestational age
Introduction
Hypertensive disorders of pregnancy and poor fetal growth often co-occur1 and may share underlying pathology.2 Hypertensive disorders of pregnancy, including gestational hypertension (GHTN) and preeclampsia (PE), occur in up to 20% of pregnancies3 and are a leading cause of maternal mortality.4 Pregnancies complicated by hypertension are at increased risk of stillbirth, poor fetal growth, and preterm birth.1,5 The prevalence and health consequences of poor fetal growth depend largely on the metric of assessment6 and the specific etiology.7 However, low birth weight is consistently associated with increased infant mortality.8 Each outcome has also been associated with later risk of cardiovascular disease in the mother.9–11
Hypertension in pregnancy and poor fetal growth may share common placental pathology including shallow or inadequate placentation.2,12 Adequate placentation requires remodeling of the spiral arteries in the decidua to allow adequate blood flow to the placenta.12,13 With PE, there is poor to absent vascular remodeling in the myometrium and decidua. For pregnancies complicated by poor fetal growth, vascular remodeling is more evident but is diminished in depth and extent.14 Placental invasion and development are dependent on tightly regulated local inflammatory processes.2,15 In particular, the interaction between fetal trophoblasts and maternal decidual cells is mediated by maternal natural killer (NK) and T-cell activity.16,17 In addition to mediating placentation, inflammatory cytokines may be released as a result of hypoxia and ischemic–reperfusion injuries which result from inadequate placentation.13,18 In fact, women with either outcome exhibit altered inflammatory biomarkers both preceding15,19,20 and at the time of diagnosis.2,15,21
The extensive placental growth and uterine wall remodeling that occur during normal gestation may additionally be influenced by cell cycle genes that regulate cell growth and division in rapidly growing tissue such as the placenta.22,23 Human and animal placental expression studies suggest that cell cycle genes may differ in expression or function in women with hypertension in pregnancy or small for gestational age (SGA).24,25
Although a fetal genetic component has been identified for both hypertension in pregnancy and fetal growth, these disorders also have a maternal genetic component.26,27 Very few candidate gene studies have examined poor fetal growth. Studies of PE, while more numerous, have largely been dominated by genes in the renin angiotensin, coagulation, and angiogenic pathways.28 Candidate gene studies for other pathways have been limited, have focused on relatively few markers per gene without accounting for up- and downstream regions, and have often lacked control for population stratification.29 To address these limitations, we conducted a study nested within the pregnancy, infection, and nutrition (PIN) cohort, a biracial pregnancy cohort in North Carolina. Our focus was on genes related to inflammation and cell cycle pathways.
Methods
Study participants were drawn from the PIN cohort, which was established to assess antenatal risk factors for a variety of reproductive outcomes.30 Women were enrolled between August 1995 and June 2005 from prenatal clinics at Wake County Human Services Department, Wake Medical Center and the University of North Carolina (UNC) Hospital. Women were ineligible if they were less than 16 years old, did not speak English, lacked a telephone, were carrying more than one fetus, or did not plan to continue care at the recruitment hospital.
Outcome and covariate data were collected through self-administered questionnaires, telephone interviews during pregnancy, and abstraction of maternal and infant medical records. Maternal blood for genetic analysis was collected during the first study visit. All participants gave consent, and the study was approved by the Institutional Review Boards of UNC School of Medicine and Wake Medical Center.
Of those enrolled, 3539 women (68.5%) consented to genetic analysis. Eligibility for genotyping in the present study also included collection of a suitable bio-specimen (N = 3289), self-reported race as White or African American (N = 3075) and recorded date of delivery (N = 3065). These eligibility criteria resulted in 3065 (59.3%) women who were eligible for selection. From the eligible population, 1646 pregnancies were selected for genotyping. Attempts were made to genotype all eligible cases. Missing or inadequate samples resulted in (N, % of eligible): 216 (90.4%) SGA, 398 (95.2%) GHTN, and 170 (91.9%) PE cases being included in the study. Although GHTN and PE cases were exclusive case definitions, pregnancies complicated by SGA could also have a diagnosis of GHTN or PE. Control births (N = 918) were selected from the remaining eligible women.
Among the 1646 samples genotyped, 11 individuals were dropped due to fewer than 95% of markers successfully called. Exclusions were also made for congenital anomalies (N = 24), stillbirths (N = 10), and unintentional duplicate samples (N = 3). As a result of these exclusions, 1598 women were available for analysis.
Outcome Assessment
Small for gestational age status was defined as birth weight below the 10th percentile for gestational age, stratified by infant race, sex, and maternal parity based on percentiles from 1989 US births.31 As a proxy measure for impaired fetal growth, SGA may not appropriately classify preterm infants6 and may misidentify constitutionally small infants as SGA. In as much as common causes may exist for preterm birth and poor intra-uterine growth, term SGA was considered as an additional phenotype.
Gestational hypertension and PE were assessed using evidence of new hypertension after 20 weeks and proteinuria abstracted from antenatal charts and discharge diagnoses. Prior to 2002, hypertension during pregnancy was defined using a relative increase of 30 mmHg in systolic blood pressure (BP) or a 15 mmHg increase in the diastolic BP from a woman's baseline blood pressure. Following American College of Obstetrics and Gynecology (ACOG) recommendations in 2002, the definition of hypertension was changed to an absolute cut point of systolic BP ≥140 mmHg or diastolic BP ≥90 mmHg.3 Diagnoses in this study reflect the clinical criteria in use at the time of the pregnancy. While the newer ACOG criteria reduces the number of women who receive a diagnosis of PE, the positive predictive value for adverse maternal and infant outcomes is similar for the two sets of criteria.32 GHTN was defined as new onset hypertension following 20 weeks in the absence of proteinuria. Pregnancies with evidence of GHTN that developed proteinuria later in pregnancy were classified as PE. PE was defined as new onset hypertension (using the criteria appropriate at the time of pregnancy) and evidence of proteinuria. Women with preexisting hypertension, or hypertension before 20 weeks, were excluded from both the case and control groups for all analyses of GHTN and PE.
DNA Extraction and Genetic Analysis
Maternal blood was drawn at a study visit in the second trimester. Buffy coat fractions were isolated from fresh whole blood and stored at –80°C in CPT tubes. DNA was extracted using Applied Biosystems automated DNA extractor and Qiagen (Gentra, Valencia, CA, USA) Puregene chemistry.
Thirty inflammatory and ten cell cycle genes (546 SNPs) were selected as candidate genes. TagSNPs were selected using TagZilla33 for two population (Eurpoean and Yoruban, HapMap build 27) with a 20 kb upstream and 10 kb downstream margin, restricting to minor allele frequencies ≥10% in at least one population and linkage disequilibrium (LD) r2 > 0.8. A custom 1536 Illumina GoldenGate plate was designed which also included SNPs from genes in the angiogenesis and apoptosis pathways.
Genotyping was conducted at the University of North Carolina Mammalian Genotyping Core (Chapel Hill, NC, USA), and genotypes were called with Illumina GenomeStudio software (Illumina, San Diego, CA, USA).
Poor genotyping quality (<95% of individuals called) resulted in the loss of 43 SNPs. Further quality control was conducted on the remaining 503 SNPs using blinded PIN samples and standardized controls from Corriel Utah family trios. There was one instance of a single base pair genotyping discrepancy found among 199 blind samples, and there were no instances of Mendelian errors among 21 trios examined. Hardy–Weinberg equilibrium was assessed using SAS 9.234 among non-cases stratified by genetic ancestry. One SNP (IL2: rs10027390) significantly violated HWE (P < 10–5) in African American non-cases and was dropped from analysis for this ancestry group.
In tagging two populations, inclusion of redundant tagSNPs with high LD in one ancestral population was necessary to adequately capture variation in the other population. In order to examine the linkage between both typed and untyped markers in this study population, heatmaps were created using Haploview,35 and long-range LD with untyped SNPs was explored using SNAP36 with the 1000 Genomes Project Pilot 1 data.
Genetic Ancestry
Differences in the allelic frequency in genes associated with inflammation have been reported among Whites and African Americans in the United States.37 Birth weight8 and the incidence of PE3 also vary by self-reported race, raising the possibility of confounding by genetic ancestry. Genetic ancestry was therefore assessed using ancestry informative markers (AIMs) (N = 148) that have been used in a similar population in North Carolina.38 STRUCTURE was used to calculate a continuous genetic ancestry variable assuming two underlying populations.39 The calculated ancestry variable was used to dichotomize the population into European American and African American ancestry groups using a cut point that corresponded with self-reported race. All analyses were stratified by genetic ancestry and also included continuous percent ancestry.
Statistical Analysis
We employed a two-stage approach to identify associations between inflammatory and cell cycle genes, and the outcomes of SGA, term SGA, GHTN and PE. Stage 1 utilized the SNP set kernel association test (SKAT) with a linear kernel.40 This approach is analogous to logistic regression with an additive genetic model. SKAT was chosen because it permits SNP interactions within a gene, allows adjustment for covariates, and accounts for SNPs with opposing effects (protective or risk) within a gene. We included all markers within a given gene as a SNP set, including markers within the upstream and downstream margins. If genes were positioned within 25 kbp of each other, they were analyzed together. Genes with single SNPs (cyclin H (CCNH), transforming growth factor beta 3 (TGFB3), and beta 1 (TGFB1), MDM2 oncogene (MDM2)) or SNPs chosen solely for replication (interleukin(IL) 1A, interleukin 1B) were not included in the Stage 1 analysis. Individuals with missing genotype information on any SNPs in a set were dropped from the Stage 1 analysis. On average, 99% of genotyped individuals were included in each SNP set, with a minimum of 91%. Analyses were conducted for each outcome with uncomplicated term births as the control group (‘Disease Free’ column in Tables I and II). For GHTN and PE, women with chronic hypertension were excluded from both the case and control group.
Table I.
Demographic Characteristics of Small for Gestational Age (SGA) Cases and Controls
| Genetic ancestrya | SGA cases |
Disease free |
Non-SGA controlsb |
|||
|---|---|---|---|---|---|---|
| European American N = 117 N (%) | African American N = 92 N (%) | European American N = 409 N (%) | African American N = 204 N (%) | European American N = 890 N (%) | African American N = 499 N (%) | |
| Maternal age (years) | ||||||
| <25 | 55 (47.0) | 27 (29.4) | 215 (52.6) | 68 (33.3) | 440 (49.4) | 182 (36.5) |
| 25–34 | 46 (39.3) | 59 (64.1) | 133 (32.5) | 132 (64.7) | 325 (36.5) | 295 (59.1) |
| 35+ | 16 (13.7) | 6 (6.5) | 61 (14.9) | 4 (2.0) | 125 (14.0) | 22 (4.4) |
| Smokingc | ||||||
| No | 68 (63.6) | 58 (72.5) | 302 (77.4) | 148 (84.6) | 588 (71.0) | 362 (83.2) |
| Yes | 39 (36.5) | 22 (27.5) | 88 (22.6) | 27 (15.4) | 240 (29.0) | 73 (16.8) |
| Missing | 10 (8.5) | 12 (13.0) | 19 (4.6) | 29 (14.2) | 62 (7.0) | 64 (12.8) |
| BMI (kg/m2)d | ||||||
| <18.5 | 11 (9.9) | 11 (13.1) | 23 (5.7) | 16 (8.3) | 44 (5.1) | 27 (5.8) |
| 18.5–24.9 | 72 (64.9) | 35 (41.7) | 250 (62.2) | 79 (40.9) | 454 (52.7) | 166 (35.8) |
| 25–29.9 | 16 (14.4) | 13 (15.5) | 71 (17.7) | 39 (20.2) | 174 (20.2) | 100 (21.6) |
| 30+ | 12 (10.8) | 25 (29.8) | 58 (14.4) | 59 (30.6) | 189 (22.0) | 171 (36.9) |
| Missing | 6 (5.1) | 8 (8.7) | 7 (1.7) | 11 (5.4) | 29 (3.3) | 35 (7.0) |
| Marital status | ||||||
| Married | 77 (65.8) | 13 (14.1) | 313 (76.5) | 32 (15.7) | 644 (72.4) | 109 (21.9) |
| Unmarried | 40 (34.2) | 79 (85.9) | 96 (23.5) | 172 (84.3) | 246 (27.6) | 389 (78.1) |
| Education (years) | ||||||
| 13+ | 61 (52.1) | 27 (29.4) | 275 (67.2) | 68 (33.3) | 542 (60.9) | 201 (40.3) |
| ≤12 | 56 (47.9) | 65 (70.7) | 134 (32.8) | 136 (66.7) | 348 (39.1) | 298 (59.7) |
| Parity | ||||||
| Nulliparous | 49 (41.9) | 48 (52.2) | 189 (46.4) | 88 (43.1) | 437 (49.2) | 220 (44.2) |
| Multiparous | 68 (58.1) | 44 (47.8) | 218 (53.6) | 116 (56.9) | 451 (50.8) | 278 (55.8) |
| Missing | 0 | 0 | 2 (0.5) | 0 | 2 (0.2) | 1 (0.2) |
| Poverty index | ||||||
| Mean (S.D.) | 310 (237) | 119 (85) | 367 (238) | 144 (123) | 345 (237) | 148 (124) |
| Missing | 10 (8.6) | 26 (28.3) | 24 (5.9) | 37 (18.1) | 70 (7.9) | 85 (17.0) |
| Additional outcomes | ||||||
| Term SGA | 96 (82.1) | 64 (69.6) | 0 | 0 | 717e | 393e |
| Gestational HTN | 19 (17.0) | 14 (16.9) | 0 | 0 | 226 (27.2) | 131 (28.5) |
| PE | 11 (9.8) | 10 (12.1) | 0 | 0 | 82 (9.9) | 62 (13.5) |
| Preterm birth | 21 (18.0) | 28 (30.4) | 0 | 0 | 173 (19.4) | 106 (21.2) |
Genetic ancestry determined using 148 ancestry informative markers and STRUCTURE.
Non-SGA controls include uncomplicated births as well as births with other pregnancy outcomes including preterm birth; GHTN, gestational hypertension; and PE, preeclampsia.
Self-reported smoking during months 1–6 of pregnancy.
Pre-pregnancy body mass index (BMI) calculated from self-reported pre-pregnancy weight and measured height.
Controls for term SGA are limited to term births.
Table II.
Demographic Characteristics of Gestational Hypertension (GHTN) and Preeclampsia (PE) Cases and Controls
| Genetic ancestrya | GHTN case |
PE case |
Disease freeb |
GHTN controlc |
PE controlc |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| European American N = 245 N (%) | African American N = 145 N (%) | European American N = 93 N (%) | African American N = 72 N (%) | European American N = 394 N (%) | African American N = 190 N (%) | European American N = 699 N (%) | African American N = 397 N (%) | European American N = 851 N (%) | African American N = 470 N (%) | |
| Age (years) | ||||||||||
| <25 | 107 (43.7) | 54 (37.2) | 52 (55.9) | 22 (30.6) | 211 (53.6) | 64 (33.7) | 355 (50.8) | 131 (33.0) | 410 (48.2) | 163 (34.7) |
| 25–34 | 101 (41.2) | 85 (58.6) | 36 (38.7) | 48 (66.7) | 123 (31.2) | 122 (64.2) | 254 (36.3) | 254 (64.0) | 319 (37.5) | 291 (61.9) |
| 35+ | 37 (15.1) | 6 (4.1) | 5 (5.4) | 2 (2.8) | 60 (15.2) | 4 (2.1) | 90 (12.9) | 12 (3.0) | 122 (14.3) | 16 (3.4) |
| Smokingd | ||||||||||
| No | 150 (65.8) | 101 (80.2) | 57 (71.3) | 56 (86.2) | 293 (77.5) | 135 (83.3) | 468 (72.0) | 283 (81.6) | 561 (70.3) | 328 (80.4) |
| Yes | 78 (34.2) | 25 (19.8) | 23 (28.8) | 9 (13.9) | 85 (22.5) | 27 (16.7) | 182 (28.0) | 64 (18.4) | 237 (29.7) | 80 (19.6) |
| Missing | 17 (6.9) | 19 (13.1) | 13 (14.0) | 7 (9.7) | 16 (4.1) | 28 (14.7) | 49 (7.0) | 50 (12.6) | 53 (6.2) | 62 (13.2) |
| BMI (kg/m2)e | ||||||||||
| <18.5 | 7 (3.0) | 6 (4.6) | 4 (4.4) | 1 (1.5) | 22 (5.7) | 16 (8.9) | 48 (7.0) | 32 (8.6) | 51 (6.2) | 37 (8.5) |
| 18.5–24.9 | 106 (46.1) | 46 (34.9) | 41 (45.6) | 23 (33.8) | 242 (62.2) | 74 (41.1) | 406 (59.5) | 149 (40.1) | 471 (57.3) | 172 (39.5) |
| 25–29.9 | 47 (20.4) | 29 (22.0) | 21 (23.3) | 15 (22.1) | 69 (17.7) | 36 (20.0) | 132 (19.4) | 74 (19.9) | 158 (19.2) | 88 (20.2) |
| 30+ | 70 (30.4) | 51 (38.6) | 24 (26.7) | 29 (42.7) | 56 (14.4) | 54 (30.0) | 96 (14.1) | 117 (31.5) | 142 (17.3) | 139 (31.9) |
| Missing | 15 (6.1) | 13 (9.0) | 3 (3.2) | 4 (5.6) | 5 (1.3) | 10 (5.3) | 17 (2.4) | 25 (6.3) | 29 (3.4) | 34 (7.2) |
| Marital status | ||||||||||
| Married | 171 (69.8) | 41 (28.3) | 58 (62.4) | 13 (18.3) | 304 (77.2) | 30 (15.8) | 504 (72.1) | 66 (16.7) | 617 (72.5) | 94 (20.0) |
| Unmarried | 74 (30.2) | 104 (71.7) | 35 (37.6) | 58 (81.7) | 90 (22.8) | 160 (84.2) | 195 (27.9) | 330 (83.3) | 234 (27.5) | 376 (80.0) |
| Education (years) | ||||||||||
| 13+ | 141 (57.6) | 62 (42.8) | 47 (50.5) | 31 (43.1) | 267 (67.8) | 65 (34.2) | 420 (60.1) | 140 (35.3) | 514 (60.4) | 171 (36.4) |
| ≤12 | 104 (42.5) | 83 (57.2) | 46 (49.5) | 41 (56.9) | 127 (32.2) | 125 (65.8) | 279 (39.9) | 257 (64.7) | 337 (39.6) | 299 (63.6) |
| Parity | ||||||||||
| Nulliparous | 136 (55.5) | 70 (48.3) | 59 (63.4) | 43 (60.6) | 181 (46.2) | 81 (42.6) | 328 (47.1) | 181 (45.7) | 405 (47.7) | 208 (44.3) |
| Multiparous | 109 (44.5) | 75 (51.7) | 34 (36.6) | 28 (39.4) | 211 (53.8) | 109 (57.4) | 369 (52.9) | 215 (54.3) | 444 (52.3) | 262 (55.7) |
| SGA | 19 (7.8) | 14 (9.7) | 11 (11.8) | 10 (13.9) | 0 | 0 | 93 (13.3) | 69 (17.4) | 101 (11.9) | 73 (15.5) |
| PTB | 19 (7.8) | 17 (11.7) | 29 (31.2) | 19 (26.4) | 0 | 0 | 160 (22.9) | 97 (24.4) | 150 (17.6) | 95 (20.2) |
BMI, body mass index.
Genetic ancestry determined using 148 ancestry informative markers and STRUCTURE.
Disease-free controls are uncomplicated term births without preexisting hypertension used in Stage 1 for both outcomes.
GHTN and PE controls include pregnancies with small for gestational age (SGA) and preterm birth (PTB), but exclude those with pre-existing hypertension. GHTN controls further exclude women with PE.
Self-reported smoking during months 1–6 of pregnancy.
Pre-pregnancy BMI calculated from self-reported pre-pregnancy weight and measured height.
Covariates considered were maternal age, body mass index (BMI), maternal smoking, parity, and a continuous measure of socioeconomic status. A false discovery rate (FDR)41 of 20% was used to identify SNP sets which advanced to Stage 2 analysis. P-values from SKAT were transformed to Q42 values using R (p.adjust, FDR)43 accounting for the number of SNP sets analyzed (N = 31).
Among the genes that advanced to Stage 2 analysis, individual SNP associations were measured. Given the relatively common outcome of GHTN (14% in the underlying PIN cohort, Table S1), associations were measured using risk ratios from a log-linear risk model. Dominant genetic models were used unless the smallest genotype cell size was >5. Each outcome was compared to a control group, which included all genotyped pregnancies without the outcome of interest (‘Control’ columns in Tables I and II). For the outcome of term SGA, only term births were included in the control group.
Inverse probability of selection weighting was used so that the estimates are generalizable to the entire eligible population. Briefly, a logistic model was used to calculate the probability that an eligible woman (N = 3065) was selected for genotyping (N = 1646) based on all covariates in Table I, and other study-related, demographic and pregnancy-related variables. The inverse of this probability was then used to weight the model. Robust variances were used, although these likely overestimate the true variance.44
Top-ranking SNPs from Stage 2 were reported based on the consistency of the observed association across related outcomes (SGA and term SGA) as well as between genetic ancestry groups. Analyses were stratified on genetic ancestry and additionally adjusted for a continuous ancestry variable.
Results
The final analysis dataset included 1598 women. Tables I and II present the demographic characteristics of the case and control groups for SGA and hypertensive disorders of pregnancy, respectively. Women who consented and were successfully geno-typed were similar to the underlying PIN cohort (Table S1) and were predominately White (62.6%), well educated (51.9% with more than high school education), non-smokers (74.4%) with a mean age of 26.1 years, and a mean BMI of 26.5 kg/m2. Although the distribution of some covariates differed by self-identified race, the differences were not as pronounced between cases and controls within strata of genetic ancestry (Tables I and II). Adjustment for any or all of the covariates in Table I failed to change the single SNP point estimates more than 10% (results not shown). Therefore, we present estimates adjusted only for genetic ancestry.
Stage 1: Gene-Set Analysis in SKAT
SNPs were combined into 31 SNP sets. Due to proximity on the genome, tumor necrosis factor (TNF) and lymphotoxin α (TNF superfamily member 1)(LTA), IL-13 (IL13) and IL-4 (IL4), and cyclin-dependent kinase inhibitor 2A (CDKN2A) and 2B (CDKN2B) were considered as single SNP sets. Four SNP sets met our FDR criteria of 20% in Stage 1. IL-6 (IL6) (q = 0.17) and killer cell lectin-like receptor subfamily D, member 1 (KLRD1) (q = 0.17) were associated with term SGA among African Americans, and T-box 21 (TBX21) (q = 0.05) and LTA/TNF (q = 0.05) were associated with PE among European Americans. No SNP sets met the FDR criteria for SGA overall or GHTN (Table III).
Table III.
Qa Values from SKAT Analysis for each SNP Set Stratified by Genetic Ancestry
| Pathway/gene | European Americanb |
African Americanb |
European American |
African American |
||||
|---|---|---|---|---|---|---|---|---|
| SGA | Term SGA | SGA | Term SGA | GHTN | PE | GHTN | PE | |
| Inflammatory | ||||||||
| TBX21 c | 1.00 | 1.00 | 0.87 | 0.83 | 0.84 | 0.05 | 0.83 | 0.99 |
| LTA&TNF c | 0.84 | 0.91 | 1.00 | 1.00 | 0.58 | 0.05 | 0.83 | 0.81 |
| IL6 c | 0.84 | 1.00 | 0.87 | 0.17 | 0.58 | 0.91 | 0.83 | 0.99 |
| KLRD1 c | 0.84 | 0.91 | 0.87 | 0.17 | 0.83 | 0.91 | 1.00 | 1.00 |
| IL12B | 0.84 | 1.00 | 0.87 | 0.83 | 0.35 | 0.91 | 0.83 | 1.00 |
| IL2 | 0.84 | 1.00 | 0.87 | 0.83 | 0.35 | 0.91 | 0.83 | 0.81 |
| IL10 | 0.84 | 1.00 | 0.87 | 0.83 | 0.35 | 0.91 | 0.83 | 0.81 |
| IFNGR2 | 0.84 | 1.00 | 1.00 | 1.00 | 0.80 | 0.39 | 0.84 | 0.81 |
| IL12A | 0.84 | 0.91 | 0.87 | 0.83 | 0.80 | 0.39 | 0.83 | 0.99 |
| KIR2DL4 | 0.84 | 1.00 | 0.87 | 0.87 | 0.53 | 0.91 | 0.83 | 0.81 |
| IL13&IL4 | 0.84 | 1.00 | 0.87 | 0.87 | 0.58 | 0.81 | 0.83 | 0.81 |
| IL6R | 0.84 | 0.91 | 0.87 | 0.83 | 0.58 | 0.91 | 0.83 | 0.81 |
| IL8 | 0.84 | 0.91 | 0.87 | 0.83 | 0.62 | 0.91 | 0.83 | 0.81 |
| K R3DL3 | 0.84 | 1.00 | 0.94 | 1.00 | 0.62 | 0.91 | 0.83 | 0.81 |
| IL18 | 0.84 | 1.00 | 0.87 | 0.83 | 0.80 | 0.81 | 0.83 | 0.81 |
| CSF2 | 0.84 | 1.00 | 0.87 | 0.83 | 0.80 | 0.91 | 0.84 | 1.00 |
| IFNG | 1.00 | 1.00 | 0.88 | 1.00 | 0.80 | 1.00 | 0.84 | 0.81 |
| NFKB1 | 0.84 | 0.91 | 0.89 | 0.83 | 0.80 | 0.81 | 0.84 | 1.00 |
| TNFRSF1B | 0.84 | 0.91 | 0.87 | 0.83 | 0.98 | 0.91 | 0.83 | 0.81 |
| IL15 | 0.84 | 1.00 | 0.87 | 0.83 | 0.80 | 0.53 | 0.83 | 0.81 |
| KIR3DL2 | 0.84 | 1.00 | 0.87 | 0.84 | 0.92 | 0.81 | 0.83 | 0.81 |
| GATA3 | 0.84 | 1.00 | 0.87 | 0.83 | 0.84 | 1.00 | 0.83 | 0.81 |
| IL8RB | 0.84 | 0.91 | 0.87 | 0.83 | 1.00 | 0.91 | 0.83 | 0.81 |
| CXCL10 | 0.84 | 1.00 | 0.87 | 0.83 | 0.80 | 0.91 | 0.84 | 0.81 |
| Cell cycle | ||||||||
| RASSF1 | 0.84 | 1.00 | 0.79 | 0.83 | 0.35 | 0.23 | 0.83 | 0.81 |
| NOV | 0.84 | 1.00 | 0.87 | 0.83 | 0.80 | 0.81 | 0.83 | 0.81 |
| CNNM2 | 0.84 | 1.00 | 0.87 | 0.83 | 0.80 | 0.81 | 0.83 | 0.81 |
| GADD45A | 0.84 | 1.00 | 0.87 | 0.83 | 0.80 | 0.91 | 0.83 | 0.88 |
| CDKN2A&CDKN2B | 0.84 | 1.00 | 0.87 | 0.83 | 0.80 | 0.91 | 0.84 | 0.81 |
| CCND1 | 0.84 | 1.00 | 0.87 | 0.83 | 1.00 | 0.91 | 0.83 | 0.81 |
| CCNA2 | 1.00 | 1.00 | 0.87 | 0.83 | 1.00 | 1.00 | 0.83 | 1.00 |
Bold values indicate significant (Q <0.20).
SKAT, SNP-set Kernel Association Test; SGA, small for gestational age; GHTN, gestational hypertension; PE, preeclampsia; AIMs, ancestry informative markers; FDR, false discovery rate.
Q values represent the proportion of false positives (number of false rejections/total number of rejections).
Genetic ancestry determined from 148 AIMS and STRUCTURE.
Meets FDR <0.20 for at least one outcome.
Stage 2: Single SNP Analysis
There were 55 individual SNPs within the SNP sets that passed FDR criteria in Stage 1, and eight additional SNPs not included in Stage 1 that were assessed in Stage 2 analyses (Tables IV and V). Variant allele frequencies and genotype frequencies for SNPs reported in Tables IV and V can be found in Tables S2–S5. Single SNP results for all genes and outcomes can be found in Tables S6 and S7.
Table IV.
Single SNP Associations, Risk Ratio, and 95% Confidence Interval for Maternal SNPs for SGA and Term SGA in Infants Stratified by Maternal Genetic Ancestry
| African Americana |
European Americana |
|||
|---|---|---|---|---|
| Gene/SNP | SGA | Term SGA | SGA | Term SGA |
| IL6 b | ||||
| rs4719711 | 1.0 (0.7, 1.6) | 0.9 (0.5, 1.5) | 0.7 (0.4, 0.9) | 0.7 (0.4, 1.0) |
| rs6963444 | 1.8 (1.2, 2.8) | 2.5 (1.5, 4.2) | 1.9 (1.0, 3.7) | 2.1 (1.0, 4.2) |
| rs1546762 | 1.1 (0.7, 1.7) | 0.8 (0.5, 1.5) | 0.6 (0.4, 0.9) | 0.6 (0.4, 1.0) |
| rs7784987 | 1.5 (1.0, 2.4) | 2.1 (1.2, 3.6) | 2.0 (1.0, 3.8) | 2.1 (1.1, 4.4) |
| rs3087221 | 1.4 (0.9, 2.3) | 1.8 (1.1, 3.1) | 2.3 (1.0, 5.4) | 2.0 (0.8, 5.4) |
| rs1800795 | 1.1 (0.6, 2.2) | 0.7 (0.3, 1.8) | 0.9 (0.6, 1.3) | 1.0 (0.7, 1.6) |
| rs1548216 | 1.6 (1.0, 2.5) | 2.1 (1.2, 3.5) | 2.3 (1.2, 4.2) | 2.4 (1.2, 4.7) |
| rs2069843c | 1.5 (1.0, 2.3) | 2.0 (1.3, 3.1) | 2.1 (1.2, 3.8) | 2.4 (1.2, 4.6) |
| rs2069849 | 1.4 (0.9, 2.3) | 1.9 (1.1, 3.2) | 2.2 (1.2, 4.1) | 2.4 (1.2, 4.6) |
| KLRD1 | ||||
| rs3759270 | 0.6 (0.4, 1.0) | 0.4 (0.3, 0.8) | 1.3 (0.9, 2.0) | 1.3 (0.8, 2.0) |
| rs3809214c | 1.3 (0.9, 1.8) | 1.6 (1.1, 2.4) | 0.8 (0.6, 1.0) | 0.8 (0.6, 1.1) |
| rs2302489c | 1.3 (0.9, 1.8) | 1.6 (1.1, 2.4) | 0.8 (0.6, 1.1) | 0.8 (0.6, 1.1) |
| rs7301562 | 0.6 (0.4, 1.0) | 0.5 (0.3, 0.9) | 1.1 (0.2, 7.9) | 1.6 (0.2, 11.8) |
| rs10772256 | 0.6 (0.4, 1.0) | 0.5 (0.3, 0.9) | 1.1 (0.2, 7.9) | 1.6 (0.2, 11.8) |
| rs2270238 | 0.8 (0.4, 1.8) | 0.8 (0.3, 2.1) | 1.5 (1.0, 2.2) | 1.5 (1.0, 2.4) |
| rs11611333c | 1.1 (0.8, 1.5) | 1.0 (0.7, 1.5) | 1.3 (1.0, 1.6) | 1.2 (0.9, 1.7) |
| rs12829155c | 1.3 (0.9, 1.9) | 1.5 (0.9, 2.4) | 0.8 (0.6, 1.0) | 0.8 (0.6, 1.1) |
SGA, small for gestational age.
Allele and genotype frequencies presented in Tables S2 and S3.
Genetic ancestry determined from 148 ancestry informative markers and STRUCTURE.
Selected results for IL6 presented. Results from all SNPs available in Table S6.
Additive model presented.
Table V.
Single SNP Associations, Risk Ratio, and 95% Confidence Interval for Maternal SNPs and PE Stratified by Genetic Ancestry
| Gene/SNP | European Americana | African Americana |
|---|---|---|
| NFKBIL1 | ||
| rs2857605 | 1.1 (0.7, 1.6) | 1.0 (0.5, 1.9) |
| rs2239707 | 1.0 (0.6, 1.5) | 1.4 (0.8, 2.4) |
| rs2230365 | 1.0 (0.6, 1.5) | 1.5 (0.8, 3.0) |
| rs3130062 | 1.2 (0.7, 2.1) | 1.7 (0.5, 5.4) |
| rs4947324 | 0.6 (0.3, 1.1) | 0.9 (0.5, 1.7) |
| rs2857709 | 1.0 (0.6, 1.6) | 0.8 (0.3, 1.9) |
| LTA | ||
| rs915654 | 1.0 (0.6, 1.5) | 1.7 (0.9, 3.1) |
| rs909253b | 1.5 (1.1, 2.0) | 1.0 (0.7, 1.4) |
| rs2229094 | 0.6 (0.4, 1.0) | 1.3 (0.8, 2.1) |
| rs1041981b | 1.4 (1.0, 2.0) | 1.0 (0.7, 1.5) |
| Intergenic | ||
| rs1799964 | 0.7 (0.4, 1.1) | 1.0 (0.6, 1.8) |
| rs1800630 | 0.8 (0.5, 1.2) | 1.4 (0.8, 2.4) |
| TNF | ||
| rs1800629 | 1.8 (1.1, 2.7) | 0.6 (0.3, 1.2) |
| rs7769073 | 0.8 (0.4, 1.8) | 0.9 (0.4, 1.7) |
| TBKBP1 | ||
| rs2013383 | 0.6 (0.4, 0.9) | 1.1 (0.7, 1.8) |
| rs1808192 | 0.6 (0.4, 0.9) | 1.0 (0.6, 1.6) |
| TBX21 | ||
| rs4461115 | 1.2 (0.8, 1.9) | 0.8 (0.3, 2.0) |
| rs16946264 | 1.1 (0.6, 1.8) | 1.0 (0.6, 1.6) |
| rs11079788 | 0.8 (0.5, 1.3) | 1.2 (0.7, 1.9) |
| rs16946878 | 1.3 (0.7, 2.4) | 1.0 (0.5, 1.9) |
| rs16947078 | 0.8 (0.5, 1.3) | 0.8 (0.5, 1.3) |
PE, preeclampsia.
All single SNP estimates are presented in Table S7.
Allele and genotype frequencies presented in Tables S4 and S5.
Genetic ancestry determined from 148 ancestry informative markers and STRUCTURE.
Additive model presented.
Term SGA
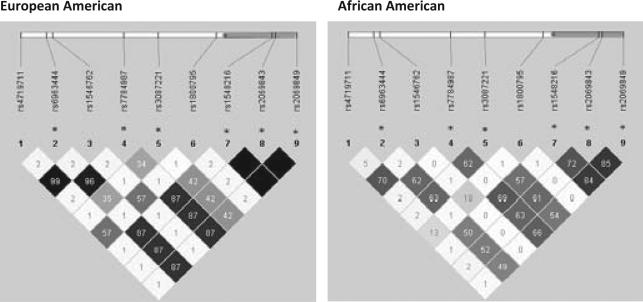
In Stage 1, IL6 and KLRD1 were associated with term SGA in African Americans. Single SNP associations within IL6, however, were generally consistent in both ancestral groups, although slightly weaker for European Americans (Table IV). The strongest single SNP association for term SGA for both ancestry groups was seen for a group of three IL6 SNPs (rs1548216, rs2069843, and rs2069849), all having risk ratios in the range of 1.9–2.4. This group of SNPs is in full LD (r2 = 1) among European Americans and partial LD in African Americans (r2 = 0.72– 0.85) (Fig. 1). An additional group of SNPs upstream of IL6 was also associated with an increased risk of term SGA of similar magnitude for both African and European Americans (rs6963444, rs7784987, and rs3087221).
Fig. 1.
Linkage disequilibrium (r2) in IL6 stratified by genetic ancestry. r2 is the amount of correlation between two SNPs with empty black cells representing 100% correlation and lighter cells representing less correlation. *SNPs are mentioned in results section. Approximate SNP location (vertical bars) and IL6 location (grey bar) noted along genome.
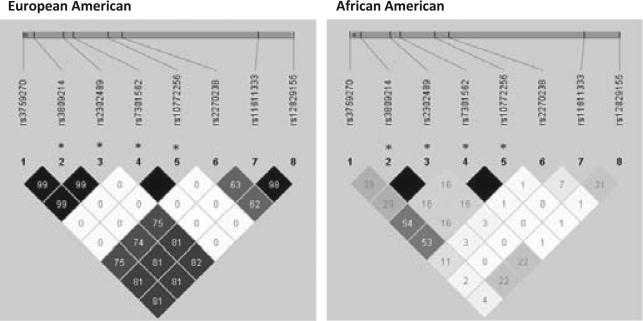
For KLRD1, however, single SNP associations were not similar across the two ancestry groups. In Euro pean Americans, the tagSNPs represent a single block of high LD, except for two SNPs which were quite rare in European Americans (rs10772256 and rs7301562) (Fig. 2, Table S2). Seen as a block, there was an overall null association with term SGA for European Americans. In contrast, among African Americans, a number of SNPs were relatively strongly associated with term SGA. In particular, rs10772256 and rs7301562 (which were in high LD, r2 = 1) were associated with a decreased risk of term SGA (RR = 0.5, 95% CI 0.3–0.9). Additionally, rs3759270 was associated with a substantially decreased risk of term SGA (RR = 0.4, 95% CI 0.3–0.8). Finally, two SNPs in perfect LD (rs3809214 and rs2302489) were associated with an increased risk of term SGA (RR = 1.6) among African Americans.
Fig. 2.
Linkage disequilibrium (r2) in KLRD1 stratified by genetic ancestry. r2 is the amount of correlation between two SNPs with empty black cells representing 100% correlation and lighter cells representing less correlation. *SNPs are mentioned in results section. Approximate SNP location (vertical bars) and KLRD1 location (grey bar) noted along genome.
Preeclampsia
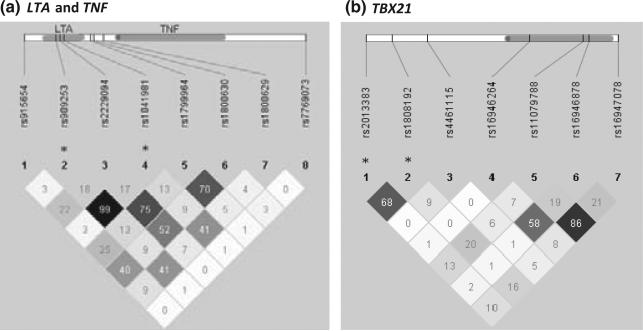
LTA/TNF and TBX21 were associated at the gene level with PE, but only for European American women. Moreover, the single SNP results were generally null for African Americans (Table V). LTA and TNF are quite close on chromosome 6 (6p21.3), and tagging for LTA also captured regions of the adjacent gene nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibition-like 1 (NFKBIL1). Single SNP results for tags within NFKBIL1 were generally null. Among European Americans, two SNPs in LTA that were in high LD (r2 = 0.99) (Fig. 3a), rs909253 and rs1041981, were associated with an increased risk of PE (RR = 1.5 and 1.4). A missense SNP in LTA rs2229094 was associated with a decreased risk of PE (RR = 0.6, 95% CI 0.4–1.0). In TNF, rs1800629, an upstream SNP that is thought to upregulate TNF expression,45 was found to increase the risk of PE (RR = 1.8, 95% CI 1.1–2.7).
Fig. 3.
Linkage disequilibrium (r2) among European Americans in (a) LTA, TNF, and (b) TBX21. r2 is the amount of correlation between two SNPs with empty black cells representing 100% correlation and lighter cells representing less correlation. *SNPs are mentioned in results section. Approximate SNP location (vertical bars) and gene location (grey bar) noted along genome.
TagSNPs for TBX21 captured two upstream SNPs which were closer to an adjacent gene TBK-binding protein 1 (TBKBP1) (rs2013383 and rs1808192). These two SNPs are in moderate LD with each other (r2 = 0.68) (Fig. 3b) and have long-range LD extending through TBKBP1 with very little pairwise or long-range LD with typed or untyped SNPs in TBX21. The two TBKBP1 SNPs were associated with a decreased risk of PE RR = 0.6. For tagSNPs within TBX21 itself, the associations were null.
Discussion
We examined 40 genes related to inflammation and cell cycle pathways, and their associations with SGA, GHTN and PE in a biracial North Carolina popula tion. There were no significant associations for any of the cell cycle genes. However, a number of interesting and novel associations were found for inflammatory gene variants. Among African American women, IL6 and KLRD1 were associated with term SGA. Single SNP associations within IL6 suggest that a similar, though weaker, association may exist for European Americans. For the outcome of PE, associations within LTA and TNF were seen for European American women. Two SNPs within a novel gene, TBKBP1, were associated with a decreased risk of PE among European American women only.
We report gene-level associations among African Americans only within the term SGA phenotype, suggesting that these associations are mediated by pathways unrelated to prematurity. It is unclear why distinctive association patterns emerged for African Americans and Whites; however, it may reflect distinct phenotypes, or possibly interactions with social or environmental exposures that are differentially distributed across these two populations. IL6 was chosen as a candidate gene due to evidence that in early pregnancy IL6 may influence the ability of the placenta to become adequately implanted.46IL6 has been examined previously in relation to SGA with null associations, although many fewer SNPs were used to tag the gene,21,47,48 and, apart from the Engel study, the populations were White.
KLRD1 (CD94) codes for a portion of a receptor on both NK- and T-cells that recognize HLA-E molecules expressed by trophoblasts during pregnancy.49 Tight regulation of maternal immunity at the time of implantation mitigates against rejection of the fetal tropho-blasts and permits adequate placental development and implantation. Dysregulation of KLRD1 might result in a heightened inflammatory response to the invading placenta resulting in inadequate placentation impairing fetal growth. In this study, polymorphisms in KLRD1 were only associated with term SGA among African Americans. Although genetic diversity of HLA-E is much lower than the diversity seen in other major histocompatibility genes, there have been haplotype differences documented between individuals of European and African descent.50 Underlying differences in HLAE haplotypes may explain these discrepant associations across racial groups.
Genes coding for TNF-α (TNF) and TNF-β (LTA) were associated with an increased risk of PE among European American women. TNF-α has been implicated in the pathogenesis of PE1 both as a modulator of placental implantation and as a response to the hypoxia created by poor placental perfusion.13 Infusion of TNF-α induces hypertension in pregnant rats.29 Despite the credibility of the biology implicating TNF-α with PE, results of genetic epidemiologic studies have been less encouraging. Meta-analyses of the 308A missense mutation (rs18000629)51,52 found null pooled estimates. The meta-analyses, however, combined estimates from populations of very different genetic ancestries. It has previously been reported that allele frequencies in TNF and its related receptors vary substantially by self-reported race. Moreover, regulation of TNF-α levels during pregnancy appears to differ between White and African American women.53 In our stratified analysis, an association at the gene level was only apparent for European Americans. Despite the null association in the meta-analysis, the complexity of TNF regulation and apparent heterogeneity based on genetic ancestry support further investigation into this gene.
TBX21 is an important regulator of TH1 immunity.54 However, the strongest single SNP association in our study may have been with a tagSNP associated with a close upstream gene TBKBP1 (aka SINTBAD). TBKBP1 was identified in 2004, and its function is still being elucidated. It appears to be related to signaling in the TNF-α/NFκB pathways and the activation of interferon.55,56 Given the role of TNF-α in PE, further investigation of this novel gene is warranted.
Preeclampsia is a heterogeneous disease and may represent a number of distinct phenotypes with a shared clinical presentation. We did not collect data on the severity of disease or the timing of clinical onset and therefore we were unable to subdivide our case group into more homogeneous phenotypes. In addition, the changing diagnostic criteria over the course of the study resulted in further heterogeneity of phenotype based on the year of diagnosis. Newly released diagnostic criteria present the possibility that there is under ascertainment of PE cases among women with hypertension and other signs of end-organ involvement who do not present with protein-uria.57 These shortcomings reduce our power to identify associations that may exist between SNPs and specific subtypes of PE (for example, early onset disease or PE with severe features). Future studies of PE will benefit from increased sample size and additional clinical information that will permit the classification of cases based on gestational age at onset and the presence of severe features.
The heterogeneity in the PE phenotype may also be the result of interactions between genetic susceptibility and environmental exposures such as smoking, infections, nutrition, or exposure to environmental toxicants. Although we were underpowered to explore gene by environment interactions in this study, the strong protective association between smoking and PE offers an opportunity for future genetic association studies to explore gene by environment interactions.
While our study comprises the largest bi-racial study of inflammatory genes in relation to these important obstetric outcomes, it suffers from some notable limitations. This study was limited by the lack of fetal DNA, which is an important genetic factor in both of these reproductive outcomes. However, the results of this study support a role of maternal genes in these outcomes. Although we incorporated genetic variation in upstream and downstream regions, our pre-specified boundaries may be too narrow to capture long-range regulatory elements.58 Finally, previous research has demonstrated that a candidate gene approach has been only modestly successful in identifying genetic associations that replicate across populations. Nonetheless, the results presented herein are consistent with current understanding of the biological underpinning of PE and SGA, although verification in an independent population is needed.
Conclusions
This study expanded coverage of candidate inflammatory genes and examined novel cell cycle and NK cell genes while carefully addressing population stratification. The results of this study reveal important differences in the genetic underpinnings of term SGA and PE in European and African Americans while also finding that there may be pathways in common. Novel associations between KLRD1 and term SGA, and TBKBP1 and PE warrant further exploration, ideally with enhanced tagging of polymorphisms within TBKBP1 and consideration of HLA-E haplotypes for KLRD1.
Supplementary Material
Acknowledgments
Funding for this study was provided by NICHD R21 HD060207 (PI, Engel SM), NIEHS 2-T32-ES07018 (Harmon), NIEHS P30ES010126 (Olshan), and the Mount Sinai Center for Children's Environmental Health. Funding for PIN was provided by NIH HD28684, HD28684A, HD37584, HD39373, and RR00046. We would also like to gratefully acknowledge the significant contributions of the following personnel Dr. Patricia Basta UNC Biospecimen Processing Facility, Kathryn Carrier UNC Project Manager, Michael Andre and Amanda Beaty for genotyping at UNC Mammalian Genomics Core and Alison Wise UNC Department of Biostatistics for help with validation of the PE outcome.
Footnotes
Citation Harmon QE, Engel SM, Wu MC, Moran TM, Luo J, Stuebe AM, Avery CL, Olshan AF. Polymorphisms in inflammatory genes are associated with term small for gestational age and preeclampsia. Am J Reprod Immunol 2014
References
- 1.Sibai B, Dekker G, Kupferminc M. Pre-eclampsia. Lancet. 2005;365:785–799. doi: 10.1016/S0140-6736(05)17987-2. [DOI] [PubMed] [Google Scholar]
- 2.Ness RB, Sibai BM. Shared and disparate components of the pathophysiologies of fetal growth restriction and preeclampsia. Am J Obstet Gynecol. 2006;195:40–49. doi: 10.1016/j.ajog.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 3.Bulletins–Obstetrics ACoP: ACOG practice bulletin. Diagnosis and management of preeclampsia and eclampsia. Obstet Gynecol. 2002 Jan;99(33):159–167. doi: 10.1016/s0029-7844(01)01747-1. 2002. [DOI] [PubMed] [Google Scholar]
- 4.Berg CJ, Callaghan WM, Syverson C, Henderson Z. Pregnancy-related mortality in the United States, 1998 to 2005. Obstet Gynecol 2010. 116:1302–1309. doi: 10.1097/AOG.0b013e3181fdfb11. [DOI] [PubMed] [Google Scholar]
- 5.Basso O, Rasmussen S, Weinberg CR, Wilcox AJ, Irgens LM, Skjaerven R. Trends in fetal and infant survival following preeclampsia. JAMA. 2006;296:1357–1362. doi: 10.1001/jama.296.11.1357. [DOI] [PubMed] [Google Scholar]
- 6.Hutcheon JA, Platt RW. The missing data problem in birth weight percentiles and thresholds for “small-for-gestational-age”. Am J Epidemiol. 2008;167:786–792. doi: 10.1093/aje/kwm327. [DOI] [PubMed] [Google Scholar]
- 7.Basso O, Wilcox AJ, Weinberg CR. Birth weight and mortality: causality or confounding? Am J Epidemiol. 2006;164:303–311. doi: 10.1093/aje/kwj237. [DOI] [PubMed] [Google Scholar]
- 8.Wilcox AJ. Fertility and Pregnancy: An Epidemiologic Perspective. Oxford Univeristy Press; New York, NY: 2010. [Google Scholar]
- 9.Bellamy L, Casas JP, Hingorani AD, Williams DJ. Pre-eclampsia and risk of cardiovascular disease and cancer in later life: systematic review and meta-analysis. BMJ. 2007;335:974. doi: 10.1136/bmj.39335.385301.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey Smith G, Hypponen E, Power C, Lawlor DA. Offspring birth weight and parental mortality: prospective observational study and meta-analysis. Am J Epidemiol. 2007;166:160–169. doi: 10.1093/aje/kwm054. [DOI] [PubMed] [Google Scholar]
- 11.Catov JM, Newman AB, Roberts JM, Kelsey SF, Sutton-Tyrrell K, Harris TB, Colbert L, Rubin SM, Satterfield S, Ness RB. the Health ABCS: Preterm delivery and later maternal cardiovascular disease risk. Epidemiology. 2007;18:733–739. doi: 10.1097/EDE.0b013e3181567f96. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann P, Black S, Huppertz B. Endovascular trophoblast invasion: implications for the pathogenesis of intrauterine growth retardation and preeclampsia. Biol Reprod. 2003;69:1–7. doi: 10.1095/biolreprod.102.014977. [DOI] [PubMed] [Google Scholar]
- 13.LaMarca BD, Ryan MJ, Gilbert JS, Murphy SR, Granger JP. Inflammatory cytokines in the pathophysiology of hypertension during preeclampsia. Curr Hypertens Rep. 2007;9:480–485. doi: 10.1007/s11906-007-0088-1. [DOI] [PubMed] [Google Scholar]
- 14.Khong TY, De Wolf F, Robertson WB, Brosens I. Inadequate maternal vascular response to placentation in pregnancies complicated by pre-eclampsia and by small-for-gestational age infants. Br J Obstet Gynaecol. 1986;93:1049–1059. doi: 10.1111/j.1471-0528.1986.tb07830.x. [DOI] [PubMed] [Google Scholar]
- 15.Bowen JM, Chamley L, Keelan JA, Mitchell MD. Cytokines of the placenta and extra-placental membranes: roles and regulation during human pregnancy and parturition. Placenta. 2002;23:257–273. doi: 10.1053/plac.2001.0782. [DOI] [PubMed] [Google Scholar]
- 16.Saito S. The role of the immune system in preeclampsia. Mol Aspects Med. 2007;28:192–209. doi: 10.1016/j.mam.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Ahn H, Park J, Gilman-Sachs A, Kwak-Kim J. Immunologic characteristics of preeclampsia, a comprehensive review. Am J Reprod Immunol. 2011;65:377–394. doi: 10.1111/j.1600-0897.2010.00913.x. [DOI] [PubMed] [Google Scholar]
- 18.Ness RB, Bass D, Hill L, Klebanoff MA, Zhang J. Diagnostic test characteristics of placental weight in the prediction of small-forgestational-age neonates. J Reprod Med. 2007;52:793–800. [PubMed] [Google Scholar]
- 19.Freeman DJ, McManus F, Brown EA, Cherry L, Norrie J, Ramsay JE, Clark P, Walker ID, Sattar N, Greer IA. Short- and long-term changes in plasma inflammatory markers associated with preeclampsia. Hypertension. 2004;44:708–714. doi: 10.1161/01.HYP.0000143849.67254.ca. [DOI] [PubMed] [Google Scholar]
- 20.Bowen JM, Chamley L, Mitchell MD, Keelan JA. Cytokines of the placenta and extra-placental membranes: biosynthesis, secretion and roles in establishment of pregnancy in women. Placenta. 2002;23:239–256. doi: 10.1053/plac.2001.0781. [DOI] [PubMed] [Google Scholar]
- 21.Engel SA, Olshan AF, Savitz DA, Thorp J, Erichsen HC, Chanock SJ. Risk of small-for-gestational age is associated with common anti-inflammatory cytokine polymorphisms. Epidemiology. 2005;16:478–486. doi: 10.1097/01.ede.0000164535.36412.6b. [DOI] [PubMed] [Google Scholar]
- 22.De Falco M, Fedele V, Cobellis L, Mastrogiacomo A, Giraldi D, Leone S, De Luca L, Laforgia V, De Luca A. Pattern of expression of cyclin D1/CDK4 complex in human placenta during gestation. Cell Tissue Res. 2004;317:187–194. doi: 10.1007/s00441-004-0880-z. [DOI] [PubMed] [Google Scholar]
- 23.Handwerger S, Aronow B. Dynamic changes in gene expression during human trophoblast differentiation. Recent Prog Horm Res. 2003;58:263–281. doi: 10.1210/rp.58.1.263. [DOI] [PubMed] [Google Scholar]
- 24.Xiong Y, Liebermann DA, Tront JS, Holtzman EJ, Huang Y, Hoffman B, Geifman-Holtzman O. Gadd45a stress signaling regulates sFlt-1 expression in preeclampsia. J Cell Physiol. 2009;220:632–639. doi: 10.1002/jcp.21800. [DOI] [PubMed] [Google Scholar]
- 25.Gellhaus A, Schmidt M, Dunk C, Lye SJ, Winterhager E. The circulating proangiogenic factors CYR61 (CCN1) and NOV (CCN3) are significantly decreased in placentae and sera of preeclamptic patients. Reprod Sci. 2007;14:46–52. doi: 10.1177/1933719107309816. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson E, Salonen Ros H, Cnattingius S, Lichtenstein P. The importance of genetic and environmental effects for pre-eclampsia and gestational hypertension: a family study. BJOG. 2004;111:200–206. doi: 10.1111/j.1471-0528.2004.00042x.x. [DOI] [PubMed] [Google Scholar]
- 27.Svensson AC, Pawitan Y, Cnattingius S, Reilly M, Lichtenstein P. Familial aggregation of small-for-gestational-age births: the importance of fetal genetic effects. Am J Obstet Gynecol. 2006;194:475–479. doi: 10.1016/j.ajog.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 28.Mutze S, Rudnik-Schoneborn S, Zerres K, Rath W. Genes and the preeclampsia syndrome. J Perinat Med. 2008;36:38–58. doi: 10.1515/JPM.2008.004. [DOI] [PubMed] [Google Scholar]
- 29.Williams PJ, Pipkin FB. The genetics of pre-eclampsia and other hypertensive disorders of pregnancy. Best Pract Res Clin Obstet Gynaecol. 2011;25:405–417. doi: 10.1016/j.bpobgyn.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savitz DA, Dole N, Williams J, Thorp JM, McDonald T, Carter AC, Eucker B. Determinants of participation in an epidemiological study of preterm delivery. Paediatr Perinat Epidemiol. 1999;13:114–125. doi: 10.1046/j.1365-3016.1999.00156.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Bowes WA., Jr Birth-weight-for-gestational-age patterns by race, sex, and parity in the United States population. Obstet Gynecol. 1995;86:200–208. doi: 10.1016/0029-7844(95)00142-e. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Klebanoff MA, Roberts JM. Prediction of adverse outcomes by common definitions of hypertension in pregnancy. Obstet Gynecol. 2001;97:261–267. doi: 10.1016/s0029-7844(00)01125-x. [DOI] [PubMed] [Google Scholar]
- 33.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.SAS Institute Inc . SAS/GeneticsTM 9.3 User's Guide. SAS Institute Inc.; Cary, NC: 2011. [Google Scholar]
- 35.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 36.Johnson AD, Handsaker RE, Pulit S, Nizzari MM, O'Donnell CJ, de Bakker PIW. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Dyke AL, Cote ML, Wenzlaff AS, Land S, Schwartz AG. Cytokine SNPs: comparison of allele frequencies by race and implications for future studies. Cytokine. 2009;46:236–244. doi: 10.1016/j.cyto.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnholtz-Sloan JS, Shetty PB, Guan X, Nyante SJ, Luo J, Brennan DJ, Millikan RC. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis. 2010;31:1417–1423. doi: 10.1093/carcin/bgq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, Hunter DJ, Lin X. Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet. 2010;86:929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol. 1995;57:289–300. [Google Scholar]
- 42.Shaffer JP. Multiple hypothesis testing. Annu Rev Psychol. 1995;46:561–584. [Google Scholar]
- 43.Team RDC . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- 44.Sato T, Matsuyama Y. Marginal structural models as a tool for standardization. Epidemiology. 2003;14:680–686. doi: 10.1097/01.EDE.0000081989.82616.7d. [DOI] [PubMed] [Google Scholar]
- 45.Haggerty CL, Ferrell RE, Hubel CA, Markovic N, Harger G, Ness RB. Association between allelic variants in cytokine genes and preeclampsia. Am J Obstet Gynecol. 2005;193:209–215. doi: 10.1016/j.ajog.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Markert UR, Morales-Prieto DM, Fitzgerald JS. Understanding the link between the IL-6 cytokine family and pregnancy: implications for future therapeutics. Expert Rev Clin Immunol. 2011;7:603–609. doi: 10.1586/eci.11.60. [DOI] [PubMed] [Google Scholar]
- 47.Stonek F, Metzenbauer M, Hafner E, Philipp K, Tempfer C. Interleukin 6-174G/C promoter polymorphism and pregnancy complications: results of a prospective cohort study in 1626 pregnant women. Am J Reprod Immunol. 2008;59:347–351. doi: 10.1111/j.1600-0897.2007.00577.x. [DOI] [PubMed] [Google Scholar]
- 48.Edwards DR, Romero R, Kusanovic JP, Hassan SS, Mazaki-Tovi S, Vaisbuch E, Kim CJ, Erez O, Chaiworapongsa T, Pearce BD, Bartlett J, Friel LA, Salisbury BA, Anant MK, Vovis GF, Lee MS, Gomez R, Behnke E, Oyarzun E, Tromp G, Menon R, Williams SM. Polymorphisms in maternal and fetal genes encoding for proteins involved in extracellular matrix metabolism alter the risk for small-for-gestational-age. J Matern Fetal Neonatal Med. 2011;24:362–380. doi: 10.3109/14767058.2010.497572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tilburgs T, van der Mast BJ, Nagtzaam NM, Roelen DL, Scherjon SA, Claas FH. Expression of NK cell receptors on decidual T cells in human pregnancy. J Reprod Immunol. 2009;80:22–32. doi: 10.1016/j.jri.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Antoun A, Jobson S, Cook M, Moss P, Briggs D. Ethnic variability in human leukocyte antigen-E haplotypes. Tissue Antigens. 2009;73:39–45. doi: 10.1111/j.1399-0039.2008.01170.x. [DOI] [PubMed] [Google Scholar]
- 51.Bombell S, McGuire W. Tumour necrosis factor (-308A) polymorphism in pre-eclampsia: meta-analysis of 16 case-control studies. Aust N Z J Obstet Gynaecol. 2008;48:547–551. doi: 10.1111/j.1479-828X.2008.00924.x. [DOI] [PubMed] [Google Scholar]
- 52.Buurma AJ, Turner RJ, Driessen JHM, Mooyaart AL, Schoones JW, Bruijn JA, Bloemenkamp KWM, Dekkers OM, Baelde HJ. Genetic variants in pre-eclampsia: a meta-analysis. Human Reprod Update. 2013;19:289–303. doi: 10.1093/humupd/dms060. [DOI] [PubMed] [Google Scholar]
- 53.Menon R, Velez DR, Morgan N, Lombardi SJ, Fortunato SJ, Williams SM. Genetic regulation of amniotic fluid TNF-alpha and soluble TNF receptor concentrations affected by race and preterm birth. Hum Genet. 2008;124:243–253. doi: 10.1007/s00439-008-0547-z. [DOI] [PubMed] [Google Scholar]
- 54.Lazarevic V, Glimcher LH. T-bet in disease. Nat Immunol. 2011;12:597–606. doi: 10.1038/ni.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B. Superti-Furga G: A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 56.Goncalves A, Burckstummer T, Dixit E, Scheicher R, Gorna MW, Karayel E, Sugar C, Stukalov A, Berg T, Kralovics R, Planyavsky M, Bennett KL, Colinge J, Superti-Furga G. Functional dissection of the TBK1 molecular network. PLoS ONE. 2011;6:e23971. doi: 10.1371/journal.pone.0023971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Task Force on Hypertension in Pregnancy: Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists. Obstet Gynecol. 2013;122:1122–1131. doi: 10.1097/01.AOG.0000437382.03963.88. [DOI] [PubMed] [Google Scholar]
- 58.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, Frazer KA. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.