Significance
Ribosomes synthesize proteins by translating mRNAs into linear chains of amino acids through the decoding of consecutive nucleotide triplets (codons). Specific mRNA signals, however, can stimulate ribosomes to shift into an alternative triplet reading frame (ribosomal frameshifting) resulting in translation of a different protein. Typically, such signals are regions of intramolecular nucleotide base-pairing in the mRNA which form structures that stall ribosome progress. Here we show that the frameshifting signal used to express the nsp2TF and nsp2N proteins of porcine reproductive and respiratory syndrome virus, an important swine pathogen, requires the action of a transacting viral protein rather than a structured RNA. This novel mechanism of gene expression may also be used by other viruses or in cellular gene expression.
Keywords: genetic recoding, translational control, nsp1beta
Abstract
Programmed −1 ribosomal frameshifting (−1 PRF) is a widely used translational mechanism facilitating the expression of two polypeptides from a single mRNA. Commonly, the ribosome interacts with an mRNA secondary structure that promotes −1 frameshifting on a homopolymeric slippery sequence. Recently, we described an unusual −2 frameshifting (−2 PRF) signal directing efficient expression of a transframe protein [nonstructural protein 2TF (nsp2TF)] of porcine reproductive and respiratory syndrome virus (PRRSV) from an alternative reading frame overlapping the viral replicase gene. Unusually, this arterivirus PRF signal lacks an obvious stimulatory RNA secondary structure, but as confirmed here, can also direct the occurrence of −1 PRF, yielding a third, truncated nsp2 variant named “nsp2N.” Remarkably, we now show that both −2 and −1 PRF are transactivated by a protein factor, specifically a PRRSV replicase subunit (nsp1β). Embedded in nsp1β’s papain-like autoproteinase domain, we identified a highly conserved, putative RNA-binding motif that is critical for PRF transactivation. The minimal RNA sequence required for PRF was mapped within a 34-nt region that includes the slippery sequence and a downstream conserved CCCANCUCC motif. Interaction of nsp1β with the PRF signal was demonstrated in pull-down assays. These studies demonstrate for the first time, to our knowledge, that a protein can function as a transactivator of ribosomal frameshifting. The newly identified frameshifting determinants provide potential antiviral targets for arterivirus disease control and prevention. Moreover, protein-induced transactivation of frameshifting may be a widely used mechanism, potentially including previously undiscovered viral strategies to regulate viral gene expression and/or modulate host cell translation upon infection.
Among the repertoire of mechanisms that viruses use to control or regulate their gene expression, noncanonical translation plays an important role, in particular for positive-strand RNA viruses whose genomic RNA serves a dual function as mRNA and genome (reviewed in ref. 1). A commonly used strategy is −1 programmed ribosomal frameshifting (−1 PRF), in which mRNA signals induce a significant proportion of translating ribosomes to change reading frame, with ribosomes slipping back (in the 5′ direction) by 1 nt into an overlapping ORF before continuing translation, generating a fusion protein composed of the products of both upstream and downstream ORFs (reviewed in refs. 1–4). PRF was first described as the mechanism by which the Gag-Pol polyprotein of the retrovirus Rous sarcoma virus is expressed from overlapping gag and pol ORFs (5, 6) and related signals have since been documented in many other viruses of medical, veterinary, and agricultural importance (7–11). PRF has also been increasingly recognized in cellular genes of both prokaryotes and eukaryotes as well as in other replicating elements, such as insertion sequences and transposons (12).
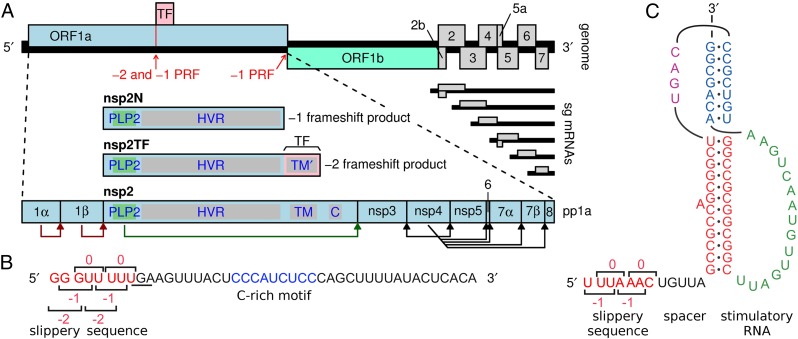
Recently, we identified an unusual −2 programmed ribosomal frameshifting (−2 PRF) event that operates during the translation of the genome of porcine reproductive and respiratory syndrome virus (PRRSV), a member of the arterivirus family in the order Nidovirales (13). PRRSV can be divided into distinct European (EU, type 1) and North American (NA, type 2) genotypes. The viral genome comprises a positive-sense RNA molecule, ∼15 kb in length (14). As in other nidoviruses, its 5′ proximal region contains two large replicase ORFs (ORF1a and ORF1b) (15), with the ORF1b product being expressed as a fusion with the ORF1a product following −1 PRF in the short ORF1a/ORF1b overlap region (Fig. 1). Four ORF1a-encoded proteinases (residing in nsp1α, nsp1β, nsp2, and nsp4) subsequently cleave the pp1a and pp1ab polyproteins into (at least) 14 different nonstructural proteins (nsps; Fig. 1A). The recently identified −2 PRF signal is located several kilobases upstream of the ORF1a/ORF1b −1 PRF signal, and maps to the part of ORF1a that encodes nsp2. This large, multifunctional replicase subunit is involved in diverse steps of the arterivirus replicative cycle, including replicase polyprotein processing (16), the formation of replication structures (17, 18), and innate immune evasion (19–22). At the PRRSV −2 PRF signal, a proportion of ribosomes back up 2 nt, to generate a transframe fusion protein (nsp2TF) comprising the N-terminal two-thirds of nsp2 and the product encoded by a conserved alternative ORF [transframe (TF)] in the −2 reading frame. Compared with full-length nsp2, the nsp2TF product is truncated, equipped with an alternative C-terminal transmembrane domain (Fig. 1A), and targeted to a different subcellular compartment (13). Mutations preventing nsp2TF expression reduce PRRSV replication efficiency in cell culture 50- to 100-fold, highlighting the biological importance of the frameshifting event and nsp2TF expression. The −2 PRF takes place at a highly conserved RG_GUU_UUU slippery sequence (R = G or A), and frameshifting is remarkably efficient (around 20% in virus-infected cells and up to 50% in expression systems) (13).
Fig. 1.
PRRSV genome organization and location of ribosomal frameshifting signals. (A) Overview of the ∼15-kb PRRSV genome. The long 5′ ORFs 1a and 1b encode nonstructural polyproteins, and at least eight shorter 3′ ORFs (2a-7) encode structural proteins. The 3′ ORFs are translated from a nested set of subgenomic mRNAs, two of which are bicistronic. ORF1a and ORF1b are translated from the genomic RNA, with translation of ORF1b depending on −1 PRF at the end of ORF1a. The TF ORF overlaps the central ORF1a region in the −2 reading frame and is accessed via −2 PRF (13). A −1 frameshift at the same site generates the nsp2N product (see details under the section “Alternative −2 and −1 PRF at the Same PRRSV Slippery Sequence”). The vertical red line indicates the location of the RG_GUU_UUU shift site (R = A or G, in different arteriviruses). Domains in nsp2/nsp2TF: C, Cys-rich domain HVR, hypervariable region; PLP2, papain-like proteinase;TM/TM′, (putative) transmembrane domains. (B) Sequence of the SD01-08 RNA in the region of the −2/−1 PRF signal, with the slippery sequence (red) and C-rich motif (blue) highlighted. The −1 reading frame stop codon is underlined and codons for each of the reading frames are indicated. (C) Features of the canonical −1 PRF signal present in the PRRSV ORF1a/ORF1b overlap region. The stimulatory RNA pseudoknot is composed of two stems connected by single-stranded loops.
As depicted in Fig. 1 B and C, the elements that promote PRF in PRRSV are quite distinct. The −1 PRF signal at the ORF1a/1b junction comprises a slippery sequence (generally U_UUA_AAC) where the ribosome changes frame, and a stimulatory RNA pseudoknot structure immediately downstream, an organization that is conserved throughout the Nidovirales order (23, 24) and widely used in other viral −1 PRF mechanisms. It is thought that interaction of the translating ribosome with the pseudoknot confounds its RNA-unwinding activity (25, 26) and may induce tension in the mRNA that assists in the uncoupling of codon–anticodon interactions at the shift site (27–29). In contrast, only a few cases of −2 PRF in mammalian cells have been documented thus far (13, 29) and the elements involved are poorly understood. Our previous computer-based RNA-folding analysis suggested that the RNA downstream of the slippery sequence (RG_GUU_UUU) used for −2 PRF in PRRSV is rather unstructured and does not fold into a structure compatible with canonical RNA-structure-stimulated PRF. However, mutations within a conserved CCCANCUCC motif located 11 nt downstream of the shift site can reduce or inhibit frameshifting, consistent with the presence of a 3′ stimulatory element of some form (13). Remarkably, our previous study also provided indications for the occurrence of efficient −1 frameshifting at (or near) the same slippery sequence. Due to the presence of a translation termination codon in the −1 reading frame immediately following the slippery sequence, this would yield a truncated form of nsp2, termed “nsp2N” (Fig. 1A).
In this report, we identify PRRSV replicase subunit nsp1β as a transactivator of efficient −2 and −1 PRF at the same slippery sequence and provide evidence that its frameshift-stimulatory activity requires interaction with the viral mRNA. In support of this, a highly conserved putative RNA-binding motif (GKYLQRRLQ), integrated into the structure of nsp1β’s papain-like autoproteinase domain, was found to be critical for the stimulation of frameshifting and for interacting with the RNA sequence of the PRRSV PRF signal. The minimal RNA sequence required to direct efficient PRF was mapped within a 34-nt region of the PRRSV nsp2-coding sequence that includes the shift site and the conserved CCCANCUCC motif. Our findings reveal an unusual noncanonical translation mechanism in which a viral protein functions as a transactivator of efficient −2 and −1 PRF. This study advances our understanding of noncanonical translation, suggests that viruses may use additional strategies to modulate viral and potentially host cell translation during infection, and has practical implications in biotechnology and the design of antiviral strategies.
Results
Alternative −2 and −1 PRF at the Same PRRSV Slippery Sequence.
Previously (13), we demonstrated expression of the PRRSV TF ORF (Fig. 1A) using a rabbit antiserum raised against the epitope on the C terminus of the polypeptide it encodes. Subsequently, the frameshift product was immunopurified from infected cells and mass spectrometry (MS) was used to identify both the site (RG_GUU_UUU) and direction (−2, rather than +1) of ribosomal frameshifting. In both PRRSV-infected cells and an ORF1a expression system, and using distantly related type 1 and type 2 PRRSV isolates, the same studies revealed an additional nsp2-related product (nsp2N) with a size consistent with −1 PRF occurring at the same site (estimated efficiency ∼7%) (13). However, a stop codon is present in the −1 frame immediately downstream of the RG_GUU_UUU slippery sequence (Fig. 1A) and consequently, if nsp2N were derived from −1 frameshifting, it would lack a unique C-terminal sequence that could be used to discriminate it from a product derived through the internal proteolytic cleavage of full-length nsp2. In an attempt to confirm the occurrence of −1 PRF by immunoprecipitation and mass spectrometric analysis (MS), we sought to extend the potential −1 frameshift product with a unique C-terminal signature. In a full-length cDNA clone of the previously used PRRSV isolate SD01-08 (a type 1 virus) (30), the −1 frame stop codon (UGA) was replaced by a tryptophan codon (UGG), extending the −1 frame by an additional 87 codons (Fig. S1, SD01-08-M1). However, this point mutation unavoidably also introduced amino acid substitutions in the overlapping 0 and −2 frames encoding nsp2 and nsp2TF (Glu→Gly and Lys→Glu, respectively), and perhaps as a consequence, the resulting recombinant virus was severely crippled [titer reduced to 103 fluorescent-focus units (FFU)/mL], preventing us from immunopurifying sufficient nsp2N for reliable MS analysis. We, therefore, reverted to a type 2 PRRSV isolate (SD95-21) (31) and introduced the same A-to-G mutation, which in this case extended the −1 ORF by 23 additional codons to generate mutant SD95-21-M1 (Fig. S1). Fortunately, despite carrying Asp→Gly and Thr→Ala mutations in the nsp2 and nsp2TF products, respectively, this recombinant virus replicated to much higher titers (106.2 FFU/mL) and the C-terminally extended nsp2N product (nsp2N*) could be immunopurified from infected MARC-145 cells. A gel slice containing the nsp2N* band was analyzed by liquid chromatography tandem MS (LC-MS/MS) and a QVFWPR tryptic peptide that spanned the frameshift site and is compatible with −1 PRF at the RG_GUU_UUU sequence was identified (Fig. S2). To verify correct identification of this peptide, a synthetic version was subjected to the same LC-MS/MS analysis. The tandem mass spectrum of this synthetic peptide was found to be identical to that of the peptide derived from the nsp2N*-containing gel slice (Fig. S2D), confirming that nsp2N is indeed translated via −1 PRF at the RG_GUU_UUU slippery sequence, which is, therefore, able to direct both −1 and −2 PRF.
PRRSV nsp1β Is Required for Efficient −1 and −2 Frameshifting in the nsp2-Coding Region.
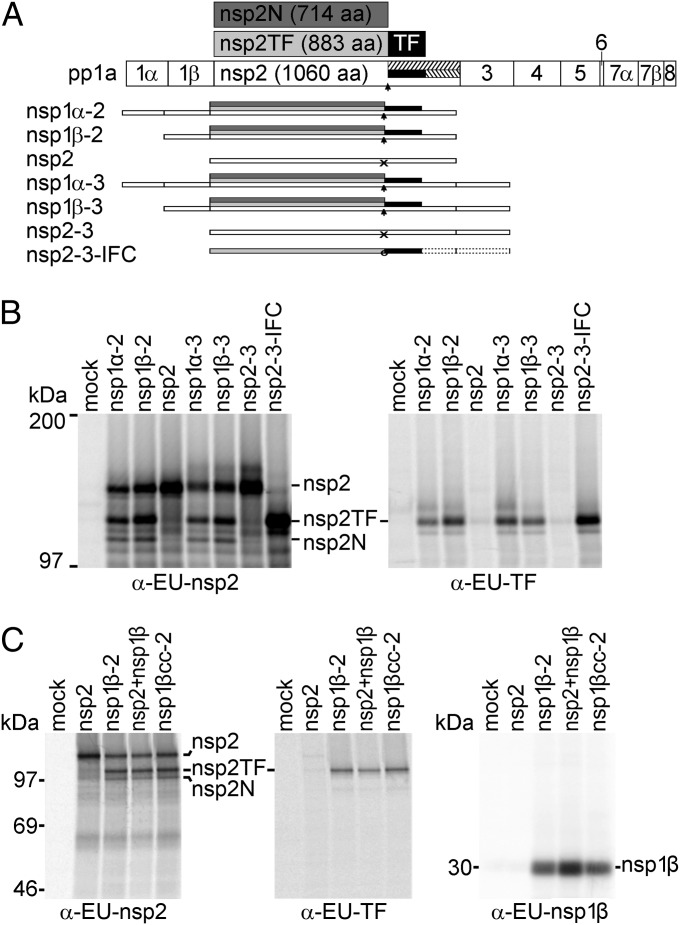
Previously we demonstrated that translation of the complete PRRSV ORF1a sequence is sufficient to allow efficient −2 PRF (13). To define the minimal sequence requirements for −2/−1 PRF in PRRSV isolate SD01-08, we focused our attention on the N-terminal half of ORF1a (the nsp1α–nsp3 region) and generated a panel of truncated ORF1a constructs (Fig. 2A) for expression in the recombinant vaccinia virus–T7 RNA polymerase system (32). Following radiolabeling of proteins synthesized in transfected RK-13 cells, expression of nsp2, nsp2TF, and nsp2N was analyzed by immunoprecipitation using monoclonal antibody (mAb) α-EU-nsp2 and rabbit antiserum α-EU-TF, recognizing all three nsp2-related products and the unique C-terminal epitope of nsp2TF, respectively (see Fig. S1B for a summary of antibody nomenclature and epitopes recognized). As shown in Fig. 2B, constructs lacking the nsp1α- and/or nsp3-coding region still efficiently expressed nsp2TF and nsp2N. In contrast, constructs lacking the nsp1β-coding region expressed nsp2 but only trace amounts of nsp2TF or nsp2N were detected. This indicates that nsp1β, or the RNA sequence encoding nsp1β, is required for efficient −2/−1 PRF at the RG_GUU_UUU slippery sequence in the nsp2-coding region, located some 2.5 kb downstream of the nsp1β-coding region.
Fig. 2.
PRRSV nsp1β transactivates −2/−1 PRF. (A) Schematic representation of expression products from vectors encoding different combinations of replicase subunits from the nsp1α–nsp3 region of type 1 PRRSV (isolate SD01-08), expressed as single nsps or self-cleaving multi-nsp polyproteins. PRRSV pp1a and its processing scheme are shown at the top and the −2 and −1 frameshift products, nsp2TF and 2N, are shown in light and dark gray with the polypeptide encoded by the TF ORF indicated in black. The arrow indicates the PRF site and untranslated parts of the −2 and −1 reading frames are hatched. The scheme below, for each expression vector used in B, shows which nsps were expressed, with arrows indicating the occurrence of −2/−1 PRF and “X” indicating lack of efficient frameshifting. Nsp2-3-IFC (13) is an engineered IFC construct that expresses nsp2TF only, due to the insertion of 2 nt at the PRF site (circle). (B) Expression of different protein combinations (A) using the recombinant vaccinia virus–T7 RNA polymerase expression system and RK-13 cells, revealing that nsp1β expression is required for efficient −2/−1 PRF. After metabolic labeling, expression products were immunoprecipitated with the antibodies indicated below each panel; mAb α-EU-nsp2 recognizes the common N-terminal domain of nsp2, nsp2TF, and nsp2N, whereas α-EU-TF recognizes the C-terminal domain of nsp2TF. Immunoprecipitated proteins were separated by SDS/PAGE and visualized by autoradiography. (C) Analysis of −2/−1 PRF transactivation by nsp1β using the recombinant vaccinia virus–T7 RNA polymerase expression system as described for B. RK-13 cells were transfected with plasmid DNAs expressing nsp2, nsp1β-2, nsp2+nsp1β (from separate plasmids), or nsp1βcc-2, with the latter containing an nsp1β-coding sequence in which the large majority of codons had been synonymously mutated (Fig. S3). Expression products were immunoprecipitated using specific antibodies indicated at the bottom of each panel and visualized by SDS/PAGE and autoradiography.
Extending this further, using the same expression system, nsp2 and nsp1β were expressed from separate, cotransfected plasmids (pLnsp2 and pLnsp1β) rather than as a self-cleaving nsp1β-2 polyprotein (pLnsp1β-2). Again, both nsp2TF and nsp2N were produced (Fig. 2C), indicating that nsp1β can stimulate −2/−1 PRF in the nsp2-coding region in trans. To establish whether this effect was mediated by the nsp1β protein or the nsp1β-coding RNA sequence, a drastically altered version of the nsp1β-2 expression vector was produced in which almost every codon of the nsp1β-coding sequence was mutated synonymously, while avoiding rare codons (mutant pLnsp1βcc-2; Fig. S3). This pLnsp1βcc-2 construct expresses an unaltered nsp1β protein, but the nucleotide sequence encoding it is changed to such an extent that we would expect to have disrupted any primary sequence or RNA secondary structure elements that might be involved in −2 PRF (for example, an element having a long-range interaction with the PRF region in the nsp2-coding sequence). Immunoprecipitation analysis revealed that nsp2TF and nsp2N were expressed with equal efficiency in cells transfected with pLnsp1βcc-2 and wild-type (WT) pLnsp1β-2 (Fig. 2C), indicating that PRF stimulation involves the nsp1β protein rather than an RNA signal in the nsp1β-coding sequence.
Minimal RNA Sequence Requirements for −2/−1 PRF.
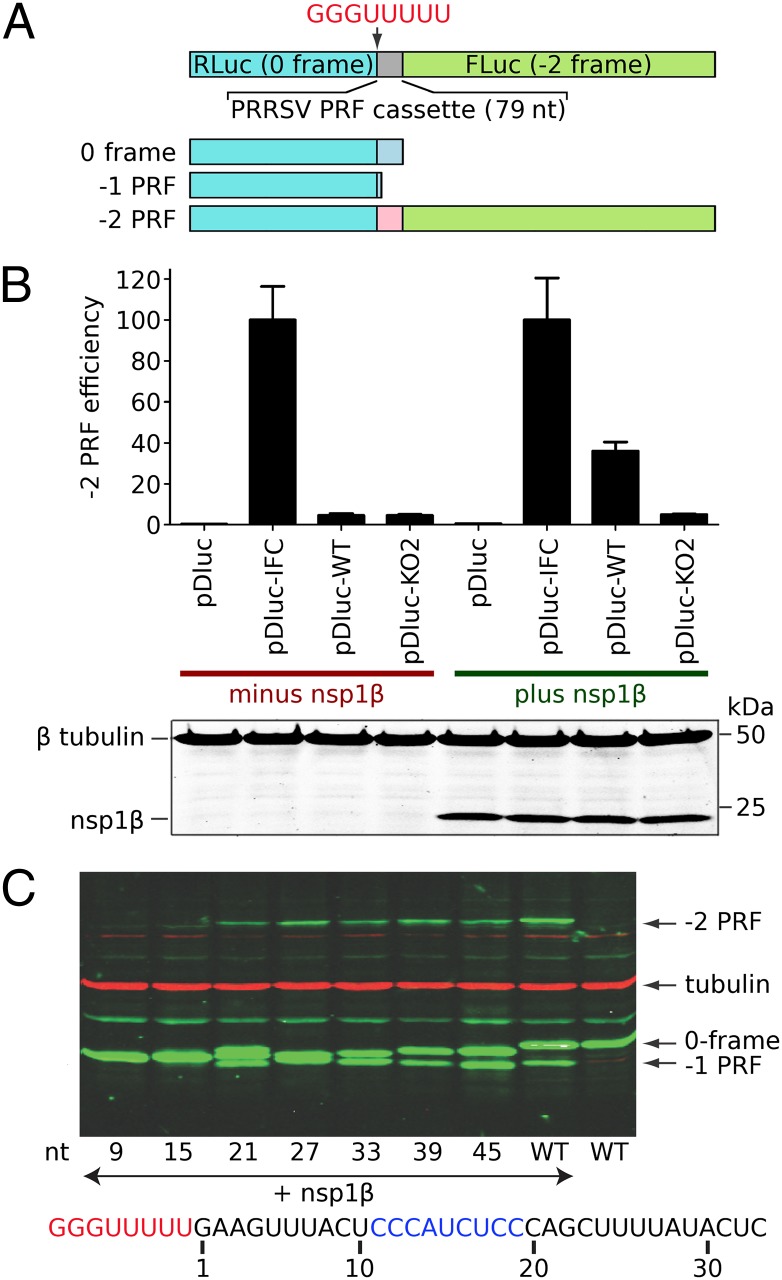
We next set out to define the minimal RNA sequences in the nsp2-coding region that are required for efficient −2/−1 PRF. To this end, we prepared a reporter gene construct in which PRRSV RNA sequences from the PRF-inducing region were placed between two luciferase genes [pDluc (33, 34); Fig. 3A]. Whereas the ORF1a frame of the PRRSV insert was placed in-frame with the upstream (Renilla) luciferase gene, the downstream (firefly) luciferase was in the −2 frame and thus its expression depended on the occurrence of −2 frameshifting. Also −1 PRF could be monitored, because the native stop codon in the −1 frame was retained and −1 PRF would, therefore, yield a polypeptide slightly shorter than the product resulting from translation termination in the zero reading frame. As controls, an in-frame control (IFC) construct was also prepared in which the two luciferase genes were aligned in the same frame by inserting two nucleotides (CU) immediately downstream of the slippery sequence. A previously described PRF knockout construct [(KO2); Fig. S1] (13) containing point mutations within the slippery sequence and downstream C-rich region was also included in the analysis.
Fig. 3.
Delineation of RNA elements required for PRRSV −2/−1 PRF. (A) Schematic representation of the pDluc dual luciferase construct. The GG_GUU_UUU shift site, 5 upstream nucleotides and 66 downstream nucleotides (79 nt in total) were inserted between the Renilla and firefly luciferase genes such that −2 PRF is required for firefly luciferase expression. (B, Upper) Dual luciferase reporter assay showing that efficient −2 PRF depends on coexpression of nsp1β. For type1 PRRSV (isolate SD01-08), nsp1β was coexpressed with dual luciferase constructs containing a WT or −2 PRF knockout (KO2) frameshift signal. Mutant KO2 (Fig. S1) (13) contains point mutations within both slippery sequence and downstream C-rich motif. The −2 PRF efficiencies were calculated by comparing the ratio of firefly and Renilla luciferase activities, using the IFC mutant (Fig. S1A) as a reference. Error bars represent the SD of three independent experiments, in which each construct was transfected in duplicate. (B, Lower) Western blot analysis confirming equal expression of nsp1β and equal loading (β-tubulin). (C) Delineation of the minimal RNA sequence requirements for efficient −2/−1 PRF. Starting from a construct containing the 66 nt downstream of the slippery sequence, a series of 3′ truncations was engineered in pDluc. Upon coexpression with nsp1β, cell lysates were analyzed by Western blot, using an antibody recognizing the common Renilla luciferase part of all pDluc translation products (A). The number below each lane represents the remaining PRRSV-specific RNA sequence downstream of the slippery sequence, of which 21 nt were sufficient for efficient −2/−1 PRF in this assay.
Initially, a 79-nt region spanning 5 nt upstream of the slippery sequence to 66 nt downstream (including the conserved CCCANCUCC motif) was cloned between the two luciferase genes (construct pDluc-WT). Frameshifting efficiencies were determined by comparing the ratio of enzymatic activities of firefly and Renilla luciferase in parallel HEK-293T cell cultures transfected with individual pDluc constructs with or without cotransfection of the plasmid expressing nsp1β. As shown in Fig. 3B, in comparison with the IFC control, the WT PRRSV −2 PRF efficiency was ∼38%, and this high level of −2 PRF was only observed in cells cotransfected with the nsp1β-expressing plasmid; in the absence of the transactivator, only low levels of −2 PRF (<5%) were observed. As expected, frameshifting was not observed in cells transfected with pDluc-KO2. Western blot analysis of transfected cell lysates revealed that both efficient −2 PRF and efficient −1 PRF could be observed with pDluc-WT provided that an nsp1β expression plasmid was cotransfected (Fig. 3C). These data indicated that the 79-nt PRRSV sequence included in pDluc-WT contains all cis-acting sequences required for efficient −2/−1 PRF, and that, as documented above, both types of frameshift depend on the presence of nsp1β. In the absence of this transactivator, only low levels of PRF were observed.
To further investigate the key RNA sequences required for PRF, in-frame deletions were introduced into pDluc-WT, starting from the 3′ end of the PRRSV insert. As shown in Fig. 3C, an initial deletion that reduced the PRRSV sequence downstream of the shift site to 45 nt (pDluc-45) led to a small reduction in −2 PRF (about twofold), albeit with a concurrent increase in −1 PRF. Subsequent deletions had no further effect until part of the conserved CCCANCUCC motif was removed (Fig. 3C; compare pDluc-21 and pDluc-15). In pDluc-15, which lacked the second half (CUCC) of the conserved motif, the capacity for transactivation of PRF by nsp1β was lost. These data provided further support for a role of the C-rich motif in PRF, and allowed us to define the functional PRRSV −2/−1 PRF cassette as a 34-nt region containing the slippery sequence and the 3′ C-rich motif.
Identification of a Conserved nsp1β Motif That Is Critical for PRF TransActivation.
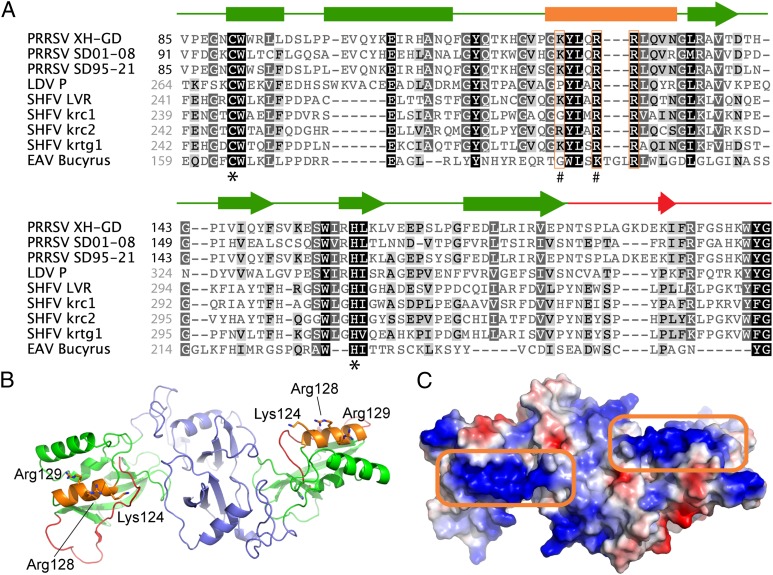
The nsp1α-nsp1β region has previously been implicated in a variety of processes in the arterivirus replicative cycle, including replicase polyprotein processing (35), transcriptional control (36, 37), and innate immune evasion (31, 38). An analysis of nsp1β sequence conservation (Fig. 4A), together with the published crystal structure of nsp1β from a type 2 PRRSV isolate (39) (Fig. 4 B and C), pointed toward a previously identified conserved sequence motif as a potential RNA interaction domain. This sequence, GKYLQRRLQ in both type 1 and type 2 PRRSV, forms one of three α-helices (labeled “α4” in ref. 39) in the region between the active site Cys and His residues of the papain-like proteinase domain (PLP1β) that constitutes the C-terminal two-thirds of nsp1β. Interestingly, compared with the active site of the PLP1β proteinase, helix α4 maps to the other side of the molecule and, in the available crystal structure, the three conserved basic residues of the GKYLQRRLQ motif are exposed on the nsp1β surface. Moreover, in the nsp1β homodimer that was the basis for structural studies, the α4 helices of both monomers map to the same side of the dimer and may form a continuous surface across the protein that binds nucleic acid (Fig. 4C; Discussion).
Fig. 4.
PRRSV nsp1β sequence and structure. (A) Amino acid sequence alignment of the PLP1β domains from selected arterivirus nsp1β proteins. Secondary structure elements (based on the published crystal structure from type 2 PRRSV isolate XH-GD) (39) are shown above the alignment and are color matched to the nsp1β structure in B. Conserved basic residues in PLP1β helix α4 are boxed in orange. #, residues mutated in mutant 1βKO (see details in Results, Identification of a Conserved nsp1β Motif That Is Critical for PRF Trans-Activation); *, PLP1β active site residues. PRRSV sequences are numbered (black) from the nsp1α/nsp1β cleavage site, whereas all other sequences are numbered (gray) starting from the N terminus of the pp1a polyprotein. The names of specific isolates used are indicated. GenBank accession nos. of sequences used are as follows: EU624117 (PRRSV XH-GD), DQ489311 (PRRSV SD01-08), KC469618 (PRRSV SD95-21), NC_001639 (LDV P), NC_003092 (SHFV LVR), HQ845737 (SHFV krc1), HQ845738 (SHFV krc2), JX473847 (SHFV krtg1), and NC_002532 (EAV Bucyrus). (B) Cartoon representation of the crystal structure of the nsp1β dimer from a type 2 PRRSV isolate (PRRSV XH-GD; PDB ID code 3MTV) (39). For both monomers, the N-terminal domain is colored purple, whereas the PLP1β domain and the C-terminal extension (leading up to the nsp1β/nsp2 site cleaved by PLP1β) are colored green and red, respectively. Helix α4 of PLP1β, containing the conserved GKYLQRRLQ motif, is colored orange with basic residues represented as sticks. (C) Electrostatic surface representation of the nsp1β dimer showing the positively charged (blue) patches on helix α4 of PLP1β (boxed in orange) created by the basic residues of the GKYLQRRLQ motif. Both patches reside on the same side of the structure, potentially allowing for RNA to bind across the entire dimer surface.
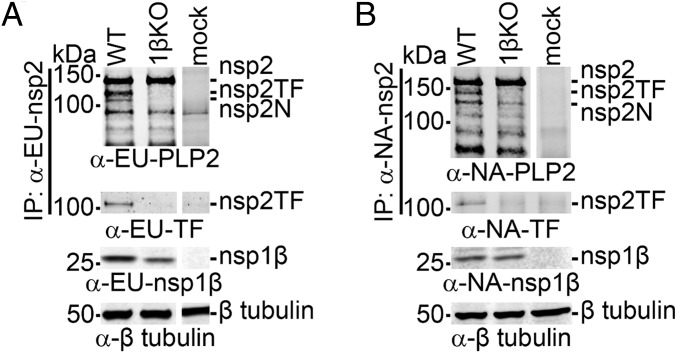
In a recent study (31), the GKYLQRRLQ motif was targeted by site-directed mutagenesis and the Lys and the first Arg of the motif were replaced with Ala (mutant 1βKO, Fig. S1). For both PRRSV genotypes, the replication of the 1βKO mutant in MARC-145 cells was found to be seriously crippled. The fact that we had observed similar defects in mutants in which the −2/−1 PRF signal had been inactivated, or in which the expression of a functional nsp2TF was prevented (13), prompted us to investigate whether this KR→AA double mutation affected nsp2TF/nsp2N expression. Strikingly, upon expression of nsp1β-nsp2 from either PRRSV genotype carrying these nsp1β mutations, neither nsp2TF nor nsp2N could be detected (Fig. 5). These data indicate that the GKYLQRRLQ motif plays a key role in PRF activation.
Fig. 5.
A conserved motif in PRRSV PLP1β is critical for transactivation of −2/−1 PRF in an expression system. The recombinant vaccinia virus–T7 RNA polymerase expression system and HEK-293T cells were used to express WT and 1βKO mutant nsp1β-nsp2 polyproteins from (A) type 1 and (B) type 2 PRRSV. The 1βKO mutant carried a double Ala substitution of basic residues in the highly conserved GKYLQRRLQ motif of nsp1β (see also Fig. 4 and Fig. S1). Expression products were immunoprecipitated with mAbs recognizing the common N-terminal domain of the nsp2-related products. Following SDS/PAGE, they were identified in Western blot analysis using antibodies recognizing the common nsp2 domain, the C terminus of nsp2TF, or nsp1β. A tubulin antiserum was used for a loading control. Samples in all panels and rows were run on the same gel, but some gel images were spliced to remove lanes derived from samples not related to this study.
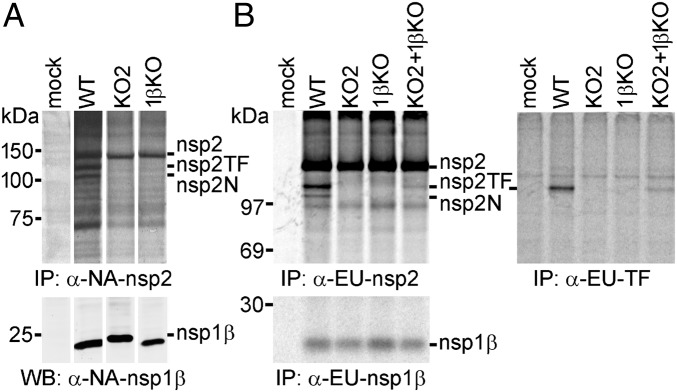
To investigate nsp1β transactivation of PRF in the context of PRRSV infection, we analyzed nsp2 expression using the 1βKO mutant of both PRRSV genotypes. As controls, we included the corresponding KO2 mutants, which carry mutations within the slippery sequence and C-rich region that eliminate frameshifting (Fig. S1) (13). Using reverse genetics, KO2 and 1βKO mutant viruses were recovered from full-length infectious clones of the two PRRSV genotypes. Both mutants replicated poorly in MARC-145 cells, but for the type 2 PRRSV isolate (SD95-21), they produced titers (105.1 and 105.3 FFU/mL for KO2 and 1βKO, respectively) that sufficed for the subsequent experiments of infection, metabolic labeling, and radioimmunoprecipitation analysis. As expected (Fig. 6A), the expression of nsp2, nsp2TF, and nsp2N was detected in SD95-21-WT-infected cells, whereas only nsp2 was recovered from cells infected with either SD95-21-KO2 or SD95-21-1βKO, whereas their nsp1β was expressed at a level similar to that observed with the WT virus.
Fig. 6.
A conserved motif in PRRSV PLP1β is critical for transactivation of −2/−1 PRF in infected cells. (A) Analysis of nsp2-related products in MARC-145 cells infected with WT type 2 PRRSV (isolate SD95-21) or mutants KO2 and 1βKO. Mutant KO2 (Fig. S1) contained PRF-inactivating point mutations in both slippery sequence and downstream C-rich motif, whereas 1βKO carried a double Ala substitution of basic residues in the highly conserved GKYLQRRLQ motif of nsp1β. Following metabolic labeling, proteins were immunoprecipitated using mAb α-NA-nsp2 and visualized by SDS/PAGE and autoradiography. The expression of nsp1β was monitored by Western blot analysis. Samples in all rows were run on the same gel, but gel/blot images were spliced to remove lanes derived from samples not related to this study and to achieve a lane order consistent with B. (B) BHK-21 cells were transfected with in vitro-transcribed full-length RNA of WT, KO2, or 1βKO PRRSV SD01-08 (type 1) or were double transfected with equal amounts of KO2 and 1βKO RNA to demonstrate complementation between these two virus mutants. Following metabolic labeling, viral proteins were immunoprecipitated using specific mAbs that recognize a common nsp2 domain (Upper Left), the polypeptide encoded by the TF ORF (Upper Right), or nsp1β (Lower). Protein products were visualized using SDS/PAGE and autoradiography.
Unfortunately, the 1βKO mutant of the PRRSV type 1 isolate (SD01-08) yielded very low titers in MARC-145 cells (102 FFU/mL). Considering the number of viral functions and properties potentially affected by nsp1β mutations (Discussion), we therefore performed a so-called first-cycle analysis of the phenotypes of SD01-08 WT, KO2, and 1βKO. The three viruses were launched by transfecting in vitro-transcribed full-length RNA into BHK-21 cells, which support replication of transfected PRRSV RNA but cannot be infected by the progeny virus released from the transfected cells, due to the lack of the appropriate receptor(s) on their surface (40). Moreover, BHK-21 cells have a defect in IFN production (41), thus minimizing the (potential) impact of host innate responses on the comparison of viral replication phenotypes. Following metabolic labeling of protein synthesis in transfected cells, a radioimmunoprecipitation analysis revealed that SD01-08-1βKO produced large amounts of nsp2, whereas the production of nsp2TF was greatly reduced and nsp2N was not detected (Fig. 6B). As previously established, SD01-08-KO2 produced only nsp2, whereas SD01-08-WT produced all three nsp2 variants. Equal expression of nsp1β in WT-, KO2-, and 1βKO-transfected cells was confirmed by immunoprecipitation with an nsp1β-specific mAb. We also investigated whether the mutations in 1βKO affected the activity of the PLP1β protease or the (potential) involvement of nsp1β in the control of viral subgenomic mRNA synthesis. Although the total amount of nsp1β and viral RNA was somewhat reduced in 1βKO-transfected cells, cleavage of the site between nsp1β and nsp2 and subgenomic mRNA production (Fig. S4 B and C) were not affected by the mutations in the GKYLQRRLQ motif nor did they affect nsp1β stability (Fig. S4D). Finally, we included a double transfection of BHK-21 cells with KO2 and 1βKO full-length RNA (Fig. 6B) and demonstrated complementation between the two PRF-negative mutants leading to reactivation of nsp2TF/nsp2N expression. As expected, the WT nsp1β expressed by mutant KO2 was able to transactivate −2/−1 PRF on the WT PRF signal in the 1βKO genome, again confirming that the GKYLQRRLQ motif plays a critical role in the PRF stimulatory activity of nsp1β in PRRSV-infected cells.
PRRSV nsp1β Interacts with the RNA Signals That Direct −2/−1 PRF.
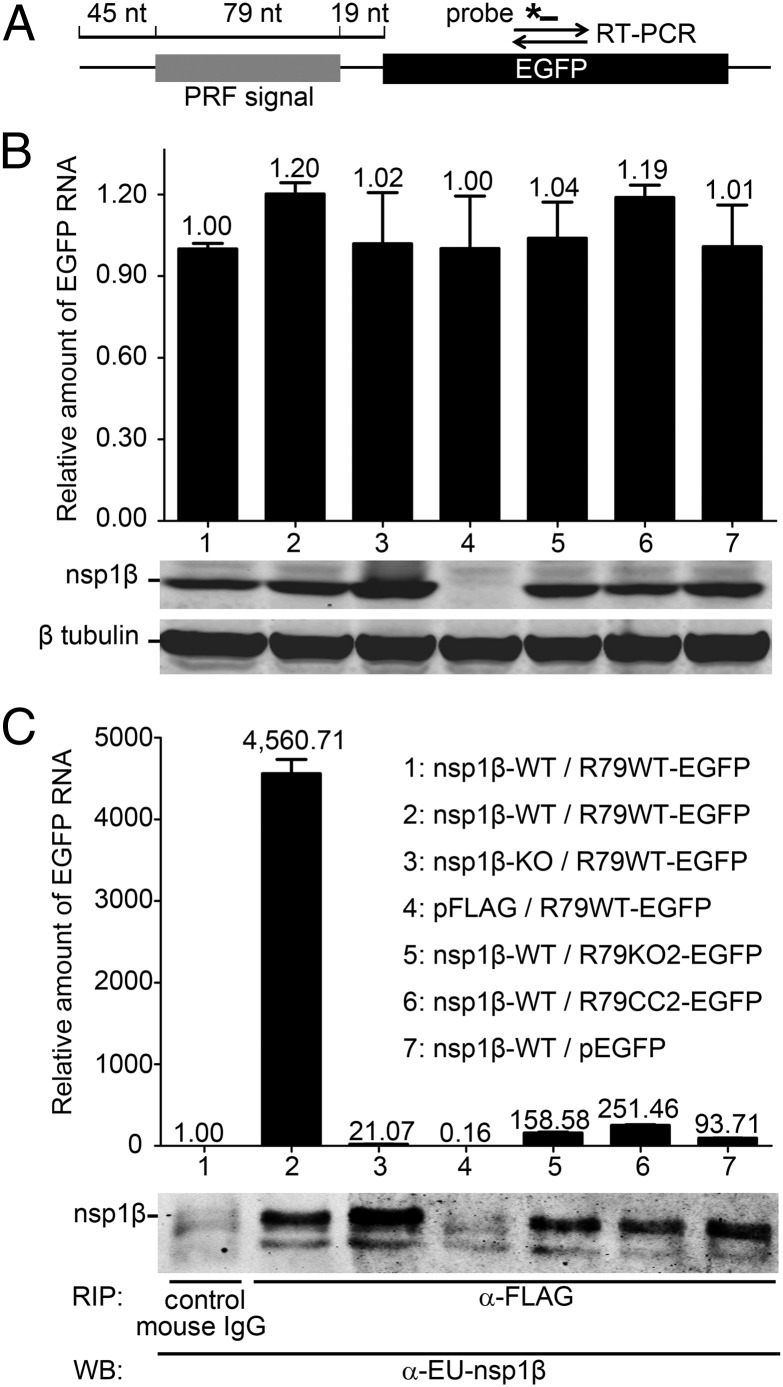
To test the hypothesis that nsp1β, and specifically its GKYLQRRLQ motif, interacts with the PRRSV RNA sequences that direct −2/−1 PRF, we developed an RNA-binding protein immunoprecipitation assay. To produce an RNA target, we engineered plasmid pR79WT-EGFP yielding an RNA in which a 79-nt PRRSV SD01-08 RNA sequence (Fig. 3A) containing the shift site and conserved CCCAUCUCC motif was fused to the EGFP ORF (Fig. 7A). The latter served as a target for quantitative RT-PCR (qRT-PCR) amplification of target RNA bound to nsp1β. As controls, we included plasmids pR79KO2-EGFP and pR79CC2-EGFP, containing combinations of point mutations in the shift site and/or CCCAUCUCC motif that were previously demonstrated to completely inactivate PRF (Fig. S1) (13). To express the nsp1β bait, we used constructs pFLAG-nsp1β-WT and pFLAG-nsp1β-KO, producing WT and mutant (K130A/R134A) nsp1β, respectively, each fused to an N-terminal triple FLAG tag. The empty vectors pFLAG and pEGFP were included as negative controls.
Fig. 7.
An RNA carrying the PRRSV −2/−1 PRF signal coimmunoprecipitates with nsp1β. A protein–RNA interaction assay was designed based on coimmunoprecipitation of nsp1β and RNA transcripts containing 79 nt of type 1 PRRSV sequence, including the −2/−1 PRF signal. (A) Schematic representation of the target RNA in which a WT 79-nt PRF signal (R79WT-EGFP), or its mutant KO2 or CC2 derivatives (Fig. S1A), was fused to the EGFP sequence. The latter served as a target for qRT-PCR amplification (primer set and TaqMan probe indicated), which was used to quantify the amount of RNA target bound to nsp1β. (B and C) HEK-293T cells were cotransfected with a plasmid expressing WT or 1βKO FLAG-tagged nsp1β and a plasmid expressing a WT or mutant target RNA. Empty vectors (pFLAG and pEGFP) were included as negative controls. (B) Detection of input levels of nsp1β bait and RNA target in cell lysates before the coimmunoprecipitation assay. qRT-PCR was used to determine the levels of WT or mutant R79-EGFP mRNA in transfected cells (Top). Western blot analysis was used to monitor the input of 1βKO or WT nsp1β bait (Middle) and to verify the use of equal amounts of cell lysate (β-tubulin control; Bottom). Lane numbers are explained in C. (C) Following FLAG-nsp1β immunoprecipitation, the amount of coprecipitating target RNA was determined by qRT-PCR (A). Western blot analysis using a mAb α-EU-nsp1β was used to monitor the amount of immunoprecipitated nsp1β. A legend explaining the cotransfected plasmids for each lane number is given on the right.
Following cotransfection of vectors expressing RNA target and nsp1β into 293T cells, cell lysates were prepared. Western blot and qRT-PCR analysis (Fig. 7B) were first used to determine the expression levels of nsp1β bait and target RNA, respectively, and confirmed the presence of similar amounts of both molecules in all cotransfection samples. Subsequently, we immunoprecipitated FLAG-nsp1β using an anti-FLAG mAb and analyzed these samples for coimmunoprecipitation of target RNA using the same qRT-PCR method, while verifying successful immunoprecipitation of nsp1β with a specific mAb (Fig. 7C). A strong and specific RNA coimmunoprecipitation signal was detected only in samples from cells cotransfected with pFLAG-nsp1β-WT and pR79WT-EGFP. In contrast, when mutant 1βKO carrying the K130A/R134A double mutation in the GKYLQRRLQ motif was used, only very low levels of target RNA were pulled down, suggesting that the K130A/R134A mutations impaired the interaction of nsp1β with PRRSV RNA. Only background signal was detected when using a negative control mouse IgG for immunoprecipitation, or when expressing the pFLAG empty vector control, demonstrating specificity for nsp1β. When the PRRSV PRF-specific sequences in the RNA target were mutated (R79KO2-EGFP or R79CC2-EGFP) or the pEGFP empty vector control was used, only trace amounts of RNA (6% or less of the R79WT-EGFP signal) could be captured, thus demonstrating that coimmunoprecipitation of the target RNA strongly depends on the presence of the CCCAUCUCC motif.
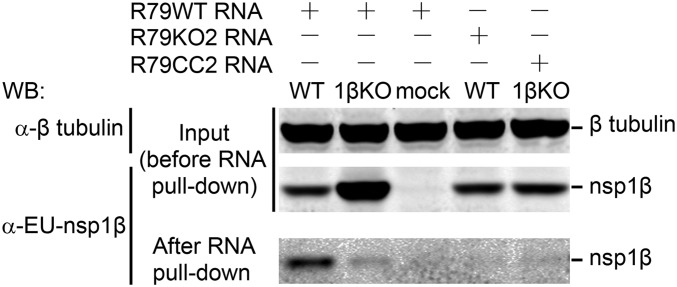
To further corroborate the interaction between nsp1β and the 79-nt RNA sequence from the PRF region, we used a complementary assay (the RiboTrap system) (42) in which RNA transcripts were labeled with 5-bromo-uridine, facilitating their immunopurification using a 5-bromo-U–specific mAb, and subsequent analysis of immunoprecipitates for the presence of proteins binding to the RNA bait. Using PCR amplicons containing the T7 promoter and 79-nt PRRSV RNA sequence (R79WT, R79KO2, or R79CC2) as a template, 5-bromo-U–labeled RNA transcripts were produced in vitro and incubated with lysates of 293T cells transfected with the plasmid expressing 1βKO or WT nsp1β. Following immunoprecipitation with the 5-bromo-U–specific mAb, samples were analyzed for the presence of nsp1β using SDS/PAGE and Western blot analysis (Fig. 8). A strong and specific nsp1β signal was detected only when the R79WT bait was incubated with cell lysates containing nsp1β-WT. When using lysates containing 1βKO of nsp1β, only a very small fraction of the available protein was bound to the RNA. Likewise, only trace amounts of WT nsp1β were pulled down when 5-bromo-U–labeled R79KO2 or R79CC2 RNA was used as bait. These data are consistent with those obtained in the RNA-binding protein immunoprecipitation assay presented in Fig. 7 and further support a key role for the GKYLQRRLQ motif in the specific transactivation of the PRRSV −2/−1 PRF by nsp1β.
Fig. 8.
PRRSV nsp1β can be pulled down using an RNA carrying the −2/−1 PRF signal. Cell lysates from 1βKO or WT nsp1β-expressing HEK-293T cells were incubated with in vitro produced BrU-labeled RNA transcripts containing WT or mutant (KO2 or CC2, Fig. S1) versions of a 79-nt sequence from the −2/−1 PRF region of type 1 PRRSV (isolate SD01-08). RNA–protein complexes were immunoprecipitated with an anti-BrU antibody and subjected to Western blot analysis using an nsp1β-specific mAb. The amount of β-tubulin in the initial samples was monitored to verify equal loading.
Discussion
In this paper, we report the remarkable discovery that efficient ribosomal frameshifting in the expression of the PRRSV nsp2TF and nsp2N proteins requires the viral nsp1β protein as a transactivator. Protein-stimulated PRF is unprecedented. It has been reported that cellular annexin A2 may interact with the −1 PRF signal of the coronavirus infectious bronchitis virus, but its role appears to be to down-regulate frameshifting through destabilization of the stimulatory pseudoknot (43), and no specific frameshift-stimulatory protein factors have been identified to date. Although down-regulation of eukaryotic translation release factor levels can lead to a low-level stimulation of −1 PRF (44, 45), this is a poorly characterized phenomenon, likely to be a rather nonspecific effect brought about by changes in translation rates (46). It is known that −1 PRF at the HIV type 1 slippery sequence can be promoted by replacing the natural stimulatory RNA with a combination of the iron-responsive element (IRE) RNA and its cognate binding partner (the IRE-binding protein), but this is a highly artificial experimental system and the stimulation of −1 PRF is very weak (47).
Exactly how nsp1β stimulates frameshifting remains to be determined. Based on the RNA-binding experiments, we propose that the region 3′ of the PRRSV PRF slippery sequence acts to recruit nsp1β, or an nsp1β-containing protein complex, which modulates ribosome function to promote frameshifting. Our evidence to date supports the view that nsp1β binds directly to the C-rich region because point mutations within this region strongly reduce RNA binding (Fig. 7). However, we cannot rule out the involvement of other factors, for example, poly C-binding proteins (PCBPs) (48), which are known to interact with C-rich regions. Moreover, certain PCBPs have been reported to bind to PRRSV nsp1β in pull-down assays (49). As the C-rich region is located only 11 nt downstream of the slippery sequence, this would likely place bound nsp1β, or an nsp1β-containing complex, in close proximity to a ribosome decoding the slippery sequence, permitting interactions that may lead to frameshifting. Although this model is speculative, there is growing evidence that proteins can modulate the elongation step of protein synthesis. The fragile X mental retardation protein reversibly stalls ribosomes on its target mRNAs (50) and the HIF-1α mRNA-associated cytoplasmic polyadenylation element-binding protein 2 binds eEF2 and slows elongation (51). Conceivable routes through which bound proteins could modulate ribosomal function include induction of ribosomal pausing by acting as a roadblock, recruitment of or localized depletion of translation factors, and direct interaction with a ribosomal component(s). It may be significant that nsp1β was reported to interact with rpS14 (49), a protein immediately adjacent to rpS3 of the ribosomal helicase (52), and PCBP1, which is known to interact with RACK1, a ribosome-associated protein located close to the mRNA entry channel (53). These features are consistent with a role for nsp1β in modulating the ribosomal helicase, the suspected target for the stimulatory RNA sequences of canonical −1 PRF signals.
An interesting aspect of this PRRSV PRF signal is that both −2 and −1 frameshifting events are promoted. Tandem slippage of ribosome-bound tRNAs on RG_GUU_UUU would allow complete A-site repairing in both −1 and −2 frames (tRNA anticodon:mRNA codon pairing in 0-frame is 3′-AAG-5′:5′-UUU-3′; single tRNAPhe isoacceptor AAG), but especially for −2 PRF, P-site repairing appears to be compromised [at least at the second and third positions (lowercase), 3′-Cai-5′:5′-Ggg-3′; “i” is inosine]. Some tolerance for P-site mispairing has been noted at certain viral −1 PRF signals, but usually, these are associated with single mismatches in the anticodon–codon interaction (1, 54). It may be that in the particular context of a −2 PRF, stable P-site repairing is not required, reminiscent of the unusual single-tRNA slippage events seen in prokaryotic systems with slippery sequences ending in AAG, where P-site repairing does not appear to be present (55, 56). In a recent study on RNA secondary structure-stimulated −1 and −2 PRF on a GU_UUU_UUA slippery sequence (29), it was noted that the length of the spacer between slippery sequence and secondary structure affected the relative utilization of −1 or −2 PRF modes, perhaps a reflection of differences in mRNA tension arising as the ribosomal helicase unwinds the secondary structure, with increased tension forcing the ribosome into the −2 rather than the −1 frame. Interestingly, the relative levels of the PRRSV −2 and −1 PRF products were seen to change as the length of the PRRSV region 3′ of the slippery sequence was shortened from 67 to 45 nt (Fig. 3C), although the sum total of PRF was similar. This may hint at the involvement of additional factors, or some subtle effect on the positioning of a bound protein (or complex), that can influence frameshift magnitude, although it remains to be determined whether this is linked to mRNA tension.
Analysis of the published structure of nsp1β from a type 2 PRRSV isolate provides insights into the mechanism of how the protein may interact with viral RNA. The PLP1β domain of nsp1β adopts a papain-like fold consisting of three α-helices that pack against a β-sheet of four antiparallel strands (39) (Fig. 4B). One of the helices (helix α4 in the overall nsp1β structure) contains a conserved GKYLQRRLQ motif that we now show plays a critical role in the transactivation of frameshifting. The crystal structure of nsp1β suggests that the protein exists as a homodimer (39) and, interestingly, helix α4 of both nsp1β monomers resides on the same side of the dimer, which may generate a continuous, positively charged surface that could bind a long single- or double-stranded RNA molecule (Fig. 4C). The involvement of an α-helix in RNA binding is consistent with the observation that nucleoproteins of many RNA viruses encapsidate the viral genome using domains of α-helical structure (57, 58). Thus, it is plausible that the GKYLQRRLQ motif of helix α4 directly binds viral RNA, although we cannot exclude the possibility that this helix may be a binding site for a cellular protein that in turn could bind to the PRF signal in the viral RNA.
Except for equine arteritis virus (EAV), the −2 PRF mechanism seems to be conserved in all currently known arteriviruses as judged by the presence of a TF ORF overlapping ORF1a and a conserved slippery sequence and downstream C-rich region (13). In PRRSV and lactate dehydrogenase-elevating virus (LDV) −1 PRF can occur, but in contrast the RG_GUC_UCU shift site in some of the recently identified simian hemorrhagic fever virus (SHFV)-like viruses (59) would preclude −1 PRF while still allowing −2 PRF. It is expected that such PRF events in LDV and SHFV would also be controlled by nsp1β, and indeed the transactivating motif in nsp1β was found to be largely conserved in these viruses (Fig. 4A). Possibly, the nsp1β component of the frameshift mechanism, which is encoded several kilobases upstream of the PRF site, evolved secondarily, for example to enhance the efficiency of nsp2TF/nsp2N expression because in the absence of nsp1β low levels of PRF could still be observed (Fig. 2A). Amino acid sequence comparisons reveal that the GKYLQRRLQ motif-containing helix is highly conserved in the PLP1β domains of PRRSV, SHFV, and LDV, but the motif is lacking in EAV. For the latter virus, the three helices of the PLP1β domain are predicted to be present, but with an insertion of 3 aa in the EAV equivalent of the α4 helix compared with the other arteriviruses. The nsp2-encoding region of EAV lacks an equivalent of the (overlapping) TF ORF and produces a substantially smaller nsp2. Assuming the TF ORF was lost at some point during the evolution of the EAV lineage, changes in this helix may have been tolerated when it was no longer required to stimulate PRF in trans. Although an alternative evolutionary scenario (i.e., a common ancestor of PRRSV, SHFV, and LDV independently acquiring a TF ORF) cannot be excluded, loss of the requirement to transactivate PRF may also explain a second remarkable difference between the nsp1 region of EAV and other arteriviruses: the inactivation of the proteolytic activity of the PLP1α proteinase, resulting in the synthesis of a single nsp1 protein rather than nsp1α and nsp1β (35, 60). In particular, the N-terminal zinc finger of nsp1 (EAV) or nsp1α (PRRSV) has been implicated in the control of viral subgenomic mRNA synthesis (37, 61–63), a function that may not be compatible with a role in PRF transactivation, thus requiring the internal cleavage of nsp1 by PLP1α in arteriviruses that use nsp1β-mediated transactivation of TF ORF expression.
The capacity of nsp1β to stimulate both −1 and −2 PRF suggests that protein transactivation could be used more widely in the induction of programmed frameshifting events in diverse systems. With regards to arteriviruses, it is possible that nsp1β might also modulate translation of host cell mRNAs containing appropriate signals. A cursory search of porcine mRNAs revealed hundreds of −1 and/or −2 frameshift-compatible shift sites followed by C-rich motifs at an appropriate spacing, although no site that is exactly identical to the PRRSV minimal PRF cassette (8-nt shift site plus the downstream 21 nt). Whether and to what extent frameshifting occurs at such sites remains to be investigated. Although the occurrence of nsp1β-responsive frameshift signals in host mRNAs would presumably be spurious, the overall effect may perturb cellular gene expression, thus adding an extra dimension to virus–host interactions.
When screening PRRSV nonstructural proteins for their capacity to suppress type I IFN expression, both nsp1β and nsp2 were found to possess such activities (20, 22, 31, 38, 49). In reporter gene-based assays, nsp1β had the strongest potential to inhibit IFN-β promoter activity and could also inhibit downstream IFN-induced signaling pathways for expression of IFN-stimulated genes (ISGs), including ISG15 (31, 38, 49, 64, 65). On the other hand, the PLP2 activity of nsp2 is able to disrupt innate immune signaling by removing ubiquitin (Ub) and Ub-like modifiers from host cell substrates, exhibiting a general deubiquitinating (DUB) activity toward cellular Ub conjugates and also cleaving the Ub homolog ISG15 (19–22). As documented here, nsp1β transactivates both nsp2TF and nsp2N expression, resulting in the synthesis of three nsp2-related proteins (nsp2, nsp2TF, and nsp2N) that have the N-terminal PLP2-DUB domain in common. Thus, it remains to be established to which extent nsp1β directly modulates the innate immune response or does so by stimulating the expression of nsp2TF and nsp2N. Furthermore, nsp1β may affect the immune response through modulation of host cell mRNA translation. The identification of viral/host elements responsible for innate immune evasion is fundamental for the development of modified live virus vaccines. As illustrated by our reverse genetics studies, mutagenesis of key residues in nsp1β and the PRF site could attenuate virus growth and improve host innate immune responses (13, 31). Because the GKYLQRRLQ motif and PRF site are highly conserved, technologies developed in this study may have broad application in the field.
Materials and Methods
Cells and Viruses.
HEK-293T, RK-13, BHK-21, and MARC-145 cells were cultured as described previously (30, 66). The US type 1 PRRSV isolate SD01-08 (GenBank accession no. DQ489311) and type 2 PRRSV isolate SD95-21 (GenBank accession no. KC469618) were used in all experiments.
Antibodies.
Antibodies recognizing PRRSV proteins (see also Fig. S1B for the nomenclature used in this paper), including mAb 22-28 (α-EU-nsp1β), mAb 123-128 (α-NA-nsp1β), mAb 36-19 (α-EU-PLP2), mAb 58-46 (α-EU-nsp2), mAb140-68 (α-NA-PLP2), mAb 148-43 (α-NA-nsp2), and a rabbit antiserum recognizing the C-terminal part of nsp2TF (α-EU-TF) were produced as described previously (13). A rabbit antiserum (α-NA-TF) recognizing the C-terminal epitope (CFLKVGVKSAGDLV) of nsp2TF of type 2 PRRSV was generated by GenScript. For detection of FLAG-tagged proteins, an anti-FLAG mAb was obtained from Sigma Life Science. Anti–β-tubulin and anti-dsRNA (J2-0601) mAbs were obtained from Lamda Biotech and English and Scientific Consulting, respectively.
DNA Constructs and Reverse Genetics.
Except for the KO2 (Fig. S1) and pLnsp1βcc-2 (Fig. S3) mutants, for which synthetic DNA was used, all other constructs were made by standard PCR-based mutagenesis and recombinant DNA techniques. Procedures for the construction of plasmids are provided in SI Materials and Methods. Methods for in vitro transcription, virus rescue from full-length cDNA clones, and virus titration were described previously (13, 30, 31).
MS.
Nsp2N was immunoprecipitated from SD95-21-M1–infected MARC-145 cell lysate using mAb α-NA-PLP2 and samples were separated on a 6% (wt/vol) SDS/PAGE gel, which was fixed and stained with Coomassie Brilliant Blue G-250 (Bio-Rad). The band expected to contain nsp2N* (based on predicted protein size) was excised. Trypsin digestion and LC-MS/MS analysis were performed as described previously (67). MS spectra were searched against a custom-made protein database containing the nsp2N* sequence. As positive control, a synthetic version of the identified frameshift peptide was made and analyzed by LC-MS/MS.
Immunoassays.
Different regions of PRRSV ORF1a were transiently expressed in RK-13 or HEK-293T cells using truncated derivatives of expression plasmid pL1a and the recombinant vaccinia virus–T7 polymerase expression system (66). Expression products were 35S labeled, immunoprecipitated, and analyzed by SDS/PAGE and autoradiography as described previously (13). Alternatively, nsp1β- and nsp2-related products were detected by consecutive immunoprecipitation of (unlabeled) proteins and Western blot analysis, using a combination of PRRSV nsp-specific mAbs as described previously (13, 31). WT and mutant SD01-08 viruses were launched by transfecting in vitro-transcribed full-length RNA into BHK-21 cells, and radioimmunoprecipitation was conducted to detect the expression of nsp1β- and nsp2-related products (see SI Materials and Methods for detailed procedures).
Dual Luciferase Assay.
Using FuGENE HD transfection reagent (Roche Molecular Biochemicals), HEK-293T cells were cotransfected with 0.2 μg dual luciferase plasmid containing the PRRSV PRF sequence and 50 ng pFLAG-nsp1β. At 24 h posttransfection, cells were harvested and luciferase expression was measured using the Dual Luciferase Stop & Glo Reporter Assay System (Promega) and a luminometer (Berthold). Frameshifting efficiencies were calculated from the ratio of firefly to Renilla luciferase activities, using the IFC control construct as the standard.
Analysis of Protein Sequences and Structure.
Sequence alignment of the PLP1β domain of PRRSV, LDV, and SHFV nsp1β and EAV nsp1 was performed using the MUSCLE algorithm in Geneious 6 (Biomatters Ltd, Auckland, NZ). Potential RNA-binding residues in nsp1β were identified using the program BindN (68). Images of the crystal structure of the PRRSV nsp1β dimer [Protein Data Bank (PDB) ID code 3MTV] (39) were created using PyMOL (69).
Assays for Detecting Interactions Between nsp1β and Viral RNA.
Immunoprecipitation assays to detect RNA-binding proteins were performed using a Magna RIP kit (Millipore) and a RiboTrap kit (Medical & Biological Laboratories) following the manufacturer’s instructions. The amount of target mRNA bound to nsp1β was determined by qRT-PCR, and the presence of nsp1β in RNA–protein complexes verified by Western blot. Detailed experimental procedures are presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Mike Howard and John Atkins (University of Utah) for providing the pDluc plasmid. This work was supported in part by Natural Sciences and Engineering Research Council of Canada Grant 311775-2010 (to B.L.M.), Wellcome Trust Grant 088789 (to A.E.F.), UK Biotechnology and Biological Sciences Research Council Grant BB/G008205/1 (to I.B.), TOP Grant 700.57.301 from the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (to E.J.S.), and US Department of Agriculture National Institute of Food and Agriculture Grant 2007-01745 (to Y.F.).
Footnotes
Conflict of interest statement: The authors have filed a patent application that relates to some aspects of this work.
This article is a PNAS Direct Submission. I.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321930111/-/DCSupplemental.
References
- 1.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J Gen Virol. 2012;93(Pt 7):1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkins JF, Björk GR. A gripping tale of ribosomal frameshifting: Extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev. 2009;73(1):178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: Structure and mechanism. Virus Res. 2009;139(2):193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brierley I, Gilbert RJ, Pennell S. Pseudoknot-Dependent Programmed -1 Ribosomal Frameshifting: Structures, Mechanisms and Models. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Heidelberg: Springer; 2010. pp. 149–174. [Google Scholar]
- 5.Jacks T, Madhani HD, Masiarz FR, Varmus HE. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacks T, Varmus HE. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230(4731):1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 7.Jacks T, et al. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331(6153):280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 8.Mador N, Panet A, Honigman A. Translation of gag, pro, and pol gene products of human T-cell leukemia virus type 2. J Virol. 1989;63(5):2400–2404. doi: 10.1128/jvi.63.5.2400-2404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nam SH, Copeland TD, Hatanaka M, Oroszlan S. Characterization of ribosomal frameshifting for expression of pol gene products of human T-cell leukemia virus type I. J Virol. 1993;67(1):196–203. doi: 10.1128/jvi.67.1.196-203.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiel V, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84(Pt 9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 11.Firth AE, Atkins JF. A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1′ may derive from ribosomal frameshifting. Virol J. 2009;6:14. doi: 10.1186/1743-422X-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atkins JF, Gesteland RF. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Heidelberg: Springer; 2010. [Google Scholar]
- 13.Fang Y, et al. Efficient -2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc Natl Acad Sci USA. 2012;109(43):E2920–E2928. doi: 10.1073/pnas.1211145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snijder EJ, Kikkert M, Fang Y. Arterivirus molecular biology and pathogenesis. J Gen Virol. 2013;94(Pt 10):2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 15.Fang Y, Snijder EJ. The PRRSV replicase: Exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 2010;154(1-2):61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wassenaar AL, Spaan WJ, Gorbalenya AE, Snijder EJ. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: Evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J Virol. 1997;71(12):9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snijder EJ, van Tol H, Roos N, Pedersen KW. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J Gen Virol. 2001;82(Pt 5):985–994. doi: 10.1099/0022-1317-82-5-985. [DOI] [PubMed] [Google Scholar]
- 18.Knoops K, et al. Ultrastructural characterization of arterivirus replication structures: Reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J Virol. 2012;86(5):2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frias-Staheli N, et al. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2(6):404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Z, Chen Z, Lawson SR, Fang Y. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J Virol. 2010;84(15):7832–7846. doi: 10.1128/JVI.00217-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Z, Li Y, Ransburgh R, Snijder EJ, Fang Y. Nonstructural protein 2 of porcine reproductive and respiratory syndrome virus inhibits the antiviral function of interferon-stimulated gene 15. J Virol. 2012;86(7):3839–3850. doi: 10.1128/JVI.06466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Kasteren PB, et al. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc Natl Acad Sci USA. 2013;110(9):E838–E847. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell. 1989;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.den Boon JA, et al. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65(6):2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120(1):49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 26.Qu X, et al. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475(7354):118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plant EP, Dinman JD. Torsional restraint: A new twist on frameshifting pseudoknots. Nucleic Acids Res. 2005;33(6):1825–1833. doi: 10.1093/nar/gki329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441(7090):244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Z, Gilbert RJ, Brierley I. Spacer-length dependence of programmed -1 or -2 ribosomal frameshifting on a U6A heptamer supports a role for messenger RNA (mRNA) tension in frameshifting. Nucleic Acids Res. 2012;40(17):8674–8689. doi: 10.1093/nar/gks629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang Y, et al. A full-length cDNA infectious clone of North American type 1 porcine reproductive and respiratory syndrome virus: Expression of green fluorescent protein in the Nsp2 region. J Virol. 2006;80(23):11447–11455. doi: 10.1128/JVI.01032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Zhu L, Lawson SR, Fang Y. Targeted mutations in a highly conserved motif of the nsp1β protein impair the interferon antagonizing activity of porcine reproductive and respiratory syndrome virus. J Gen Virol. 2013;94(Pt 9):1972–1983. doi: 10.1099/vir.0.051748-0. [DOI] [PubMed] [Google Scholar]
- 32.Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4(4):479–486. [PMC free article] [PubMed] [Google Scholar]
- 34.Fixsen SM, Howard MT. Processive selenocysteine incorporation during synthesis of eukaryotic selenoproteins. J Mol Biol. 2010;399(3):385–396. doi: 10.1016/j.jmb.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.den Boon JA, et al. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: Identification of two papainlike cysteine proteases. J Virol. 1995;69(7):4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tijms MA, van Dinten LC, Gorbalenya AE, Snijder EJ. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc Natl Acad Sci USA. 2001;98(4):1889–1894. doi: 10.1073/pnas.041390398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nedialkova DD, Gorbalenya AE, Snijder EJ. Arterivirus Nsp1 modulates the accumulation of minus-strand templates to control the relative abundance of viral mRNAs. PLoS Pathog. 2010;6(2):e1000772. doi: 10.1371/journal.ppat.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z, et al. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology. 2010;398(1):87–97. doi: 10.1016/j.virol.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue F, et al. The crystal structure of porcine reproductive and respiratory syndrome virus nonstructural protein Nsp1beta reveals a novel metal-dependent nuclease. J Virol. 2010;84(13):6461–6471. doi: 10.1128/JVI.00301-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meulenberg JJ, et al. Localization and fine mapping of antigenic sites on the nucleocapsid protein N of porcine reproductive and respiratory syndrome virus with monoclonal antibodies. Virology. 1998;252(1):106–114. doi: 10.1006/viro.1998.9436. [DOI] [PubMed] [Google Scholar]
- 41.Clarke JB, Spier RE. An investigation into causes of resistance of a cloned line of BHK cells to a strain of foot-and-mouth disease virus. Vet Microbiol. 1983;8(3):259–270. doi: 10.1016/0378-1135(83)90078-0. [DOI] [PubMed] [Google Scholar]
- 42.Beach DL, Keene JD. Ribotrap: Targeted purification of RNA-specific RNPs from cell lysates through immunoaffinity precipitation to identify regulatory proteins and RNAs. Methods Mol Biol. 2008;419:69–91. doi: 10.1007/978-1-59745-033-1_5. [DOI] [PubMed] [Google Scholar]
- 43.Kwak H, Park MW, Jeong S. Annexin A2 binds RNA and reduces the frameshifting efficiency of infectious bronchitis virus. PLoS ONE. 2011;6(8):e24067. doi: 10.1371/journal.pone.0024067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park HJ, Park SJ, Oh DB, Lee S, Kim YG. Increased -1 ribosomal frameshifting efficiency by yeast prion-like phenotype [PSI+] FEBS Lett. 2009;583(4):665–669. doi: 10.1016/j.febslet.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi Y, Zhuang J, Peltz S, Dougherty J. Identification of a cellular factor that modulates HIV-1 programmed ribosomal frameshifting. J Biol Chem. 2010;285(26):19776–19784. doi: 10.1074/jbc.M109.085621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gendron K, et al. The presence of the TAR RNA structure alters the programmed -1 ribosomal frameshift efficiency of the human immunodeficiency virus type 1 (HIV-1) by modifying the rate of translation initiation. Nucleic Acids Res. 2008;36(1):30–40. doi: 10.1093/nar/gkm906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kollmus H, Hentze MW, Hauser H. Regulated ribosomal frameshifting by an RNA-protein interaction. RNA. 1996;2(4):316–323. [PMC free article] [PubMed] [Google Scholar]
- 48.Makeyev AV, et al. HnRNP A3 genes and pseudogenes in the vertebrate genomes. J Exp Zoolog A Comp Exp Biol. 2005;303(4):259–271. doi: 10.1002/jez.a.164. [DOI] [PubMed] [Google Scholar]
- 49.Beura LK, Dinh PX, Osorio FA, Pattnaik AK. Cellular poly(c) binding proteins 1 and 2 interact with porcine reproductive and respiratory syndrome virus nonstructural protein 1β and support viral replication. J Virol. 2011;85(24):12939–12949. doi: 10.1128/JVI.05177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen PJ, Huang YS. CPEB2-eEF2 interaction impedes HIF-1α RNA translation. EMBO J. 2012;31(4):959–971. doi: 10.1038/emboj.2011.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 53.Nahar-Gohad P, Sultan H, Esteban Y, Stabile A, Ko JL. RACK1 identified as the PCBP1-interacting protein with a novel functional role on the regulation of human MOR gene expression. J Neurochem. 2013;124(4):466–477. doi: 10.1111/jnc.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brierley I, Jenner AJ, Inglis SC. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992;227(2):463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mejlhede N, Atkins JF, Neuhard J. Ribosomal -1 frameshifting during decoding of Bacillus subtilis cdd occurs at the sequence CGA AAG. J Bacteriol. 1999;181(9):2930–2937. doi: 10.1128/jb.181.9.2930-2937.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Napthine S, Vidakovic M, Girnary R, Namy O, Brierley I. Prokaryotic-style frameshifting in a plant translation system: Conservation of an unusual single-tRNA slippage event. EMBO J. 2003;22(15):3941–3950. doi: 10.1093/emboj/cdg365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albertini AA, et al. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313(5785):360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 58.Ruigrok RW, Crépin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol. 2011;14(4):504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 59.Lauck M, et al. Exceptional simian hemorrhagic fever virus diversity in a wild African primate community. J Virol. 2013;87(1):688–691. doi: 10.1128/JVI.02433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Snijder EJ, Wassenaar AL, Spaan WJ. The 5′ end of the equine arteritis virus replicase gene encodes a papainlike cysteine protease. J Virol. 1992;66(12):7040–7048. doi: 10.1128/jvi.66.12.7040-7048.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Y, et al. Crystal structure of porcine reproductive and respiratory syndrome virus leader protease Nsp1alpha. J Virol. 2009;83(21):10931–10940. doi: 10.1128/JVI.02579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tijms MA, Nedialkova DD, Zevenhoven-Dobbe JC, Gorbalenya AE, Snijder EJ. Arterivirus subgenomic mRNA synthesis and virion biogenesis depend on the multifunctional nsp1 autoprotease. J Virol. 2007;81(19):10496–10505. doi: 10.1128/JVI.00683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kroese MV, et al. The nsp1alpha and nsp1 papain-like autoproteinases are essential for porcine reproductive and respiratory syndrome virus RNA synthesis. J Gen Virol. 2008;89(Pt 2):494–499. doi: 10.1099/vir.0.83253-0. [DOI] [PubMed] [Google Scholar]
- 64.Kim O, Sun Y, Lai FW, Song C, Yoo D. Modulation of type I interferon induction by porcine reproductive and respiratory syndrome virus and degradation of CREB-binding protein by non-structural protein 1 in MARC-145 and HeLa cells. Virology. 2010;402(2):315–326. doi: 10.1016/j.virol.2010.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patel D, et al. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J Virol. 2010;84(21):11045–11055. doi: 10.1128/JVI.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Snijder EJ, Wassenaar AL, Spaan WJ. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J Virol. 1994;68(9):5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van den Akker J, et al. The redox state of transglutaminase 2 controls arterial remodeling. PLoS ONE. 2011;6(8):e23067. doi: 10.1371/journal.pone.0023067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Brown SJ. BindN: A web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34(Web Server issue):W243–248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeLano WL. 2002. The PyMOL molecular graphics system, version 1.7.0.3 (Schrödinger, LLC, Portland, OR). Available at www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.