Significance
Polymicrobial synergy occurs when infections caused by more than one species are more severe than the sum of the individual species acting alone. Here, we show that a bacterial fight-and-flight response to an antimicrobial, hydrogen peroxide (H2O2), is required for a pathogen to display synergy with a commensal bacterium in vivo. H2O2 is generated by the commensal, and in response, the pathogen either enzymatically destroys (fights) the antimicrobial or disperses away (takes flight) from the antimicrobial-producing commensal. Remarkably, both behaviors are critical for the pathogen to display synergy during coinfection. Moreover, when the pathogen is unable to disperse, the community loses spatial organization, trapping the pathogen next to the commensal.
Keywords: synergy, microbiota, microbial ecology, microenvironment, multispecies
Abstract
The oral pathogen Aggregatibacter actinomycetemcomitans (Aa) resides in infection sites with many microbes, including commensal streptococci such as Streptococcus gordonii (Sg). During infection, Sg promotes the virulence of Aa by producing its preferred carbon source, l-lactate, a phenomenon referred to as cross-feeding. However, as with many streptococci, Sg also produces high levels of the antimicrobial hydrogen peroxide (H2O2), leading to the question of how Aa deals with this potent antimicrobial during coinfection. Here, we show that Aa possesses two complementary responses to H2O2: a detoxification or fight response mediated by catalase (KatA) and a dispersion or flight response mediated by Dispersin B (DspB), an enzyme that dissolves Aa biofilms. Using a murine abscess infection model, we show that both of these responses are required for Sg to promote Aa virulence. Although the role of KatA is to detoxify H2O2 during coinfection, 3D spatial analysis of mixed infections revealed that DspB is required for Aa to spatially organize itself at an optimal distance (>4 µm) from Sg, which we propose allows cross-feeding but reduces exposure to inhibitory levels of H2O2. In addition, these behaviors benefit not only Aa but also Sg, suggesting that fight and flight stimulate the fitness of the community. These results reveal that an antimicrobial produced by a human commensal bacterium enhances the virulence of a pathogenic bacterium by modulating its spatial location in the infection site.
Bacteria rarely live in isolation but instead, form mixed-species communities in most environments, including the human body (1). Interactions within these communities can be cooperative and competitive, and bacteria have evolved highly defined responses to sense and adapt to cues from neighboring species (2). These relationships are relevant to human health, because they can have significant consequences on the outcome and etiology of microbial infections (3). Although many well-studied infections, such as cholera and tuberculosis, are often described as being caused by a single organism, interactions between pathogens and the commensal flora in addition to pathogen–host dynamics shape disease outcomes (4). Furthermore, many infections, such as otitis media (5), chronic wounds (6), and periodontitis (7–9), only manifest themselves or are exacerbated if more than one microbe colonizes the infection site. The etiology of such infections can be attributed to interspecies interactions that enhance microbial colonization and persistence at the infection site. Although bacterial interactions have been observed since Pasteur’s era, our knowledge of specific mechanisms that mediate these interactions is limited (10).
To better understand how interspecies interactions impact virulence, we are investigating a two-species model community containing a pathogenic and commensal member of the human oral microbiome. The pathogen, the nonmotile Gram-negative facultative anaerobe Aggregatibacter actinomycetemcomitans (Aa), is a contributing agent of periodontitis (gum disease) (11), one of the most prevalent infectious diseases worldwide (12). Aa elaborates an array of virulence factors that enhance its survival and directly harm the host during infection, including leukotoxin (13), cytolethal distending toxin (14), CagE (15), fimbriae (16), and LPS (17). Several of these virulence factors contribute to bone loss, a signature of aggressive periodontitis, in various animal models (17, 18). However, despite Aa living in a microbe-rich environment, Aa–host interactions are less frequently investigated in the context of other oral microbes. In our two-species model community, a commensal, the nonmotile Gram-positive facultative anaerobe Streptococcus gordonii (Sg), serves as a representative of the genus Streptococcus, a numerically abundant group within oral microbial populations (19). In the oral cavity, Aa and Sg inhabit the gingival crevice (20), a niche defined as the narrow pocket between the gum and the tooth; however, these organisms can also spread systemically and contribute to extraoral infections, including polymicrobial abscesses (21, 22). Similar to other oral streptococci, Sg rapidly consumes carbohydrates and excretes two primary metabolic end products, l-lactate and hydrogen peroxide (H2O2), which can accumulate to high (millimolar) concentrations within mixed-species biofilms (23, 24). Rapid sugar consumption and H2O2 production render the streptococci extremely competitive by respectively limiting carbon source availability and causing oxidative stress to surrounding microbes. The question thus arises of how Aa is able to persist and cause disease in the context of highly competitive streptococci.
We are pursuing the idea that Aa displays defined responses to l-lactate and H2O2 that are crucial for it to proliferate with Sg in the host. In previous studies, we showed that Aa can cross-feed on Sg-produced l-lactate and actually prefers this carbon source over sugars (25). In a murine abscess infection model, coculture of Aa and Sg results in enhanced Aa cell numbers in the abscess compared with monoculture infections, and the ability of Aa to catabolize Sg-produced l-lactate is required for this benefit (26). Although it is clear from these data that Aa benefits from Sg production of its preferred carbon source (l-lactate) during coinfection, the question remains as to how this interaction is accomplished in the presence of high levels of the Sg-produced antimicrobial H2O2.
In this study, we sought to better understand how Aa has evolved to tolerate the presence of the antimicrobial H2O2 in coculture infections. Because it is well-established that streptococci require oxygen to produce H2O2 (27), we reasoned that identifying genes that respond to oxygen would lead to the discovery of factors used by Aa to cope with H2O2. Although previous in vitro studies showed that Aa responds to H2O2 by expressing catalase (KatA), an H2O2-detoxifying enzyme (28), we hypothesized that KatA may not be sufficient to promote Aa coculture benefits because of the extremely high levels of H2O2 produced by Sg. Here, we show that, in addition to katA, a gene encoding the enzyme Dispersin B (dspB) is also highly induced under oxic conditions. DspB is an extracellular enzyme that promotes dispersal of Aa biofilms (29) by hydrolyzing the polysaccharide poly-GlcNAc, a major component of the extracellular matrix of Aa biofilms. Additional studies showed that Sg-produced H2O2 induces expression of dspB during coculture and that DspB ameliorates oxidative stress by allowing Aa to disseminate farther away from Sg in vivo. We propose a model where Aa senses H2O2 from streptococci to modulate its position within an infection site, such that its exposure to H2O2 is minimized while its access to l-lactate, its preferred carbon source, is maximized.
Results
dspB Is Induced by Oxygen.
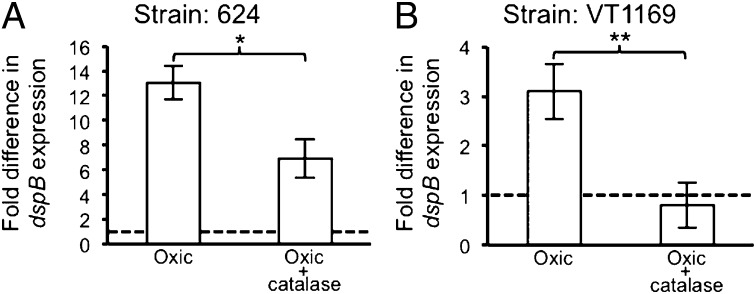
Aa experiences fluctuating oxygen levels in the gingival crevice (30) and in abscess infections (8, 26, 31, 32). Furthermore, oxygen is a major factor influencing Aa–Sg metabolic interactions (26, 27), and it is required for H2O2-mediated Aa growth inhibition by Sg (Fig. S1). Because reactive oxygen species (e.g., H2O2) are generated by aerobic respiration (33) in respiring bacteria, such as Aa, we characterized the Aa transcriptome of Aa strain VT1169 during oxic and anoxic growth using DNA microarrays, providing insight into how this bacterium responds to oxygen (Tables S1 and S2). As expected, genes known to respond to H2O2, including katA (encoding catalase) and apiA (encoding an adhesin) (28), were induced by oxygen. Interestingly, the gene (dspB) encoding the Aa biofilm-dispersing enzyme Dispersin B (29) was coregulated with katA and apiA. To verify our microarray results, we constructed a dspB transcriptional reporter by cloning the dspB promoter (Fig. S2A) upstream of the reporter gene lacZ (encoding β-gal). Based on recent RNA-seq data from our laboratory (31), the promoter controlling dspB was mapped upstream of pepB, the first gene in an operon including pepB, ndk, and dspB (Fig. S2 C and D). The dspB reporter construct was introduced into two strains of Aa: 624 and VT1169. When grown as colony biofilms, both reporter strains showed significant β-gal activity under oxic conditions but little activity under anoxic conditions (Fig. 1 and Dataset S1). Interestingly, induction in both strains could be mitigated by exogenous addition of the H2O2-detoxifying enzyme catalase (Fig. 1 and Dataset S1). These data indicate that dspB transcription is activated during growth with oxygen and that the activating factor is likely H2O2.
Fig. 1.
dspB is induced by oxygen. (A) β-galactosidase activity was quantified for anoxically grown colony biofilms of Aa strain 624 that were transferred to media alone or media supplemented with exogenous catalase (10,000 U mL−1) and incubated under anoxic or oxic conditions for 6 h. Fold difference in dspB expression (y axis) was determined by dividing the expression value for each condition by the corresponding anoxic value. The dashed line represents anoxic expression (normalized to one within each replicate). Error bars represent SEM (n = 3). *P < 0.03 by a two-tailed Student t test. (B) Same as A except with Aa strain VT1169. **P < 0.01 by a two-tailed Student t test.
In Aa, the transcriptional regulator OxyR activates katA and apiA gene expression in response to H2O2 (28). Based on our observations that catalase mitigates dspB induction and that the dspB promoter contains a DNA sequence similar to the OxyR consensus binding site (34) (Fig. S2B), we hypothesized that dspB is regulated by OxyR. Supporting this hypothesis, inactivation of oxyR eliminated activation of dspB under high oxygen growth conditions, and expression could be restored by expressing oxyR in trans (Fig. S3).
dspB Mediates Biofilm Dispersal in Response to Oxygen and H2O2.
To determine whether dspB expression results in dispersal of Aa biofilms, we developed assays to monitor this phenotype. For these assays onward, we used only strain 624, because unlike VT1169, 624 is highly fimbriated and as a result, forms much more robust biofilms. Strains of Aa that are freshly isolated from infections, such as 624, are typically fimbriated; however, repeated passage results in loss of fimbrial expression, such as in VT1169 (35). Thus, 624-like strains are more relevant for Aa biofilm studies. In addition, our expression studies (Fig. 1) indicate that dspB in strain 624, compared with VT1169, is more sensitive to oxygen, providing additional rationale for using only this strain in the following biofilm experiments.
We first used a qualitative test tube assay to examine whether oxygen can trigger dispersal (Fig. S4A). This assay takes advantage of the fact that, in shaking liquid cultures, 624-like strains grow solely attached to surfaces (35), forming a ring of attached aggregates (ring biofilm) at the maximum height reached by the shaking media. Knowing this about Aa biofilms, we hypothesized that, if a ring biofilm was initially formed in a test tube culture under anoxic conditions and subsequently grown under oxic conditions in the presence of a larger volume of media, a second biofilm ring would develop above the initial ring because of the detachment and reattachment of Aa aggregates. As expected, introducing oxygen into anoxic Aa ring biofilms led to biofilm dispersal, and importantly, this behavior depended on dspB (Fig. S4B).
Because our major focus was on H2O2, we next developed a quantitative microtiter dish assay to determine whether H2O2 can enhance Aa biofilm dispersal (Fig. 2A). This assay compares dispersal after oxic culture with or without H2O2 treatment. A key component of this assay is to use a subinhibitory concentration of H2O2 that does not kill Aa. Because H2O2 is rapidly decomposed by Aa catalase activity (24), biofilms received regular pulses of H2O2 to simulate continuous H2O2 exposure, and with each pulsing, we refreshed the liquid medium to remove any detached cells from the supernatant. At the end of treatment, remaining biofilm mass was assessed through crystal violet staining. Decreased crystal violet staining was observed for H2O2-treated biofilms relative to untreated biofilms, indicating that dispersal occurred upon H2O2 addition (Fig. 2B and Dataset S1). Importantly, H2O2 addition did not cause dispersal of Aa dspB− biofilms (Fig. 2B and Dataset S1). Collectively, these results indicate that H2O2 induces Aa biofilm dispersal in a dspB-dependent manner.
Fig. 2.
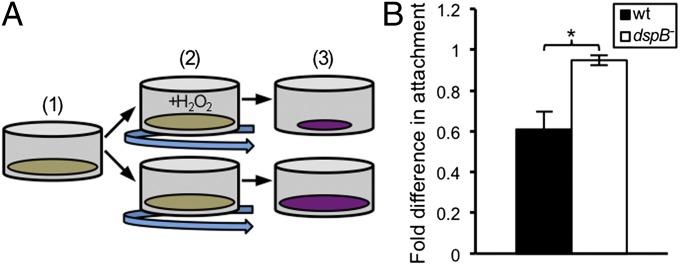
dspB mediates dispersal in response to H2O2. (A) Quantitative microtiter dish assay for measuring biofilm dispersal. (1) Aa is incubated statically under anoxic conditions to promote attachment to the bottom of microtiter dish wells, (2) biofilms are incubated under oxic shaking conditions either with (+H2O2) or without H2O2 addition, and (3) biofilm mass is quantified using crystal violet. (B) Crystal violet incorporated into biofilms was solubilized and quantified by measuring absorbance. The y axis represents the absorbance reading for H2O2-treated biofilms divided by the reading for untreated biofilms. A value less than one thus indicates that dispersal occurred, whereas a value close to one indicates that dispersal was absent. Error bars represent SEM (n = 4). *P < 0.015 by a two-tailed Student t test.
dspB Transcription Is Enhanced During Coculture with Sg.
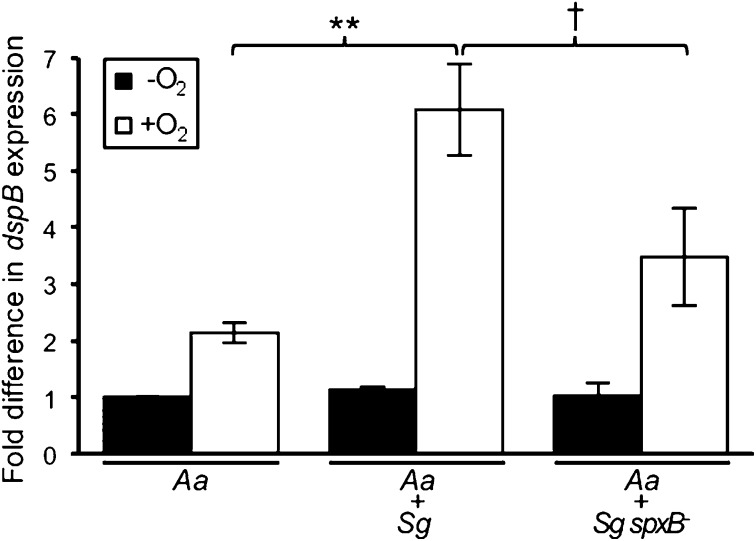
After establishing environmental stimuli and genetic factors that control dspB activity, we next set out to explore the role that this regulation may have in the context of other oral microbes. Because Sg produces high (millimolar) levels of H2O2 (24), we reasoned that dspB, like the similarly regulated gene katA (28), is up-regulated during coculture with Sg. To examine this hypothesis, we measured dspB transcription after growing Aa with Sg in mixed-species biofilms. Transcription of dspB was significantly enhanced during coculture growth compared with monoculture growth under oxic but not anoxic conditions (Fig. 3). This enhanced transcription was primarily caused by H2O2 production by Sg, as an Sg spxB mutant, in which the pyruvate oxidase gene responsible for H2O2 production has been inactivated (36), did not show a statistically significant increase in transcription during coculture (Fig. 3). These data implicate the Sg-produced antimicrobial H2O2 as an inducer of Aa biofilm dispersal.
Fig. 3.
dspB transcription is enhanced during coculture with Sg. Anoxically grown mono- and coculture colony biofilms were transferred to fresh media and incubated for 1 h under oxic or anoxic conditions before measuring β-gal activity. Coculture biofilms were formed by thoroughly mixing equal volumes of exponential-phase cultures of either Aa and Sg or Aa and Sg spxB− that were each adjusted to an OD = 1. Fold difference in dspB expression (y axis) was calculated by dividing the expression value for each condition by the anoxic monoculture value (normalized to one). Error bars represent SEM. The oxic Aa + Sg spxB− coculture value was not significantly larger than the oxic Aa monoculture value (P > 0.13 by a two-tailed t test). **P < 0.002 (n = 5); †P < 0.07 [Aa + Sg (n = 5); Aa + Sg spxB− (n = 4)] by a two-tailed Student t test.
dspB Is Required for Synergy with Sg in Vivo.
H2O2 has been shown to be a potent antimicrobial against Aa both in vitro (Fig. S1) and in vivo (37); thus, to successfully receive a benefit from coculture with Sg, Aa must have mechanisms to resist killing by H2O2. We hypothesized that both katA and dspB are important for this resistance during coculture infection. To test this hypothesis, we used a murine abscess infection model (26) to assess the ability of Aa and Sg to persist in an infection site. This model is biologically relevant because oral bacteria initiate polymicrobial abscesses both inside (22) and outside (38, 39) the mouth and because both Aa (38) and Sg (39) have been cultured from these infections. A concern with this infection model may be that abscesses are too anoxic to permit oxygen-mediated bacterial interactions; however, many studies support the viewpoint that these infections contain multiple oxygen environments (8, 26, 31, 32). Significantly, Aa can only consume l-lactate when oxygen is present, and previous work (26) showed that this process is essential for Aa to grow to higher yields during coinfection with Sg, indicating that abscesses contain sufficient oxygen to support aerobic metabolism in Aa. To confirm these observations, we used RNA-seq to measure katA and dspB transcription in the abscess. We found that both genes are expressed in single-species infections and up-regulated in mixed-species infections containing Sg (Fig. S5). Because these genes respond to oxygen (Table S1) and H2O2 (Fig. 1) (28) and because H2O2 production by Sg requires oxygen (27), the fact that they are expressed in vivo suggests that the abscess is at least partially oxic. Moreover, katA (28) and dspB (Fig. 3) are activated by H2O2 during coculture, and therefore, these data also suggest that Sg causes oxidative stress to Aa in our infection model.
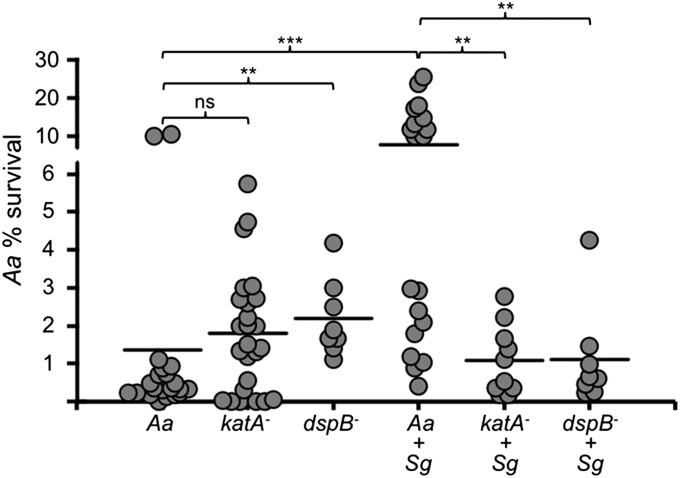
Abscesses were formed with Aa WT, Aa katA−, or Aa dspB− alone or combined with Sg. After 3 d of infection, the number of bacteria within abscesses was assessed. As noted previously, Aa substantially increased in cell number during coinfection, but strikingly, this enhanced persistence no longer occurred when either katA or dspB was absent (Fig. 4 and Fig. S6A). Importantly, the katA and dspB mutants established similar or better single-species infections compared with WT (Fig. 4 and Fig. S6A), eliminating a monoinfection defect as a possible explanation for the loss of synergy. Although not the major focus of this report, we also measured Sg fitness in the abscess, and to our surprise, we found that Sg is also attenuated in coinfections with either Aa katA− or dspB− (Fig. S6B). One explanation for this defect may be that Aa cross-protects Sg, which does not make its own catalase, from self-inflicted oxidative stress. In support of this idea, Sg was most impaired for cell survival when cocultured with Aa katA− (Fig. S6B). Together, these results suggest that the response of Aa to H2O2 imparts community-wide fitness benefits during synergistic infections with Sg.
Fig. 4.
dspB and katA are required for polymicrobial synergy with Sg. Murine abscesses were formed using Aa WT (Aa), katA−, dspB−, and Sg. At 3 d postinfection, abscesses were harvested for enumerating viable bacteria on selective media. Each data point represents a single animal, and at least two biological replicates were performed for each condition. Aa percent survival (y axis) was calculated by dividing the number of viable bacteria recovered from the abscess by the number of viable bacteria measured in the inoculum (107–108 cfu). Horizontal bars represent the mean. ns, not significant by a two-tailed Mann–Whitney U test. **P < 0.01; ***P < 0.001 by a two-tailed Mann–Whitney U test.
DspB Modulates Community Spatial Structure in Vivo.
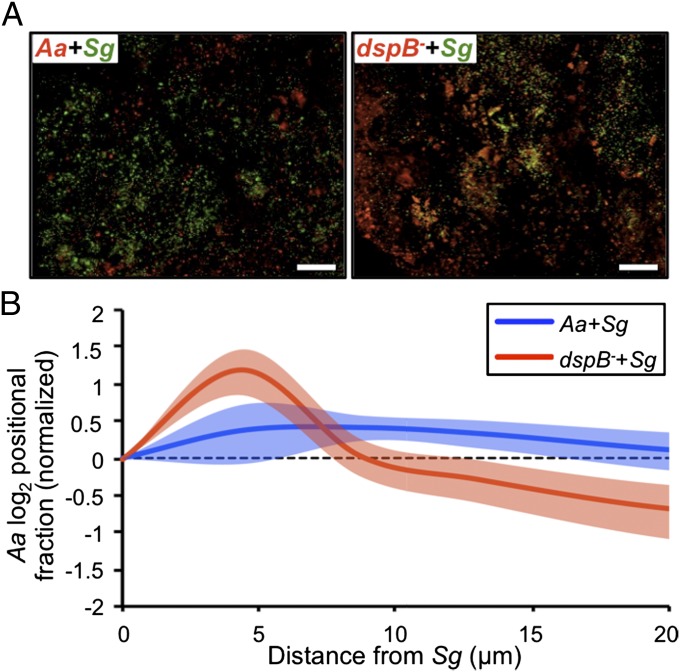
Although the role of KatA during coculture is to protect Aa from H2O2 through detoxification, the role of DspB in mediating synergy is unknown. We hypothesized that the function of DspB is to allow Aa to move away from H2O2-producing bacteria, such as Sg; thus, the dspB mutant fails to exhibit synergy, because it cannot spatially segregate itself from Sg. If so, we predicted that the average distance between Aa dspB− and Sg would be smaller than that between Aa WT and Sg. To test this hypothesis, we formed abscesses using strains of Aa and Sg expressing the fluorescent proteins GFP and mCherry, respectively, and conducted 3D spatial analyses of intact, unfixed abscesses after imaging with confocal microscopy (Fig. 5A). A distinct advantage of this approach is that it removes the need to chemically fix abscesses before imaging, a procedure that may disrupt native spatial structure. For spatial analyses, we used the digital image analysis software daime (40). In short, daime allowed us to determine the size of bacterial populations within the abscess and quantify the spatial organization of the microbial community (Materials and Methods). Examination of confocal images revealed that both WT Aa and Sg grow in small (<0.5 pL or ∼250–1,000 cells), highly dense aggregates in coculture abscesses, whereas Aa dspB− aggregates are about two times as large as WT Aa aggregates (Fig. S7A). Examination of the spatial organization of these aggregates revealed that, although Aa WT aggregates range between 4 and 13 µm away from Sg aggregates, Aa dspB− aggregates are, on average, either immediately adjacent to or within 4 µm of Sg (Fig. 5B and Fig. S7B). These results show that Aa responds to an antimicrobial produced by commensal Sg by changing its spacing, ultimately impacting the organization of the community in the infection site.
Fig. 5.
DspB modulates community spatial structure in vivo. (A) Overlays of representative confocal Z stacks from the indicated coinfections. Aa WT (Aa) or dspB− expressing mCherry is red, and Sg expressing GFP is green. Colocalization appears yellow. (Scale bars: 100 µm.) (B) Aa positional fraction is plotted as a function of distance from Sg. Positional fraction here is defined as the proportion of total Aa biomass that is found at each distance away from Sg. Positional fractions have been normalized such that fractions above one (log2 = 0) occur more than expected, whereas fractions below one (log2 = 0) occur less than expected. Continuous lines represent means, shaded regions represent 95% confidence intervals, and the dashed line represents random spatial distribution. Data represent the results for two biological replicates. Each replicate consisted of two to three abscesses, where two to four Z stacks were acquired from each abscess [Aa + Sg (n = 12); dspB− + Sg (n = 16)].
Discussion
Bacteria more often than not exist in polymicrobial communities in which interactions between community members impact individual as well as group fitness and virulence. Such interactions are especially prevalent in the human oral cavity, where the microbiome assorts itself into spatially structured, mixed-species biofilms. Many pairs/groups of species in this community are known to form close physical associations that ultimately benefit one or all interaction partners (41). A closest neighbor proximity analysis of dental plaque recently revealed that the Pasteurellaceae family (the taxonomic family that includes Aa) strongly colocalizes with streptococci (42). Similarly, we found that, during an abscess infection, Aa localizes within a 4- to 13-µm radius around Sg (Fig. 5B). Because l-lactate catabolism enhances Aa cell yield during coculture (26), the benefit that Aa receives from physically associating with Sg is likely an abundant source of carbon, l-lactate.
The goal of this research was to define the role of Aa H2O2-responsive genes during synergistic infections with Sg. Prior in vitro studies pointed to the likely importance of catalase. Catalase is strongly up-regulated during coculture (28) and responsible for locally depleting H2O2 levels in mixed Aa–Sg biofilms (24). In light of these previous findings, we anticipated that catalase would be an essential Aa fitness determinant during coinfections with Sg. As expected, Aa lacking katA no longer exhibited coinfection synergy (Fig. 4 and Fig. S6A). Similarly, Aa fails to display synergy when it cannot catabolize l-lactate. However, an l-lactate catabolic mutant not only is deficient for synergy but also shows a >100-fold reduction in persistence relative to the WT strain when the WT is singly infecting the host (26). In contrast, Aa katA− is equally fit as the WT in both mono- and coinfection environments (Fig. 4 and Fig. S6A). To us, this observation indicates that, although cross-feeding on l-lactate is the leading nutritional requirement for Aa to achieve synergy, its resistance to Sg-produced H2O2 is a multifactorial phenotype that does not depend solely on detoxification by KatA.
A major finding of this report is that the response of Aa to H2O2 encompasses not only detoxification but also dspB-mediated dispersion. Heretofore, signals and genetic factors controlling dspB expression were unknown. We found that, similar to katA (28), dspB is induced by H2O2 during coculture with Sg (Fig. 3), and when exposed to H2O2, Aa biofilms disperse in a dspB-dependent manner (Fig. 2). Remarkably, this dispersion response is essential for Aa–Sg polymicrobial synergy (Fig. 4 and Fig. S6A). Because biofilms containing streptococci develop highly acidic (23) and/or oxidative microenvironments (24), we hypothesize that, when it cannot disperse, Aa becomes trapped within oxidative zones surrounding Sg, decreasing its fitness and abolishing synergy. Supporting our hypothesis, we showed that WT Aa localizes farther away from Sg than Aa dspB− (Fig. 5B and Fig. S7B). Because Aa is not known to be motile, it is unclear how dispersal is accomplished after DspB production; however, studies have shown that, even without a mechanism for motility, microbial communities can acquire unique spatial patterns based on the degree of cooperation or competition between community members (43–46). In such cases, cell movement can be caused by the effect of cell division and crowding (46). Alternatively, dispersal may rely on convection (47) or a source of flow. Various host tissue compartments are known to contain flow (1, 48, 49), and in some cases, the flow rate has been directly measured (e.g., <10 µL h−1 in the gingival crevice) (50). Furthermore, poly-GlcNAc is known to mediate intercellular adhesion during biofilm formation (51), and therefore, disassembly of this molecule by DspB could increase the likelihood of Aa being transported by flow or other possible mechanisms.
Many streptococci are known to produce H2O2 at amounts sufficient to inhibit the growth of surrounding microbes (24). For instance, H2O2 production by Streptococcus pneumoniae reduces the prevalence of other pathogens within the upper respiratory tract, including Haemophilus influenzae (52). H2O2-mediated antagonism has also been documented between peroxidogenic oral streptococci and Streptococcus mutans (36), Actinomyces naeslundii (53), and Aa (37). Unlike S. pneumoniae, most oral streptococci are commensals that are not associated with pathogenesis. Instead, they are highly abundant primary colonizers of tooth surfaces that lay a foundation for later colonizers of the community (54). H2O2 is, thus, ubiquitous in this ecosystem, and it has likely driven oral bacteria to evolve highly specific behaviors for sensing and responding to H2O2-induced stress.
Often, bacterial regulatory circuits are wired to anticipate an impending stress (28), and therefore, in addition to H2O2 itself, oral microbes may express responses to environmental conditions known to stimulate streptococcal H2O2 production. Notably, oxic conditions favor higher H2O2 production, whereas anoxic conditions favor higher l-lactate production by streptococci (27). Oxygen-induced dispersion in Aa (Fig. 1 and Fig. S4) may therefore represent a preemptive measure to avoid regions containing oxygen and potentially, streptococci-produced H2O2. However, migration to regions lacking oxygen, despite containing presumably higher levels of streptococci-produced l-lactate (27), would prevent Aa from consuming this beneficial carbon source because of the strict oxygen requirement for l-lactate consumption by Aa (26). A possible solution to this problem may be KatA. By converting H2O2 to water and oxygen, KatA may provide Aa with the cosubstrate that it requires for l-lactate consumption. Thus, H2O2 may not be a purely inhibitory molecule in the context of Aa–Sg interactions. We propose a model (Fig. S8) where inhibitory H2O2 levels trigger KatA-mediated detoxification as well as DspB-mediated dispersion to low-oxygen environments, where Sg-produced l-lactate is abundant and where enough Sg-produced H2O2 is present to be converted to oxygen by Aa KatA activity and enhance Aa growth within synergistic infections with Sg.
In summary, our findings show that Aa survives H2O2 stress through detoxification of the antimicrobial by KatA and dispersal away from antimicrobial-producing bacteria by DspB, responses reminiscent of the fight-or-flight response observed when animals are exposed to threats. Although this microbial response is more accurately a fight-and-flight response, it provides a previously unidentified mechanism by which a pathogen responds to an antimicrobial produced by commensal microbiota to modulate its spatial orientation and enhance its virulence. Similar behaviors may underlie the development of synergy and spatial structure in other infections and natural habitats (55).
Materials and Methods
Strains and Media.
Strains and plasmids are listed in Table S3, primers are listed in Table S4, and growth conditions are described in SI Materials and Methods.
Colony Biofilms.
Aa biofilms for monoculture experiments were formed by spotting 100 µL dense cell suspensions (OD = 0.5) onto 0.2-µm-pore–size polycarbonate filters (Whatman) placed onto the surface of solid rich media plates. Colony biofilms were incubated under anoxic conditions, and after 24 h, they were moved to a fresh location on the plate and further incubated for a total of 48 h before conducting experiments.
DNA Manipulations.
DNA manipulations were performed according to standard procedures (56). DNA was sequenced at The University of Texas Institute for Cellular and Molecular Biology DNA Sequencing Facility.
RNA Manipulations.
RNA was isolated and assessed for DNA contamination and integrity as previously described (57).
Microarray Analysis.
RNA for transcriptional profiling was isolated from Aa VT1169 grown to midexponential phase (OD = 0.4) in oxic or anoxic rich liquid cultures. Microarrays were performed in duplicate using a custom Affymetrix GeneChip as previously described (25).
Strain Construction.
Procedures for constructing the dspB transcriptional reporter, constitutive mCherry strain, and Aa mutants are described in SI Materials and Methods.
Microtiter Dish Dispersal Assay.
Logarithmic Aa 624 was adjusted to an OD of 0.5 in rich media (TSBYE), and 500 µL were inoculated into individual wells of a 12-well microtiter dish. Dishes were incubated anoxically for 2 h without shaking to promote cell attachment and biofilm formation. Biofilms were then replaced with 1 mL fresh media and incubated oxically with shaking for a total of 4 h. During this period, the media was refreshed at each 30-min interval either with or without the addition of 1 mM H2O2. Then, biofilms were rinsed with water to remove loosely attached cells and stained for 10 min with 1 mL 0.1% (wt/vol) crystal violet solution. Unbound crystal violet was removed by repeated rinsing in water. Bound crystal violet was solubilized for 30 min with 1 mL 95% (vol/vol) ethanol. The absorbance (A620) was measured two times for each biofilm on a Synergy Mx microplate reader (Biotek). Two technical replicates of each strain (WT or dspB−) and condition (0 or 1 mM H2O2 addition) combination were performed and averaged within each individual experiment.
Murine Abscess Infection Model.
Abscesses were formed as previously described (26) by inoculating each animal with 107–108 cfu. In vivo persistence was assessed by serial dilutions and plate counts on selective media as previously described (26). Experimental protocols involving mice were approved by the Texas Tech University Health Sciences Center Institutional Animal Care and Use Committee.
Spatial Analysis.
daime (40, 58) can determine whether one population (the analyzed population) colocalizes with another population (the reference population) by calculating the proportion of total analyzed population biomass that is contained within defined micrometer ranges away from the reference population. These proportions are referred to as positional fractions. To account for sample-to-sample differences in biomass, positional fractions are normalized by the fractions obtained when the analyzed population has been randomly distributed. Thus, fractions that are above one at small distances indicate that colocalization occurs more than randomly at small distances, whereas fractions that are above one at large distances indicate that colocalization occurs more than randomly at large distances.
Images were analyzed using daime version 2.0 (59). Raw images were converted to the tagged image file format and imported into daime as Z stacks. The red and green channels of each Z stack were combined using the addition operation of the Image calculator. Background was removed using the Blur and subtract function (filter radius = 122 pixels), faint pixels were eliminated by stretching the intensity histogram of each stack (minimum threshold = 5, maximum threshold = 255) using 3D filtering options, and noise was reduced (nonzero voxel neighbors = 5). Large clearly nonbacterial objects were manually defined as 2D regions of interest for each slice within a given Z stack and removed by permanently deleting these specified regions from the entire stack. Images were segmented semiautomatically in three dimensions using the color-based magic wand tool (red object tolerance = 70%, green object tolerance = 75%). 3D spatial arrangement was analyzed using the Inflate Algorithm. A reference mask was generated for spatial analysis of each Z stack by extracting the object layer from the Z stack after it was processed to remove background and segmented using automatic 3D local thresholding. Mean aggregate size was measured for each Z stack by using the Measure Objects function to calculate total volume of each red and green segmented object. Results were exported and analyzed further in Excel.
Supplementary Material
Acknowledgments
We thank members of the laboratory of M.W. for critical discussion of this manuscript. We also thank Dr. Matthew Ramsey for assistance with microarray data acquisition and Dr. Jens Kreth for the gift of the constitutive GFP Streptococcus gordonii strain. This work was supported by National Institutes of Health Grant 1R01DE020100 (to M.W.), and M.W. is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Array Express database (accession no. E-MTAB-2213).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400586111/-/DCSupplemental.
References
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: Surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8(1):15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: Impact on pathogenesis and human disease. Clin Microbiol Rev. 2012;25(1):193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci USA. 2013;110(3):1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armbruster CE, et al. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio. 2010;1(3):e00102–10. doi: 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalton T, et al. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS ONE. 2011;6(11):e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebersole JL, Kesavalu L, Schneider SL, Machen RL, Holt SC. Comparative virulence of periodontopathogens in a mouse abscess model. Oral Dis. 1995;1(3):115–128. doi: 10.1111/j.1601-0825.1995.tb00174.x. [DOI] [PubMed] [Google Scholar]
- 8.Kesavalu L, Holt SC, Ebersole JL. Virulence of a polymicrobic complex, Treponema denticola and Porphyromonas gingivalis, in a murine model. Oral Microbiol Immunol. 1998;13(6):373–377. doi: 10.1111/j.1399-302x.1998.tb00694.x. [DOI] [PubMed] [Google Scholar]
- 9.Yoneda M, et al. Mixed infection of Porphyromonas gingivalis and Bacteroides forsythus in a murine abscess model: Involvement of gingipains in a synergistic effect. J Periodontal Res. 2001;36(4):237–243. doi: 10.1034/j.1600-0765.2001.036004237.x. [DOI] [PubMed] [Google Scholar]
- 10.Pasteur L, Joubert J. Charbon et septicemie. Compt Rend Acad. 1877;85:101–105. [Google Scholar]
- 11.Henderson B, Nair SP, Ward JM, Wilson M. Molecular pathogenicity of the oral opportunistic pathogen Actinobacillus actinomycetemcomitans. Annu Rev Microbiol. 2003;57:29–55. doi: 10.1146/annurev.micro.57.030502.090908. [DOI] [PubMed] [Google Scholar]
- 12.Darveau RP. Periodontitis: A polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8(7):481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 13.Kachlany SC. Aggregatibacter actinomycetemcomitans leukotoxin: From threat to therapy. J Dent Res. 2010;89(6):561–570. doi: 10.1177/0022034510363682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, et al. A new functional site W115 in CdtA is critical for Aggregatibacter actinomycetemcomitans cytolethal distending toxin. PLoS ONE. 2013;8(6):e65729. doi: 10.1371/journal.pone.0065729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teng YT, Zhang X. Apoptotic activity and sub-cellular localization of a T4SS-associated CagE-homologue in Actinobacillus actinomycetemcomitans. Microb Pathog. 2005;38(2-3):125–132. doi: 10.1016/j.micpath.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Schreiner HC, et al. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Natl Acad Sci USA. 2003;100(12):7295–7300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madeira MF, et al. MyD88 is essential for alveolar bone loss induced by Aggregatibacter actinomycetemcomitans lipopolysaccharide in mice. Mol Oral Microbiol. 2013;28(6):415–424. doi: 10.1111/omi.12034. [DOI] [PubMed] [Google Scholar]
- 18.Schreiner H, Li Y, Cline J, Tsiagbe VK, Fine DH. A comparison of Aggregatibacter actinomycetemcomitans (Aa) virulence traits in a rat model for periodontal disease. PLoS ONE. 2013;8(7):e69382. doi: 10.1371/journal.pone.0069382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bik EM, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4(8):962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroes I, Lepp PW, Relman DA. Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci USA. 1999;96(25):14547–14552. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan AH, Weber DJ, Oddone EZ, Perfect JR. Infection due to Actinobacillus actinomycetemcomitans: 15 cases and review. Rev Infect Dis. 1989;11(1):46–63. doi: 10.1093/clinids/11.1.46. [DOI] [PubMed] [Google Scholar]
- 22.Williams BL, McCann GF, Schoenknecht FD. Bacteriology of dental abscesses of endodontic origin. J Clin Microbiol. 1983;18(4):770–774. doi: 10.1128/jcm.18.4.770-774.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao J, et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8(4):e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, et al. Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc Natl Acad Sci USA. 2011;108(7):2668–2673. doi: 10.1073/pnas.1018391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown SA, Whiteley M. A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J Bacteriol. 2007;189(17):6407–6414. doi: 10.1128/JB.00554-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7(3):e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barnard JP, Stinson MW. Influence of environmental conditions on hydrogen peroxide formation by Streptococcus gordonii. Infect Immun. 1999;67(12):6558–6564. doi: 10.1128/iai.67.12.6558-6564.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci USA. 2009;106(5):1578–1583. doi: 10.1073/pnas.0809533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaplan JB, et al. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol. 2004;186(24):8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loesche WJ, Gusberti F, Mettraux G, Higgins T, Syed S. Relationship between oxygen tension and subgingival bacterial flora in untreated human periodontal pockets. Infect Immun. 1983;42(2):659–667. doi: 10.1128/iai.42.2.659-667.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorth P, Trivedi U, Rumbaugh K, Whiteley M. Probing bacterial metabolism during infection using high-resolution transcriptomics. J Bacteriol. 2013;195(22):4991–4998. doi: 10.1128/JB.00875-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fetiye K, et al. Comparison in a rat thigh abscess model of imipenem, meropenem and cefoperazone-sulbactam against Acinetobacter baumannii strains in terms of bactericidal efficacy and resistance selection. Ann Clin Microbiol Antimicrob. 2004;3:2. doi: 10.1186/1476-0711-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 34.Toledano MB, et al. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: A mechanism for differential promoter selection. Cell. 1994;78(5):897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 35.Kachlany SC, Planet PJ, DeSalle R, Fine DH, Figurski DH. Genes for tight adherence of Actinobacillus actinomycetemcomitans: From plaque to plague to pond scum. Trends Microbiol. 2001;9(9):429–437. doi: 10.1016/s0966-842x(01)02161-8. [DOI] [PubMed] [Google Scholar]
- 36.Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 2008;190(13):4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hillman JD, Shivers M. Interaction between wild-type, mutant and revertant forms of the bacterium Streptococcus sanguis and the bacterium Actinobacillus actinomycetemcomitans in vitro and in the gnotobiotic rat. Arch Oral Biol. 1988;33(6):395–401. doi: 10.1016/0003-9969(88)90196-3. [DOI] [PubMed] [Google Scholar]
- 38.Rahamat-Langendoen JC, et al. Brain abscess associated with Aggregatibacter actinomycetemcomitans: Case report and review of literature. J Clin Periodontol. 2011;38(8):702–706. doi: 10.1111/j.1600-051X.2011.01737.x. [DOI] [PubMed] [Google Scholar]
- 39.Esposito S, et al. Streptococcus gordonii extensive multiple subcutanoeus abscesses. Infez Med. 2011;19(3):189–193. [PubMed] [Google Scholar]
- 40.Daims H, Lücker S, Wagner M. daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol. 2006;8(2):200–213. doi: 10.1111/j.1462-2920.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- 41.Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8(7):471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 42.Valm AM, et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci USA. 2011;108(10):4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Momeni B, Brileya KA, Fields MW, Shou W. Strong inter-population cooperation leads to partner intermixing in microbial communities. eLife. 2013;2:e00230. doi: 10.7554/eLife.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nadell CD, Foster KR, Xavier JB. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput Biol. 2010;6(3):e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xavier JB, Foster KR. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci USA. 2007;104(3):876–881. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Estrela S, Brown SP. Metabolic and demographic feedbacks shape the emergent spatial structure and function of microbial communities. PLoS Comput Biol. 2013;9(12):e1003398. doi: 10.1371/journal.pcbi.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaplan JB, Fine DH. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl Environ Microbiol. 2002;68(10):4943–4950. doi: 10.1128/AEM.68.10.4943-4950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishikawa T, Sato T, Mohit G, Imai Y, Yamaguchi T. Transport phenomena of microbial flora in the small intestine with peristalsis. J Theor Biol. 2011;279(1):63–73. doi: 10.1016/j.jtbi.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 49.Ymele-Leki P, Ross JM. Erosion from Staphylococcus aureus biofilms grown under physiologically relevant fluid shear forces yields bacterial cells with reduced avidity to collagen. Appl Environ Microbiol. 2007;73(6):1834–1841. doi: 10.1128/AEM.01319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodson JM. Gingival crevice fluid flow. Periodontol 2000. 2003;31:43–54. doi: 10.1034/j.1600-0757.2003.03104.x. [DOI] [PubMed] [Google Scholar]
- 51.Heilmann C, et al. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20(5):1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 52.Pericone CD, Overweg K, Hermans PW, Weiser JN. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun. 2000;68(7):3990–3997. doi: 10.1128/iai.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jakubovics NS, Gill SR, Vickerman MM, Kolenbrander PE. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol Ecol. 2008;66(3):637–644. doi: 10.1111/j.1574-6941.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kolenbrander PE, Andersen RN, Moore LV. Intrageneric coaggregation among strains of human oral bacteria: Potential role in primary colonization of the tooth surface. Appl Environ Microbiol. 1990;56(12):3890–3894. doi: 10.1128/aem.56.12.3890-3894.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng HY, Alcorn TM, Cohen MS. Effects of H2O2-producing lactobacilli on Neisseria gonorrhoeae growth and catalase activity. J Infect Dis. 1994;170(5):1209–1215. doi: 10.1093/infdis/170.5.1209. [DOI] [PubMed] [Google Scholar]
- 56.Ausubel F, et al. Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. New York: Wiley; 1997. [Google Scholar]
- 57.Korgaonkar AK, Whiteley M. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol. 2011;193(4):909–917. doi: 10.1128/JB.01175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daims H, Wagner M. In situ techniques and digital image analysis methods for quantifying spatial localization patterns of nitrifiers and other microorganisms in biofilm and flocs. Methods Enzymol. 2011;496:185–215. doi: 10.1016/B978-0-12-386489-5.00008-7. [DOI] [PubMed] [Google Scholar]
- 59.Schillinger C, et al. Co-localized or randomly distributed? Pair cross correlation of in vivo grown subgingival biofilm bacteria quantified by digital image analysis. PLoS ONE. 2012;7(5):e37583. doi: 10.1371/journal.pone.0037583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.