Abstract
Escherichia coli nucleoside diphosphate kinase (Ndk) catalyzes ATP-dependent synthesis of ribo- and deoxyribonucleoside triphosphates from the cognate diphosphate precursor. Recently, the Ndk polypeptide was reported to be a multifunctional base excision repair nuclease that processed uracil residues in DNA by acting sequentially as a uracil-DNA glycosylase inhibitor protein (Ugi)-sensitive uracil-DNA glycosylase, an apurinic/apyrimidiniclyase, and a 3′-phosphodiesterase [Postel, E. H. & Abramczyk, B. M. (2003) Proc. Natl. Acad. Sci. USA 100, 13247–13252]. Here we demonstrate that the E. coli Ndk polypeptide lacked detectable uracil-DNA glycosylase activity and, hence, was incapable of acting as a uracil-processing DNA repair nuclease. This finding was based on the following observations: (i) uracil-DNA glycosylase activity did not copurify with Ndk activity; (ii) Ndk purified from E. coli ung- cells showed no detectable uracil-DNA glycosylase activity; and (iii) Ndk failed to bind to a Ugi-Sepharose affinity column that tightly bound E. coli uracil-DNA glycosylase (Ung). Collectively, these observations demonstrate that the E. coli Ndk polypeptide does not possess inherent uracil-DNA glycosylase activity.
Escherichia coli nucleoside diphosphate kinase (Ndk) catalyzes the transfer of γ-phosphate from nucleoside triphosphates to the 5′ position of ribo- and deoxyribonucleoside diphosphates (1). The primary role of Ndk is to maintain the cellular concentration of nucleoside triphosphates for RNA and DNA synthesis (2). E. coli mutants lacking Ndk activity exhibit a mutator phenotype characterized predominantly by A·T to G·C transition mutations (3, 4). This finding is consistent with the observation that E. coli ndk- cells have elevated dCTP and dGTP pools, which could facilitate misincorporation of deoxynucleotides during DNA synthesis (4). Recently, an alternative explanation was proposed to account for the increased mutation rate of E. coli ndk- cells; namely, that Ndk functions as an essential uracil-processing DNA repair activity (5).
Lehman et al. (6) initially isolated E. coli Ndk (“deoxynucleotide kinase”) for the purpose of synthesizing radioactively labeled precursors for studying the enzymatic synthesis of DNA. More recently, Ndk has been purified to apparent homogeneity as a small (143 aa) polypeptide that forms a tetramer in solution (1). The amino acid sequence of Ndk is highly conserved among the E. coli and human forms (2). Although the E. coli genome encodes but one ndk gene, eight orthologous NM23/NDP kinase genes have been identified in the human genome (7). Human NM23-H2/NDP kinase (NM23-H2) shares 43% identical amino acids with E. coli Ndk, including essential residues at the active site (2). Postel and coworkers (8) characterized the DNA-binding properties of NM23-H2 after recognizing that the protein was identical to c-myc purine-binding transcription factor, a presumed regulator of tumor metastasis in humans (9). Subsequently, it was reported that NM23-H2 contained a site-specific endonuclease activity that produced a 3′-hydroxy terminus and a covalent NM23-H2–DNA complex at the 5′-phosphoryl incision site (8, 10). Site-directed mutagenesis studies provided evidence that NM23-H2 Lys-12 was essential for the covalent attachment of NM23-H2 to the incised 5′ end after DNA phosphodiester bond cleavage (8, 11). This finding led to speculation that NM23-H2 mediated DNA cleavage by a mechanism similar to that used by some DNA glycosylase/apurinic/apyrimidinic (AP)-lyase enzymes that use the ε-amino group of Lys as a nucleophile to form a covalent Schiff base intermediate with DNA (8). Recently, E. coli Ndk was reported to cleave both supercoiled pUC19myc DNA and duplex oligonucleotides that contained the c-myc promoter sequence element (12). Moreover, Postel and Abramczyk (5) reported that Ndk was a multifunctional base excision repair nuclease that acted sequentially to excise uracil residues from DNA, cleave the resulting apyrimidinic-site, and remove the 3′-sugar phosphate from the incision site. Thus, three novel activities [uracil-DNA glycosylase, AP-lyase, and 3′-phosphodiesterase] have been attributed to the Ndk polypeptide (5).
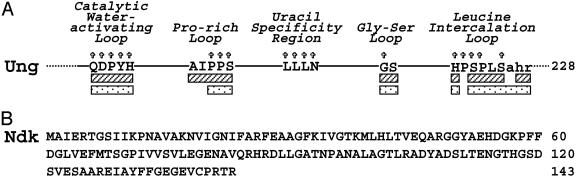
Previously, two genetically distinct forms of E. coli uracil-DNA glycosylase were purified to apparent homogeneity, characterized, and demonstrated to initiate uracil-mediated base excision repair (BER) (13, 14). E. coli Ung is a monofunctional single polypeptide with a molecular mass of 25,558 (15, 16). Significant amino acid homology exists between Ung and uracil-DNA glycosylases from other biological sources, including human, which shares 56% amino acid identity with Ung (17). Five highly conserved structural motifs serve as a signature for the Ung family of enzymes (18). These motifs include the catalytic water-activating loop (63-QDPYH-67), proline-rich loop (84-AIPPS-88), uracil-specificity region (120-LLLN-123), gly–ser loop (165-GS-166), and leucine intercalation loop (187-HPSPLSAHR-195), which facilitate DNA binding and uracil excision (Fig. 1A). Double-stranded uracil-DNA glycosylase (Dug, also termed Mug) was the second uracil-DNA glycosylase identified in E. coli cells (19). Dug is a small (18,672 Da) polypeptide that lacks amino acid sequence identity (<10%) with Ung and does not exhibit the Ung-family DNA-binding motifs (13). However, Dug and Ung do share tertiary structural features as described by x-ray crystallography (20).
Fig. 1.
Conserved motifs of E. coli Ung and amino acid sequence of E. coli Ndk. (A) Five structural motifs, previously implicated by Putnam et al. (18) as critical to Ung activity, are illustrated. Capitalized amino acids indicate structurally equivalent residues in E. coli, human, and herpes simplex virus uracil-DNA glycosylases as determined by high-resolution crystal structures (18, 34, 35). Starred residues correspond to amino acids that are absolutely conserved in E. coli, human, herpes simplex virus, and B. subtilis uracil-DNA glycosylase polypeptides. Striped and speckled bars correspond to residues that contact Ugi and DNA containing an AP-site, respectively. (B) The amino acid sequence of E. coli Ndk, as described by Hama et al. (2), is shown.
Ung and Dug can also be differentiated according to substrate specificity and sensitivity to the PBS-1/-2 uracil-DNA glycosylase inhibitor protein Ugi (13). Whereas Ung excises uracil residues from both single- and double-stranded (i.e., U·G and U·A) target sites, Dug lacks detectable activity on single-stranded uracil-DNA and preferentially recognizes uracil residues in U·G mispairs (13). In addition, although Ugi inactivates Ung by forming an essentially irreversible Ung·Ugi complex, Dug is not inhibited by Ugi (13). The Ugi protein relies on DNA mimicry for recognition by Ung, using the same amino acid contacts involved in Ung-DNA binding (Fig. 1 A). Sung et al. (14) observed that the rate of uracil-initiated BER detected in cell-free extracts was 5-fold greater in E. coli (ung+ dug-) than in E. coli (ung- dug+) extracts. Moreover, uracil-initiated repair was not observed in reactions with E. coli (ung- dug-) cell extracts (14). Therefore, Ung and Dug appeared to be responsible for most, if not all, uracil-mediated BER in E. coli.
The uracil-DNA glycosylase activity reported to be associated with E. coli Ndk was shown to excise uracil residues from both single- and double-stranded DNA substrates and to be inhibited by Ugi (5). Thus, Ndk appeared to exhibit the hallmarks of a Ung-family enzyme. However, inspection of the Ndk amino acid sequence revealed no significant similarities with Ung (Fig. 1). This apparent inconsistency prompted us to investigate whether the putative uracil-DNA glycosylase activity of Ndk was in fact inherent to the Ndk polypeptide.
Materials and Methods
Materials. E. coli BL21(DE3)/pndkec was provided by M. Konrad (Max Planck Institute for Biophysical Chemistry, Gottingen, Germany). E. coli strain BW504 (ung-151::Tn10 thi-1 relA1 spoT1) was provided by B. Weiss (Emory University, Atlanta). E. coli Ung (Fraction IV) and Ugi (Fraction V) were purified as described (21). E. coli endonuclease IV (Fraction V) was provided by B. Demple (Harvard University, Boston). Oligonucleotides 5′-CCTGCCCTGUGCAGCTGTGGG-3′ (U-21-mer), which contained a site-specific uracil residue at position 10, and the complementary strand, 5′-CCCACAGCTGCACAGGGCAGG-3′ (A-21-mer), were obtained from Integrated DNA Technologies (Coralville, IA). The U-21-mer was 5′-end 32P-labeled with [γ-32P]ATP [6,000 Ci/mmol (1 Ci = 37 GBq)] and T4 polynucleotide kinase as described (22); A-21-mer was 5′-end phosphorylated with ATP. Duplex [32P]U·A-21-mer was prepared by annealing [32P]U-21-mer to A-21-mer in a 1:2 ratio followed by buffer exchange against 10 mM Tris·HCl (pH 7.5).
Purification of E. coli Ndk. E. coli Ndk was purified as described (5). E. coli BL21(DE3)/pndkec cells were grown at 37°C in 1 liter of 2× YT medium to mid-log phase, and Ndk overproduction was induced by the addition of IPTG to 1 mM. Incubation was continued for 3 h, and the cells were harvested by centrifugation, resuspended in 50 ml of lysis buffer (50 mM Tris·HCl, pH 8.0/2 mM EDTA, pH 8.0/100 μg/ml lysozyme), and stored at -80°C. Cells were thawed on ice and lysed by sonification, and the lysate was clarified by centrifugation (20,000 × g for 20 min at 4°C); the supernatant fraction was designated Fraction I. Pulverized ammonium sulfate was slowly added to Fraction I to achieve a final concentration of 60% (saturation), and the precipitated protein was removed by centrifugation as described above. The protein pellet was resuspended in 10 ml of buffer A (20 mM Tris·HCl, pH 7.5/1 mM EDTA/1 mM DTT) and dialyzed extensively against the same buffer; the retentate constituted Fraction II. Fraction II (18 ml) was applied to a DEAE-Sepharose (Fast Flow, Amersham Biosciences) column (4.9 cm2 × 5 cm) equilibrated in buffer A. The column was washed with 100 ml of buffer A and eluted with a 300-ml linear gradient of 0–0.5 M NaCl in buffer A. Fractions (5 ml) were collected and assayed for both Ndk and uracil-DNA glycosylase activity. Samples (1 ml) containing Ndk activity were pooled (fractions 41–50), dialyzed against buffer A, and designated Fraction III. Fraction III was applied to a hydroxyapatite (Bio-Gel HTP, Bio-Rad) column (1.8 cm2 × 2.6 cm) equilibrated in buffer A. The column was washed with 20 ml of equilibration buffer and eluted with a 45-ml linear gradient of 0–300 mM potassium phosphate (pH 7.5) in buffer A. Fractions (1.2 ml) were collected and assayed for Ndk and uracil-DNA glycosylase activity.
Purification of E. coli Ndk from BL21(DE3)ung-151 Cells. The wild-type ung gene present in E. coli BL21(DE3) was insertionally inactivated by using standard P1 phage transduction techniques (23) and BW504 (ung-151::Tn10) as the donor strain to produce BL21(DE3)ung-151. Inactivation of ung was confirmed by PCR. Furthermore, the BL21(DE3)ung-151 strain acquired resistance to tetracycline, and uracil-DNA glycosylase activity was not detected in cell-free extracts as measured by using the standard uracil-DNA glycosylase assay described below. BL21(DE3)ung-151 was transformed with pndkec, and Ndk overproduction and purification were conducted as described above for E. coli BL21(DE3)/pndkec.
Ugi-Sepharose Affinity Chromatography. Ugi-Sepharose was prepared and affinity chromatography performed as described (24). Ugi-Sepharose was poured into a Micro Bio-Spin (Bio-Rad) column (250-μl bed volume) and equilibrated in buffer B (30 mM Tris·HCl, pH 7.4/1 mM EDTA/1 mM DTT/5% wt/vol glycerol). A sample (125 μl) of Ndk (Fraction IV) in buffer B was applied to the column, which was then washed with five aliquots (125 μl) of equilibration buffer. The sample and eluted fractions were assayed for Ndk and Ung activity. In a control experiment, E. coli Ung (25 units) was similarly chromatographed and the fractions (125 μl) analyzed for activity.
Ndk Assay. The assay for Ndk activity was conducted as described (25). Standard reaction mixtures (400 μl) were prepared from equal volumes (50 μl) of 0.8 M Tris·acetate (pH 7.5), 0.03 M phosphoenolpyruvate, 0.02 M ATP, pyruvate kinase (12,500 units/ml), lactate dehydrogenase (22.5 units/ml), 0.1 M MgCl2, 0.25 M KCl, and 0.003 M NADH (freshly prepared). Reactions were initiated by the addition of dTDP and the Ndk sample (50 μl each), and the rate of decrease in absorbance at 340 nm was followed. One unit of Ndk activity corresponded to the amount of enzyme that catalyzed the formation of 1 μmol of dTTP per minute under the standard conditions of the assay (25).
Uracil-DNA Glycosylase Assays. Standard uracil-DNA glycosylase reaction mixtures (100 μl) contained 70 mM Hepes-KOH (pH 7.9), 1 mM DTT, 1 mM EDTA, 2.4 μg of calf thymus [uracil-3H]DNA (4.6 μM uracil; 175 cpm/pmol of uracil), and 25 μl of sample. When appropriate, enzyme dilutions were made in 50 mM Hepes-KOH (pH 7.9), 1 mM DTT, 1 mM EDTA, and 100 μg/ml acetylated BSA. After incubation at 37°C for 30 min, reactions were terminated on ice with 250 μl of 10 mM ammonium formate (pH 4.2). [3H]Uracil was resolved from the [3H]DNA substrate by using Bio-Rad AG 1-×8 resin (formate form) and measured in a liquid scintillation spectrometer, as described (26). One unit of uracil-DNA glycosylase is defined as the amount of enzyme that releases 1 nmol of uracil per hour under standard conditions.
Uracil-DNA glycosylase activity was also detected by using the 5′-end 32P-labeled duplex oligonucleotide substrate ([32P]U·A-21-mer) described above. Reaction mixtures (40 μl) contained 10 mM Hepes-KOH (pH 7.4), 100 mM KCl, 2 pmol of [32P]U·A-21-mer, and 10 μl of sample. Samples of the DEAE-Sepharose column fractions were diluted 1/125 in 50 mM Hepes-KOH (pH 7.4), 1 mM EDTA, 1 mM DTT, and 100 μg/ml acetylated BSA. Samples of the hydroxyapatite column fractions were assayed without dilution (fractions 7–18) or at 1/10 dilution (fractions 35–50). Incubation was conducted at 37°C for 30 min and terminated by heating at 70°C for 3 min. Apyrimidinic sites generated by uracil excision were quantitatively cleaved by the addition of 0.1 units of E. coli endonuclease IV and incubation at 30°C for 30 min. Reactions were terminated by the addition of an equal volume of denaturing formamide sample buffer and heating at 95°C for 5 min. Portions (10 μl) of the reaction mixtures were analyzed by using 12% polyacrylamide/8.3 M urea gel electrophoresis as described (13).
Protein Detection. Protein concentration was determined by the Bradford reaction (27), using the Bio-Rad Protein Assay in 96-well microtiter plates. BSA was used to produce a standard curve, and absorbance was monitored at 595 nm. SDS/PAGE was carried out as described (28). Samples of various fractions from the DEAE-Sepharose column were detected by staining with 0.05% Coomassie brilliant blue G-250.
Results
Uracil-DNA Glycosylase Activity Associates with E. coli Ndk During DEAE-Sepharose Chromatography. Postel and Abramczyk (5) recently reported that a Ugi-sensitive uracil-DNA glycosylase activity was associated with the Ndk polypeptide based on copurification during DEAE ion-exchange chromatography. However, it was perplexing that neither the primary amino acid sequence (Fig. 1) nor the tertiary structure of Ndk had identifiable similarities to Ung (1, 2, 5, 18, 29). Accordingly, further investigation was warranted to determine whether Ndk exhibited an inherent uracil-processing DNA repair nuclease activity.
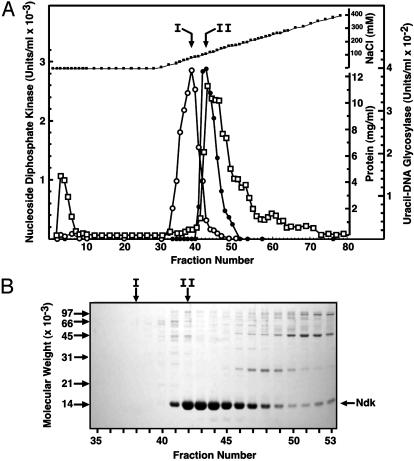
To determine whether the Ndk polypeptide was capable of acting as a uracil-DNA glycosylase, the ndk gene was overexpressed in E. coli BL21(DE3)/pndkec cells and partially purified as described (5). Ndk (Fraction II) was applied to a DEAE-Sepharose column and eluted with a linear gradient of 0–0.5 M NaCl (Fig. 2A). The Ndk activity peak eluted at about 115 mM NaCl and coeluted with the major protein peak. SDS/PAGE of samples across the Ndk activity peak revealed several protein bands (Fig. 2B); however, the predominant band corresponded to a polypeptide with an apparent molecular mass of ≈15 kDa. This molecular mass is in good agreement with the molecular mass (15,462 Da) deduced from the Ndk amino acid sequence. A single peak of uracil-DNA glycosylase activity eluted at ≈80 mM NaCl, preceding the Ndk and major protein peak (Fig. 2 A). Although the uracil-DNA glycosylase and Ndk peaks were not coincident, some uracil-DNA glycosylase activity trailed into fractions containing Ndk. Thus, the DEAE chromatography results were somewhat ambiguous as to whether Ndk possessed inherent uracil-DNA glycosylase activity. Nevertheless, the results clearly indicated that the majority of uracil-DNA glycosylase activity detected was not associated with Ndk.
Fig. 2.
DEAE-Sepharose column elution profile of uracil-DNA glycosylase and Ndk activity. (A) DEAE-Sepharose column chromatography and standard assays for uracil-DNA glycosylase (open circles) and Ndk (filled circles) were performed as described in Materials and Methods. Protein concentration (open squares) and NaCl concentration (filled squares) were determined as described in Materials and Methods. (B) 12% SDS/PAGE of DEAE-Sepharose column fractions was conducted as described in Materials and Methods. The molecular weight standards are indicated by arrows on the left side of the gel; the location of the Ndk band is shown on the right side. Fractions corresponding to the peak of uracil-DNA glycosylase (I) and Ndk (II) activity are indicated by vertical arrows.
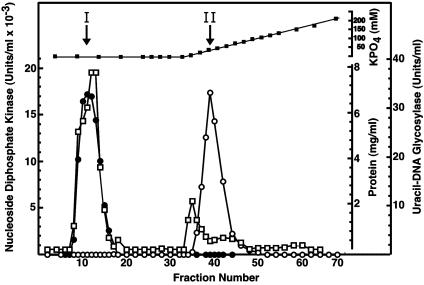
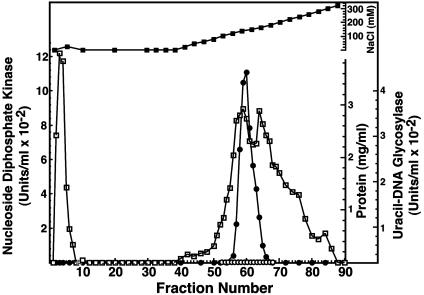
Hydroxyapatite Chromatography of the Ndk/Uracil-DNA Glycosylase Preparation Resolves the Two Activities. To investigate whether the uracil-DNA glycosylase activity detected in the Ndk peak eluting from the DEAE-Sepharose column (Fraction III) was the result of a contaminant or was associated with the Ndk polypeptide, Ndk active fractions were pooled and subjected to hydroxyapatite chromatography (Fig. 3). Ndk activity was detected in the flow-through fractions, whereas uracil-DNA glycosylase activity was bound to the hydroxyapatite matrix, eluting at ≈35 mM potassium phosphate. Uracil-DNA glycosylase activity was not detected in the fractions containing Ndk activity, nor was Ndk activity detected in the fractions containing uracil-DNA glycosylase activity. These results strongly suggest that the uracil-DNA glycosylase activity detected in the Ndk preparation was attributable to contaminating E. coli Ung and not to a uracil-DNA glycosylase activity inherently associated with Ndk.
Fig. 3.
Hydroxyapatite column elution profile of E. coli Ung and Ndk activity. Hydroxyapatite column chromatography of Fraction III and standard assays for Ung (open circles) and Ndk (filled circles) were performed as described in Materials and Methods. Protein concentration (open squares) and potassium phosphate concentration (filled squares) were determined as described in Materials and Methods. Fractions corresponding to the peak of Ung (I) and Ndk (II) activity are indicated by vertical arrows.
Detection of Uracil-DNA Glycosylase Activity by Using a Uracil-Containing Oligonucleotide Substrate. Previously, the proposed uracil-processing DNA repair nuclease of E. coli Ndk was detected with a duplex oligonucleotide substrate (5). To eliminate the possibility that DNA sequence context might have played a role in the inability to detect Ndk-associated uracil-DNA glycosylase activity in the standard uracil-DNA glycosylase assay, the eluted fractions from both the DEAE-Sepharose and the hydroxyapatite columns were reexamined (Fig. 4) by using the identical 32P-labeled oligonucleotide substrate described by Postel and Abramczyk (5).
Fig. 4.
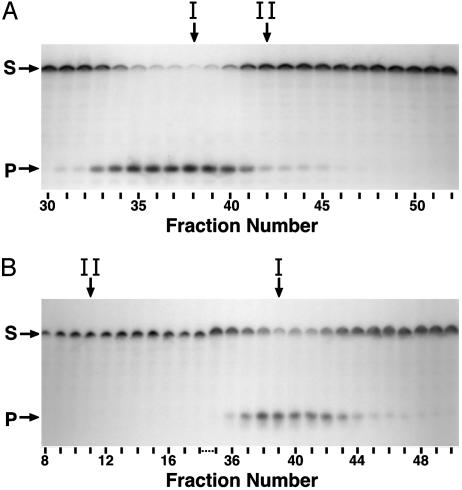
Detection of uracil-DNA glycosylase activity by using a uracil-containing oligodeoxynucleotide substrate. Reaction mixtures (40 μl) containing 2 pmol of 5′-end 32P-labeled duplex oligonucleotide ([32P]U·A 21-mer) and 10 μl of various DEAE-Sepharose (A) or hydroxyapatite (B) fractions were prepared and processed as described in Materials and Methods. Samples (10 μl) of the reaction products were resolved by using denaturing 12% polyacrylamide/8.3 M urea gel electrophoresis. Arrows indicate the location on the autoradiogram of unreacted [32P]21-mer substrate (S) and [32P]9-mer products (P). Fractions corresponding to the peak of Ung (I) and Ndk (II) activity are indicated by vertical arrows.
This duplex 21-mer oligonucleotide substrate contained a uracil residue situated 10 nucleotides from the 5′ end of the 32P-labeled strand and a complementary oligonucleotide (A-21-mer) that contained an adenine residue opposite the uracil, forming a U·A target site. Excision of the uracil residue followed by 5′ cleavage of the AP-site results in a 32P-labeled 9-mer oligonucleotide fragment (product band) that can be resolved from the intact uracil-containing 32P-labeled 21-mer oligonucleotide (substrate band) by denaturing polyacrylamide electrophoresis and visualized by autoradiography. Analysis of the DEAE-Sepharose fractions showed that the peak intensity of the 32P-labeled 9-mer product band was coincident with the peak of uracil-DNA glycosylase activity detected by using the standard assay (Fig. 4A, arrow I). In contrast, the peak of Ndk activity exhibited significantly lower amounts of product, which corresponded to the approximate 9-fold decrease in uracil-DNA glycosylase activity as detected by using the standard assay (Fig. 4A, arrow II). Thus, the results of the 32P-labeled duplex oligonucleotide-based uracil-DNA glycosylase assay corresponded very well to those produced by using the standard uracil-DNA glycosylase assay. Examination of the reaction products produced by the hydroxyapatite column fractions showed that the flow-through fractions containing Ndk did not produce the 9-mer product band indicative of uracil-DNA glycosylase activity (Fig. 4B, arrow II). Notably, excision of uracil from the 32P-labeled oligonucleotide substrate was not detected in the peak fraction of Ndk activity, which contained in excess of 17,000 units/ml and was assayed without dilution. In contrast, the fractions that constituted the peak of uracil-DNA glycosylase activity, as judged by using the standard assay, produced 32P-labeled 9-mer product. Thus, the results of the oligonucleotide-based uracil-DNA glycosylase activity assay reinforced the conclusion reached above; specifically, that hydroxyapatite chromatography separated uracil-DNA glycosylase activity from Ndk activity. These results did not support the proposition that the Ndk polypeptide possessed an inherent Ung-like activity.
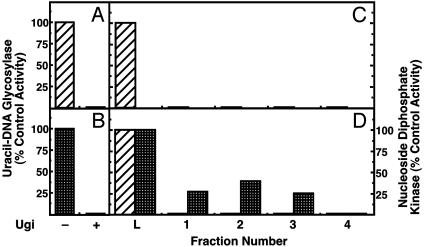
Uracil-DNA Glycosylase Activity Is Not Physically Associated with E. coli Ndk. To further examine the nature of the uracil-DNA glycosylase activity associated with the Ndk (Fraction III) preparation, inhibition studies were conducted with Ugi. The molecular mechanism of Ugi-mediated inhibition of Ung has been elucidated in detail by using both biochemical and structural approaches (18, 26, 28, 30, 31). By using the mechanism of DNA mimicry, Ugi specifically targets the conserved DNA-binding domain of Ung-family enzymes to form a tightly bound noncovalent Ung:Ugi complex with 1:1 stoichiometry (18, 26). When Ndk (Fraction III) was assayed for uracil-DNA glycosylase activity in the presence and absence of Ugi, the results showed that, like the control E. coli Ung, the uracil-DNA glycosylase activity present in the Ndk preparation was completely inhibited by Ugi (Fig. 5 A and B). This result recapitulates the observation made by Postel and Abramczyk (5) but, when taken alone, does not provide sufficient evidence to determine whether the uracil-DNA glycosylase activity detected in the Ndk (Fraction III) preparation is Ndk-associated or the product of a contaminating protein.
Fig. 5.
Ugi-Sepharose column chromatography removes the uracil-DNA glycosylase activity from the Ndk preparation. Standard uracil-DNA glycosylase assays were performed with 1.3 units of E. coli Ung (hatched bars) (A) or 127 units of Ndk (Fraction IV, checkered bars) containing 0.16 units of Ung activity with (+) and without (-) 9 units of Ugi (B). The amount of uracil-DNA glycosylase activity detected in the absence of Ugi was set to 100%. (C) E. coli Ung (25 units in 125 μl, column L) was applied to a Ugi-Sepharose column (125 μl), and fractions were collected as described in Materials and Methods. Column fractions were assayed for uracil-DNA glycosylase activity (striped bars); the amount of uracil-DNA glycosylase activity present in the 125-μl sample (L) was set to 100%. (D) Ndk (Fraction IV containing 3.2 units of uracil-DNA glycosylase activity) was applied to a Ugi-Sepharose column and fractions were collected as described for C. Column fractions were assayed for uracil-DNA glycosylase activity (hatched bars) and Ndk activity (checkered bars). The amount of uracil-DNA glycosylase or Ndk activity present in the sample applied to column (L) was set to 100%.
To determine whether the detected uracil-DNA glycosylase activity was physically associated with E. coli Ndk, the Ndk (Fraction III) preparation was further purified by using a Ugi-Sepharose affinity column. This matrix contained covalently bound Ugi-protein that acts as a ligand to bind specifically and tightly Ung-family enzymes (24). It was anticipated that if the Ugi-sensitive uracil-DNA glycosylase activity detected in the Ndk preparation were cognate to the Ndk polypeptide, then Ndk would bind to the Ugi-Sepharose column. Alternatively, if the uracil-DNA glycosylase activity were a contaminant, then Ndk would not bind to the Ugi-Sepharose matrix and would elute in the wash fractions, which would be devoid of uracil-DNA glycosylase activity. To validate this approach, purified E. coli Ung was applied to the Ugi-Sepharose column and observed to bind, because uracil-DNA glycosylase activity was undetectable in the wash fractions (Fig. 5C). When Ndk was subjected to Ugi-Sepharose affinity chromatography, Ndk activity, but not uracil-DNA glycosylase activity, was found to flow through the column (Fig. 5D). These results again demonstrated that the Ndk protein was not physically associated with the uracil-DNA glycosylase activity observed in the Ndk preparation.
E. coli Ndk Overproduced in Bacteria Lacking Detectable Uracil-DNA Glycosylase Activity. To provide genetic evidence that E. coli Ndk did not possess uracil-DNA glycosylase activity, the wild-type ung gene of the E. coli strain BL21(DE3) was insertionally inactivated. The resulting ung- strain, BL21(DE3)ung-151, was transformed with the pndkec vector, and E. coli Ndk was overproduced and partially purified as described for Ndk isolated from E. coli BL21(DE3)/pndkec. The elution profile from DEAE-Sepharose showed that the peak of Ndk activity coeluted (≈130 mM NaCl) with the major protein peak (Fig. 6). These results were consistent with those reported above (Fig. 2) for the Ndk preparation produced in an ung+ genetic background. However, when the DEAE-Sepharose fractions of the BL21(DE3)ung-151/pndkec preparation that comprised the Ndk activity peak were assayed for uracil-DNA glycosylase activity, no activity was detected. The fact that no detectable uracil-DNA glycosylase activity was observed in BL21(DE3)ung-151/pndkec DEAE-Sepharose fractions provided additional evidence against the proposition that E. coli Ndk possessed inherent uracil-DNA glycosylase activity.
Fig. 6.
DEAE-Sepharose column elution profile of E. coli Ndk activity over-produced in ung- E. coli Cells. DEAE-Sepharose column chromatography and standard assays for uracil-DNA glycosylase (open circles), Ndk (filled circles), protein concentration (open squares), and NaCl concentration (filled squares) were performed as described in Materials and Methods.
Discussion
This investigation was prompted by a recent report that the E. coli Ndk polypeptide possessed an inherent uracil-DNA glycosylase activity (5). The results of this investigation demonstrate that (i) the peak of Ndk activity and that of uracil-DNA glycosylase activity were not coincident during DEAE-Sepharose chromatography. The peak of uracil-DNA glycosylase activity eluted several fractions before the Ndk peak, which did contain the trailing fractions of the uracil-DNA glycosylase peak. (ii) Ndk activity was separated entirely from uracil-DNA glycosylase activity by hydroxyapatite chromatography. (iii) When the fractions constituting the DEAE-Sepharose peak of Ndk activity were subjected to Ugi-Sepharose affinity chromatography, Ndk activity was devoid of uracil-DNA glycosylase activity. (iv) When the Ndk protein was overproduced in E. coli ung- cells, uracil-DNA glycosylase activity was undetectable in the Ndk preparation. These results provide convincing evidence against the proposition that E. coli Ndk possessed inherent uracil-DNA glycosylase activity and suggest that the Ndk preparation of Postel and Abramczyk (5) contained a contaminant, which was very likely E. coli Ung.
E. coli Ndk has been reported to be a multifunctional BER nuclease (5). The 15,462-Da Ndk polypeptide was purported to act sequentially as a uracil-DNA glycosylase, an AP-lyase, and a 3′-phosphodiesterase (5). In addition, it was reported that Ndk exhibited a weak 3′ to 5′ exonuclease activity on duplex DNA when the glycosylase activity was not required or when the protein concentration of the Ndk preparation was high (5). Interestingly, the hypothesis was advanced that E. coli Ndk was a uracil-processing DNA repair nuclease essential for a DNA repair pathway. Moreover, it was proposed that the mutator phenotype of E. coli ndk- cells was caused by the loss of the multiple BER activities of Ndk and its associated 3′ to 5′ exonuclease activity, rather than to deoxynucleotide pool abnormalities characterized by elevated dCTP (20-fold) and dGTP (7-fold) pool sizes (3–5, 32). Lastly, it has been proposed that Ndk, by virtue of its U·A and U·G repair activities, plays a role as proofreader for errors produced by DNA polymerase during replication and DNA repair. Thus, a rather large number of functions have been ascribed to the relatively small Ndk polypeptide. In this investigation, compelling evidence has been presented that E. coli Ndk lacks uracil-DNA glycosylase activity. Because excision of uracil is the initiating event in uracil-mediated BER, E. coli Ndk cannot function in E. coli uracil-DNA repair as proposed. Interestingly, when ndk- E. coli were infected with T4 phage, Mathews and coworkers (32) observed that dNTP pools and spontaneous mutation frequencies returned to near normal, presumably because of a robust dCTPase activity expressed by the phage. These results controvert the hypothesis that the mutator phenotype of E. coli ndk- cells is attributable to the essential Ndk uracil-processing DNA repair nuclease. Postel and Abramczyk reported that the uracil-DNA glycosylase activity of the E. coli Ndk preparation (Fraction III) was inhibited by Ugi, and interpreted this result to mean that Ndk, with respect to its uracil-specific binding pocket, was structurally related to the Ung-family of enzymes. This conclusion was premature, because Ndk was not demonstrated to bind directly to Ugi. This result was not surprising, because the uracil-specificity region of E. coli Ung, as well as the conserved Ugi:Ung interface (18) shared by the Ung-family of uracil-DNA glycosylases (Fig. 1), were not present in the E. coli Ndk primary amino sequence.
When the fractions across the Ndk activity peak were analyzed by SDS/PAGE, numerous protein bands, in addition to the predominant Ndk band, were detected (Fig. 2 A). This result was consistent with that obtained by Postel and Abramczyk (5) and is not unusual, because the peak of Ndk activity corresponded to the major protein peak. Given the relatively high turnover number of the Ung enzyme (≈800 min-1) (15), it is quite possible that a low-concentration Ung contaminant would produce readily detectable activity. Established procedures exist to assess whether a polypeptide exhibits two or more activities. Kornberg and coworkers (33) suggested that the DNA polymerase activity as well as the (3′ to 5′) exonuclease activity of E. coli DNA polymerase I resided in the same polypeptide, because the ratio of the two catalytic functions was constant throughout purification. This test of ratio constancy is a powerful tool that can be used during protein purification to discriminate between the inherent multifunctionality of a polypeptide and contaminating activities that appear to copurify. Application of the ratio test to the Ndk and uracil-DNA glycosylase activities detected in the DEAE-Sepharose elution fractions (Fig. 2 A) clearly indicated that the rule of ratio constancy was not observed and, hence, that the two activities were not coresident in the Ndk polypeptide. Whether the other activities reported (5) as inherent to the Ndk polypeptide, i.e., apyrimidinic endonuclease lyase, 3′-phosphodiesterase, and 3′ to 5′ exonuclease, will pass the test of ratio constancy remains to be demonstrated.
Acknowledgments
This article is dedicated to the memory of Dale W. Mosbaugh. We thank M. Konrad for the pndkec plasmid. This work was supported by National Institutes of Health Grants GM32823 (to D.W.M.) and GM66245 (to S.E.B.) and National Institute of Environmental Health Sciences Grant P30-ES00210.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ndk, Escherichia coli nucleoside diphosphate kinase; BER, base excision repair; Ung, E. coli uracil-DNA glycosylase; Dug, E. coli double-stranded uracil-DNA glycosylase; Ugi, uracil-DNA glycosylase inhibitor protein; AP, apurinic/apyrimidinic.
References
- 1.Almaula, N., Lu, Q., Delgado, J. S. B. & Inouye, M. (1995) J. Bacteriol. 177, 2524-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hama, H., Almaula, N., Lerner, C. G., Inouye, S. & Inouye, M. (1991) Gene 105, 31-36. [DOI] [PubMed] [Google Scholar]
- 3.Miller, J. H., Funchain, P., Clendenin, W., Huang, T., Nguyen, A., Wolff, E., Yeung, A., Chiang, J. H., Garibyan, L., Slupska, M. M. & Yang, H. (2002) Genetics 162, 5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu, Q., Zhang, X., Almaula, N., Mathews, C. K. & Inouye, M. (1995) J. Mol. Biol. 254, 337-341. [DOI] [PubMed] [Google Scholar]
- 5.Postel, E. H. & Abramczyk, B. M. (2003) Proc. Natl. Acad. Sci. USA 100, 13247-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehman, I. R., Bessman, M. J., Simms, E. S. & Kornberg, A. (1958) J. Biol. Chem. 233, 163-170. [PubMed] [Google Scholar]
- 7.Lacombe, M. L., Milon, L., Munier, A., Mehus, J. G. & Lambeth, D. O. (2000) J. Bioenerg. Biomembr. 32, 247-258. [DOI] [PubMed] [Google Scholar]
- 8.Postel, E. H., Abramczyk, B. M., Levit, M. N. & Kyin, S. (2000) Proc. Natl. Acad. Sci. USA 97, 14194-14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Postel, E. H., Berberich, S. J., Flint, S. J. & Ferrone, C. A. (1993) Science 261, 478-480. [DOI] [PubMed] [Google Scholar]
- 10.Postel, E. H. (1999) J. Biol. Chem. 274, 22821-22829. [DOI] [PubMed] [Google Scholar]
- 11.Postel, E. H., Abramczyk, B. A., Gursky, S. K. & Xu, Y. (2002) Biochemistry 41, 6330-6337. [DOI] [PubMed] [Google Scholar]
- 12.Levit, M. N., Abramczyk, B. M., Stock, J. B. & Postel, E. H. (2002) J. Biol. Chem. 277, 5163-5167. [DOI] [PubMed] [Google Scholar]
- 13.Sung, J.-S. & Mosbaugh, D. W. (2000) Biochemistry 39, 10224-10235. [DOI] [PubMed] [Google Scholar]
- 14.Sung, J.-S., Bennett, S. E. & Mosbaugh, D. W. (2001) J. Biol. Chem. 276, 2276-2285. [DOI] [PubMed] [Google Scholar]
- 15.Lindahl, T., Ljungquist, S., Siegert, W., Nyberg, B. & Sperens, B. (1977) J. Biol. Chem. 252, 3286-3294. [PubMed] [Google Scholar]
- 16.Bennett, S. E., Jensen, O. N., Barofsky, D. F. & Mosbaugh, D. W. (1994) J. Biol. Chem. 269, 21870-21879. [PubMed] [Google Scholar]
- 17.Olsen, L. C., Aasland, R., Wittwer, C. U., Krokan, H. E. & Helland, D. E. (1989) EMBO J. 8, 3121-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Putnam, C. D., Shroyer, M. J., Lundquist, A. J., Mol, C. D., Arvai, A. S., Mosbaugh, D. W. & Tainer, J. A. (1999) J. Mol. Biol. 287, 331-346. [DOI] [PubMed] [Google Scholar]
- 19.Gallinari, P. & Jiricny, J. (1996) Nature 383, 735-738. [DOI] [PubMed] [Google Scholar]
- 20.Barrett, T. E., Savva, R., Panayotou, G., Barlow, T., Brown, T., Jiricny, J. & Pearl, L. H. (1998) Cell 92, 117-129. [DOI] [PubMed] [Google Scholar]
- 21.Sanderson, R. J. & Mosbaugh, D. W. (1998) J. Biol. Chem. 273, 24822-24831. [DOI] [PubMed] [Google Scholar]
- 22.Bennett, S. E., Sanderson, R. J. & Mosbaugh, D. W. (1995) Biochemistry 34, 6109-6119. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 24.Bennett, S. E., Shroyer, M. J. N., Sung, J.-S. & Mosbaugh, D. W. (2002) Methods Mol. Biol. 197, 211-225. [DOI] [PubMed] [Google Scholar]
- 25.Agarwal, R. P., Robison, B. & Parks, R. E., Jr. (1978) Methods Enzymol. 51, 376-386. [DOI] [PubMed] [Google Scholar]
- 26.Bennett, S. E. & Mosbaugh, D. W. (1992) J. Biol. Chem. 267, 22512-22521. [PubMed] [Google Scholar]
- 27.Bradford, M. M. (1976) Anal. Biochem. 72, 248-254. [DOI] [PubMed] [Google Scholar]
- 28.Bennett, S. E., Schimerlik, M. I. & Mosbaugh, D. W. (1993) J. Biol. Chem. 268, 26879-26885. [PubMed] [Google Scholar]
- 29.Janin, J., Dumas, C., Morera, S., Xu, Y., Meyer, P., Chiadmi, M. & Cherfils, J. (2000) J. Bioenerg. Biomembr. 32, 215-225. [DOI] [PubMed] [Google Scholar]
- 30.Lundquist, A. J., Beger, R. D., Bennett, S. E., Bolton, P. H. & Mosbaugh, D. W. (1997) J. Biol. Chem. 272, 21408-21419. [DOI] [PubMed] [Google Scholar]
- 31.Sanderson, R. J. & Mosbaugh, D. W. (1996) J. Biol. Chem. 271, 29170-29181. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, X., Lu, Q., Inouye, M. & Mathews, C. K. (1996) J. Bacteriol. 178, 4115-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson, C. C., Schildkraut, C. L., Aposhian, H. V. & Kornberg, A. (1964) J. Biol. Chem. 239, 222-232. [PubMed] [Google Scholar]
- 34.Mol, C. D., Arvai, A. S., Sanderson, R. J., Slupphaug, G., Kavli, B., Krokan, H. E., Mosbaugh, D. W. & Tainer, J. A. (1995) Cell 82, 701-708. [DOI] [PubMed] [Google Scholar]
- 35.Savva, R. & Pearl, L. H. (1995) Nat. Struct. Biol. 2, 752-757. [DOI] [PubMed] [Google Scholar]