Significance
We show that the phenotype of an organism can be affected both by chromosomal and nonchromosomal information. The influence of the cytoplasmic, nonchromosomal information can have a profound effect on the phenotype and can be great enough to mask the effect of a chromosomal mutation. Our ability to quantify the contribution of this cytoplasmic component shows that it could comprise a significant portion of the “missing heritability.” In addition, our findings highlight the possibility of mitochondrial/nuclear incompatibility, which could be an important consideration in evaluating the feasibility of mitochondrial replacement therapy in humans.
Keywords: genetic interactions, extrachromosomal, nonlinear model
Abstract
The measurement of any nonchromosomal genetic contribution to the heritability of a trait is often confounded by the inability to control both the chromosomal and nonchromosomal information in a population. We have designed a unique system in yeast where we can control both sources of information so that the phenotype of a single chromosomal polymorphism can be measured in the presence of different cytoplasmic elements. With this system, we have shown that both the source of the mitochondrial genome and the presence or absence of a dsRNA virus influence the phenotype of chromosomal variants that affect the growth of yeast. Moreover, by considering this nonchromosomal information that is passed from parent to offspring and by allowing chromosomal and nonchromosomal information to exhibit nonadditive interactions, we are able to account for much of the heritability of growth traits. Taken together, our results highlight the importance of including all sources of heritable information in genetic studies and suggest a possible avenue of attack for finding additional missing heritability.
A fundamental problem in genetics is unraveling the link between genotype and phenotype. Ascertaining the heritability of a trait is a key step toward harnessing the predictive capacity of genetic information for human disease risk assessment and therapy (1). Knowledge of all of the elements contributing to heritability would facilitate the establishment of a causal relationship between the information that is passed down from generation to generation and the resulting phenotype. Genome-wide association studies (GWASs) have successfully identified many human polymorphisms that are associated with traits such as height, eye color, or susceptibility to common diseases, but these variants typically explain only a small proportion of the observed heritability of a trait (2, 3).
A number of explanations for missing heritability have been suggested (2), including the existence of many weak variants with effects too small to achieve statistical significance (4), interactions between variants that cannot be identified with current studies (5), rare variants that were not identified by GWAS, and epigenetic effects (6–8). The contribution of nonchromosomal information to the missing heritability is rarely considered, despite the fact that there is a long history documenting the effect in many organisms of diverse cytoplasmic elements on phenotype. Recent work on a mouse model of Crohn disease supports a combinatorial model of complex disease traits in which the pathology requires the interaction between a specific mutation in the mouse and a specific strain of virus (9). Another recent study showed strong effects on the plant metabolome stemming from variation in mitochondrial and chloroplast genomes (10). In humans, the importance of nonchromosomal information has been supported by targeted analyses, but these studies have not analyzed its impact on heritability in a well-controlled context (11–13). Such nonchromosomal interactions might help explain why shared mutations in humans do not always produce the same phenotype, thus reducing the apparent heritability of a trait (14, 15).
We sought to characterize explicitly how nonchromosomal modifiers collectively influence the heritability of a trait, colony size, in a system unique to yeast where we use a defined chromosomal genotype and vary the cytoplasmic genetic information. Yeast has at least four well-studied sources of inherited, nonchromosomal information: mitochondrial DNA, an endogenous dsRNA virus (16, 17), prions (18, 19), and a 2µ plasmid (20, 21).
Our results show that the nonchromosomal contribution to heritability can be large, adding another dimension to the estimation of heritability in wild populations. Nonchromosomal information is not under the usual constraints of the nuclear genome. These nonchromosomal elements are extremely unstable: they mutate at higher frequencies than the DNA of the chromosomal genome, may be lost at high frequencies without loss of viability, and can vary in copy number from cell to cell. Thus, careful controls and measurements are necessary to characterize the effects of nonchromosomal modifiers.
Results
We studied the effect of two inherited sources of cytoplasmic information on the phenotype of selected chromosomal mutations in identical chromosomal genomic backgrounds: one was the presence or absence of the yeast killer dsRNA virus and the other was varying mitochondria among two backgrounds with distinct differences in their genome sequence. The two mitochondrial genomes we selected show considerable variation, with about two to three SNPs per kilobase between them and 10 times as many insertions and deletions per kilobase between them as found in the chromosomal genome. These differences are typical of those found in mitochondrial genomes sequenced from a broad collection of wild and laboratory strains (Table S1).
The protocol we use enabled us to study the effects of nonchromosomal information on the phenotype of chromosomal mutations in the same Sigma chromosomal background but with different cytoplasmic backgrounds. Strains with identical chromosomal information but different cytoplasmic information can be constructed using the kar1 mutant of yeast. The kar1 mutants are yeast strains that are deficient in nuclear fusion during mating and thus exchange cytoplasmic but not nuclear content (22) (Methods). The chromosomal mutations we studied were strain-specific: single gene deletions that confer growth defects in the Sigma strain but not in the reference strain S288c (23). Fig. 1 presents our strategy of generating four haploid strains to test all combinations of chromosomal and nonchromosomal variants. In each test of a particular chromosomal and nonchromosomal variant, meiotic spores from each of the four strains were dissected on the same plate and the size of the spore clones determined.
Fig. 1.
Experimental design for assessing nonchromosomal interactions with chromosomal variants. To determine whether particular nonchromosomal factors (red and blue represent different cytoplasmic information, e.g., mitochondrial DNAs with polymorphisms) interact with a gene deletion variant (denoted by + and ∆ symbols in the nucleus), we construct all four possible haploid strains combining these two nuclear and nonchromosomal factors as described in the main text and Methods. Phenotypic measurements of the four controlled genotypes are then compared to understand the effect the nonchromosomal element has on the phenotype. A genetic mechanism controlled only by the chromosomal gene deletion will result in strains C and D showing a similar growth defect relative to strains A and B, whereas an interaction between chromosomal and nonchromosomal genotype could yield a growth defect confined to strain D. In our study, replicate growth measurements from each controlled genotype are analyzed using a series of statistical models that vary the underlying genetic model, and the goodness of fit is compared across models.
To determine the prevalence of chromosomal and nonchromosomal interactions, we analyzed 17 single gene deletions with a growth defect in Sigma. Although these constructed deletion variants are not observed in natural populations, we note that hundreds of putative natural loss-of-function variants are observed across multiple wild and laboratory yeast strains (Table S1) (24, 25). We found that 6 of the 17 gene deletions (Fig. 2 A and B) (∆pep7, ∆pep12, ∆pho88, ∆ski8, ∆vps16, and ∆ski7) grew more slowly when the strain contained the dsRNA virus. The inhibitory effect of the dsRNA virus varied from total (∆ski7) to slight (∆vps16). The mutant clones containing the virus showed more variation in size than did their otherwise isogenic nonmutant sister clones, a phenotype that could be caused by variation in copy number of the dsRNA virus (Table S2). The dsRNA encodes a toxin that is secreted and kills strains lacking the dsRNA virus, but it has not been known to kill cells that carry it under our conditions. Strains carrying the dsRNA are resistant to killing by the toxin (26, 27).
Fig. 2.
The phenotype of a mutation is dependent upon the presence of a cytoplasmic dsRNA virus and mitochondrial genotype. For A–C, each row is the result of a dissection of meiotic products from a diploid. The four spores from a single meiosis were placed from left to right in each row. In all tetrads, the larger two colonies are those with the wild-type chromosomal allele. (A) The ∆ski7 mutation is viable in the absence of dsRNA virus [kil-0] (Right) and lethal in the presence of dsRNA virus [kil-k] (Left). All of the colonies in the right panel lack dsRNA virus ([kil-0]). In the panel on the left the two spore clones that failed to grow in each quartet contain the deletion. The two viable clones are wild type (+) and contain the dsRNA virus ([kil-k]). (B) The presence of dsRNA virus inhibits the growth of several mutations in the Sigma background. Some of the mutants with the dsRNA virus grew extremely slowly and were visible only after 10 d of incubation. For each mutation the meiotic spores with the dsRNA virus and the one without it were dissected on the same plate. (C) Variation in mitochondrial genotype influences the growth phenotypes of multiple chromosomal variants. Each heterozygous gene deletion strain (+/∆) was constructed with its native mitochondria ([rho+]Sigma) and with another mitochondrial background ([rho+]S288c).
The most extreme chromosomal mutation–dsRNA interaction was with the ∆ski7 allele, which is lethal in the presence of the dsRNA (the heterozygous diploid +/∆ski7 gives rise to two viable and two dead ∆ski7 haploid progeny; Fig. 2A) and viable in the absence of the nonkiller dsRNA. Thus, the lethality or viability of the ∆ski7 deletion in the Sigma background is completely dependent on the nonchromosomal information despite having the same chromosomal DNA sequences.
The effect of the dsRNA virus on the phenotype of chromosomal mutations raised the possibility that other sources of nonchromosomal genetic information could also alter their phenotype. Again using kar1 mutant strains, we were able to create strains with identical chromosomal complements that varied only in mitochondrial backgrounds. To test whether mitochondrial genotype altered the phenotype of these polymorphisms, we compared the phenotype of each deletion in the context of the Sigma nuclear genome in the presence of its native mitochondria or in the presence of mitochondria derived from the closely related strain, S288c . The source of mitochondria profoundly affected the phenotype of both ∆pho88 and ∆ski8 strains (Fig. 2C). Sigma ∆pho88 clones with native mitochondria containing the dsRNA virus grew faster than clones that were identical but had S288c mitochondria . This growth difference for ∆pho88 was not present in a strain lacking the dsRNA virus. In contrast, a comparison of Sigma ∆ski8 strains containing their own mitochondria with those with S288c mitochondria showed that the mitochondria from S288c enhanced the growth of ∆ski8 in the presence or absence of the dsRNA virus. Under these culture conditions none of the other mutations tested showed an influence of the mitochondrial genome on growth.
The observed differences in colony size show that the mitochondrial genome can affect not only the phenotypic consequence of a chromosomal mutation but also that independent nonchromosomal elements (dsRNA virus and mitochondrial DNA) can interact to affect the phenotype of a chromosomal mutation.
The chromosomal and nonchromosomal interactions that we observe that affect cellular growth require the specific ensemble of polymorphisms intrinsic to the Sigma genome. None of the gene deletions we examined causes a severe growth defect in S288c, with or without the dsRNA virus or, of course, with its own S288c mitochondria (Fig. S1). Therefore, the phenotypic consequence of a mutation in Sigma depends upon the complex interplay between at least three sources of information: the presence of the chromosomal mutation itself, its interactions with strain-specific polymorphisms, and nonchromosomal elements.
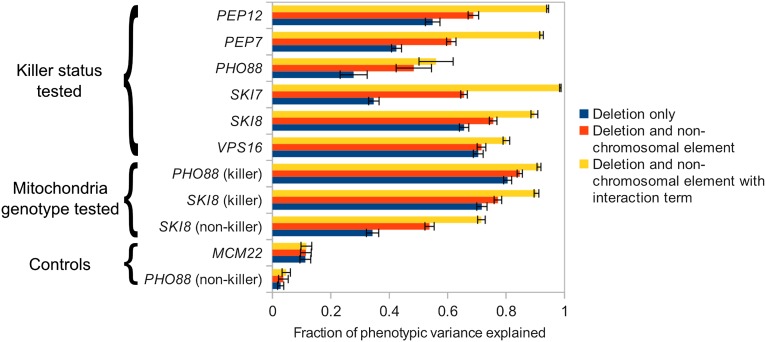
To understand the implications of our findings for trait mapping studies and related applications, we sought to interpret our results from a quantitative perspective using the lens of heritability. We used automated image analysis to measure colony size from replicates of defined chromosomal and nonchromosomal genotypes (Methods). Fig. S2 shows an illustrative example for one growth trait of computing the fraction of the phenotypic variance accounted for by models that (i) consider only the chromosomal mutation and ignore nonchromosomal information, (ii) consider the effect of the chromosomal mutation and the nonchromosomal element assuming no interaction between the two, and (iii) consider the effect of chromosomal mutation, the nonchromosomal element, and their nonadditive interaction. Across almost all traits we found that a large fraction of the heritability of colony size in a Sigma-only background is attributable to nonchromosomal modifiers, along with their interactions with single gene deletions. Fig. 3 shows the heritability results for all analyzed growth traits. Interaction effects contribute substantially to the heritability of colony size (Table S3) and represent nonadditive terms that would not be adequately modeled using conventional linear models.
Fig. 3.
Nonchromosomal elements explain an increased fraction of heritability. Model fit calculations were applied to data from 10 gene deletion traits, as described in Methods and illustrated in Fig. S2. The coefficient of determination (R2), the fraction of observed variance explained by the model, was used as the metric of recovered heritability. Error bars were calculated using 10-fold cross-validation, where 1/10 of the data are reserved for unbiased evaluation while the model is trained on the remaining points. The error bars show the SD across the 10 sampled test sets. Aside from the control MCM22 and the PHO88 nonkiller experiment, all experiments showed a strong increase in modeling accuracy by including nonchromosomal effects and interaction terms (Table S3).
Discussion
Our study shows that the phenotype of a mutation may be modified by inherited viral state and by nonchromosomal elements such as mitochondria that are transmitted through germ-line meiosis. We have further shown that the heritability of a trait can depend on nonlinear interactions between chromosomal and nonchromosomal information that is transmitted from generation to generation. The nonchromosomal information can interact with the various chromosomal alleles at a locus to modify phenotype significantly. Our ability to vary the nonchromosomal information while keeping the chromosomal constitution and environment constant permits an accurate measurement of the nonchromosomal contribution to heritability. Our results show that the nonchromosomal contribution to heritability can be large and, in some cases, can completely mask the effect of a chromosomal mutation. Nonchromosomal elements may have affected previous yeast studies (28–32) that crossed a strain carrying a dsRNA virus, as many feral yeast strains do (33), with a virus-free strain such as the reference strain S288c (34) (Supporting Information, Note 1).
Previous yeast studies analyzing the basis of quantitative traits (quantitative trait locus mapping) have either not carefully controlled nonchromosomal modifiers or have fixed them so that their influence is eliminated. Our results complement one such study that recovered the chromosomal determinants accounting for almost all of the additive portion of heritability of several traits by dramatically expanding study sizes (4). In this previous study, potentially confounding nonchromosomal effects were mitigated by standardizing on a single mitochondrial background and by using only dsRNA virus-free strains (Supporting Information, Note 2, Fig. S3, and Table S4). The control of nonchromosomal factors in model organism experiments and the inability to do so in “wild” human populations could account for part of the recent success gap between model- and human-focused genetic studies.
Our findings on the relative ubiquity of nonchromosomal genetic effects have profound implications for the association between disease susceptibility and genetic variation in humans. For the viral interaction case, these elements are not currently captured by genotyping assays, and therefore current studies cannot measure their impact. However, they may be inherited or manifest as a shared environmental factor. Both cases could contribute to the complexity of modeling disease heritability. The inclusion of nonchromosomal interactions adds another dimension to the estimation of heritability in wild populations and susceptibility to common diseases in humans.
Methods
Strain Construction.
A list of originating strains used in this paper is given in Table S5. Specific gene deletion strains created from this set of parental strains are not named in the table.
Following the strategy outlined in Fig. 1, we constructed strains that varied in chromosomal and nonchromosomal genotypes. Because the selected gene deletion mutations had growth defects in Sigma they had to be constructed in a diploid Sigma strain by transformation. This Sigma diploid contained the killer virus dsRNA and, for comparison, a nonkiller diploid lacking killer virus dsRNA was derived as a spontaneous mitotic segregant (∼1%) from that original Sigma strain. The S288c [kil-k] strain was isolated by cytoduction using the kar1-∆13 mutant. In crosses between a kar1-∆13 Sigma [kil-k] strain and an S288c [kil-0] strain where the S288c nucleus was marked with canavanine resistance marker, haploid S288c [kil-k] cytoductants can be easily isolated. Deletion mutations were introduced by transformation of the parental diploid strains. We verified the absence of dsRNA in the nonkiller segregants by gel electrophoresis, Petri plate tests, and high-throughput RNA sequencing (Fig. S4 and Supporting Information, Note 1). This procedure, which used the same original diploid parental strain or mitotic segregants of that strain (Fig. S5), ensured that the chromosomal genetic background was the same in the strain with dsRNA and the one without dsRNA. The two diploids, killer and nonkiller, both heterozygous for a given mutation, were then induced to undergo meiosis and the resulting meiotic spores were separated and grown at 30 °C. For a graphical overview of the procedure used to construct the Sigma killer comparison strains, see Fig. S5.
We also created strains with two different mitochondrial backgrounds with the same nuclear genomic information from Sigma (Fig. S5) (22). Mitochondrial DNA in yeast is designated [rho+], and we designed experiments that used either S288c mitochondria or Sigma mitochondria . Construction of a Sigma strain with was made possible by using an S288c kar1- mutant, which permits transfer of mitochondria to a Sigma strain lacking mitochondrial DNA [rho0] by conjugation without nuclear fusion. We derived an isogenic haploid canr Sigma strain lacking mitochondrial DNA ([rho0]) by growing the original Sigma strain used above in EtBr. The resulting Sigma exconjugant is a cell with the Sigma nuclear genome and mitochondria (Fig. S6). We verified the mitochondrial genotype of the and strains using PCR primers that targeted strain-specific DNA sequences in the two mitochondrial genomes. These diploids were transformed with the same deletions used in the dsRNA comparison and the two diploids heterozygous for the deletion, and either Sigma or Sigma , were dissected on the same plate and the size of the spore clones determined. For a graphical overview of the procedure used to construct the mitochondrial strains, see Fig. S5.
The growth phenotype of the deletion in +/∆ski7 (two viable:two nonviable observed in each tetrad) was not due to a closely linked mutation because replacement of the deletion in the +/∆ski7 (kil-k) strain with the wild-type SKI7 gene by transformation restored viability (4:0 viable to nonviable).
Quantitative Analysis of Growth Phenotypes and Heritability.
Images of dissection plates were analyzed with the software package ImageJ to yield measurements of colony size. Table S2 gives a summary of the colonies analyzed for each controlled genotype. A variance-stabilizing Box–Cox transform was applied to all raw colony area measurements (pixel counts). The Box–Cox exponent was 0.25, which was chosen to give the least-correlated means and variances in the analyzed set of experiments.
The joint modeling was performed by a two-way ANOVA implemented with the lm method in R version 3.0.1. Concretely, the three considered models were
In each case, the “effect” model terms and the constant were selected to minimize the squared noise term observed when comparing model predictions to a set of observed phenotypes (that is, least squares regression).
Significance tests were computed using F statistics with the anova.lm function and results are reported in Table S3. Fig. S2 gives a graphical overview of the analysis strategy and its implementation for the killer virus interacting with ∆pep7.
Recovered heritability as shown in Fig. 3 is obtained by comparing the squared differences between the model predictions and the observed data to the squared differences between all points and the mean (the total observed variance). Predictions were assessed using held-out samples (1/10 of the data) that were not used to train the model.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM035010 (to G.R.F.), the Qatar Computing Research Institute (D.K.G.), and a National Science Foundation Graduate Research Fellowship under Grant 0645960 (to M.D.E.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407126111/-/DCSupplemental.
References
- 1.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era—concepts and misconceptions. Nat Rev Genet. 2008;9(4):255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 2.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eichler EE, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11(6):446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloom JS, Ehrenreich IM, Loo WT, Lite T-LV, Kruglyak L. Finding the sources of missing heritability in a yeast cross. Nature. 2013;494(7436):234–237. doi: 10.1038/nature11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci USA. 2012;109(4):1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182(3):845–850. doi: 10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nadeau JH. Transgenerational genetic effects on phenotypic variation and disease risk. Hum Mol Genet. 2009;18(R2):R202–R210. doi: 10.1093/hmg/ddp366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rassoulzadegan M, et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441(7092):469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 9.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141(7):1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joseph B, Corwin JA, Li B, Atwell S, Kliebenstein DJ. Cytoplasmic genetic variation and extensive cytonuclear interactions influence natural variation in the metabolome. eLife. 2013;2:e00776. doi: 10.7554/eLife.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Hera B, et al. Role of the human endogenous retrovirus HERV-K18 in autoimmune disease susceptibility: Study in the Spanish population and meta-analysis. PLoS ONE. 2013;8(4):e62090. doi: 10.1371/journal.pone.0062090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nexø BA, et al. The etiology of multiple sclerosis: Genetic evidence for the involvement of the human endogenous retrovirus HERV-Fc1. PLoS ONE. 2011;6(2):e16652. doi: 10.1371/journal.pone.0016652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nissen KK, et al. Endogenous retroviruses and multiple sclerosis-new pieces to the puzzle. BMC Neurol. 2013;13:111. doi: 10.1186/1471-2377-13-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Man PYW, et al. The epidemiology of Leber hereditary optic neuropathy in the North East of England. Am J Hum Genet. 2003;72(2):333–339. doi: 10.1086/346066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prezant TR, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4(3):289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 16.Magliani W, Conti S, Gerloni M, Bertolotti D, Polonelli L. Yeast killer systems. Clin Microbiol Rev. 1997;10(3):369–400. doi: 10.1128/cmr.10.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt MJ, Breinig F. Yeast viral killer toxins: Lethality and self-protection. Nat Rev Microbiol. 2006;4(3):212–221. doi: 10.1038/nrmicro1347. [DOI] [PubMed] [Google Scholar]
- 18.Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol. 2003;4(11):878–890. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- 19.Halfmann R, et al. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482(7385):363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunge N. Yeast DNA plasmids. Annu Rev Microbiol. 1983;37:253–276. doi: 10.1146/annurev.mi.37.100183.001345. [DOI] [PubMed] [Google Scholar]
- 21.Chen XL, Reindle A, Johnson ES. Misregulation of 2 micron circle copy number in a SUMO pathway mutant. Mol Cell Biol. 2005;25(10):4311–4320. doi: 10.1128/MCB.25.10.4311-4320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conde J, Fink GR. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci USA. 1976;73(10):3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowell RD, et al. Genotype to phenotype: A complex problem. Science. 2010;328(5977):469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458(7236):337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458(7236):342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagé N, et al. A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics. 2003;163(3):875–894. doi: 10.1093/genetics/163.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wickner RB. Double-stranded and single-stranded RNA viruses of Saccharomyces cerevisiae. Annu Rev Microbiol. 1992;46:347–375. doi: 10.1146/annurev.mi.46.100192.002023. [DOI] [PubMed] [Google Scholar]
- 28.Sinha H, et al. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics. 2008;180(3):1661–1670. doi: 10.1534/genetics.108.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinmetz LM, et al. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 2002;416(6878):326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 30.Ben-Ari G, et al. Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet. 2006;2(11):e195. doi: 10.1371/journal.pgen.0020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deutschbauer AM, Davis RW. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat Genet. 2005;37(12):1333–1340. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- 32.Kim HS, Fay JC. A combined-cross analysis reveals genes with drug-specific and background-dependent effects on drug sensitivity in Saccharomyces cerevisiae. Genetics. 2009;183(3):1141–1151. doi: 10.1534/genetics.109.108068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333(6049):1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fink GR, Styles CA. Curing of a killer factor in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1972;69(10):2846–2849. doi: 10.1073/pnas.69.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.