Significance
In flowering plants, double fertilization gives rise to an embryo and the endosperm, an absorptive storage structure that supports embryogenesis and seedling germination. In cereal grains, endosperm comprises a large proportion of the mature seed, contains large amounts of carbohydrates and proteins, and is an important source of food, feed, and industrial raw materials. This study provides a comprehensive profile of the genes expressed in the early developing endosperm in maize. We also show how a series of temporal programs of gene expression correlate with progressive functional and cellular specializations.
Keywords: mRNA localization, time series
Abstract
Endosperm is a filial structure resulting from a second fertilization event in angiosperms. As an absorptive storage organ, endosperm plays an essential role in support of embryo development and seedling germination. The accumulation of carbohydrate and protein storage products in cereal endosperm provides humanity with a major portion of its food, feed, and renewable resources. Little is known regarding the regulatory gene networks controlling endosperm proliferation and differentiation. As a first step toward understanding these networks, we profiled all mRNAs in the maize kernel and endosperm at eight successive stages during the first 12 d after pollination. Analysis of these gene sets identified temporal programs of gene expression, including hundreds of transcription-factor genes. We found a close correlation of the sequentially expressed gene sets with distinct cellular and metabolic programs in distinct compartments of the developing endosperm. The results constitute a preliminary atlas of spatiotemporal patterns of endosperm gene expression in support of future efforts for understanding the underlying mechanisms that control seed yield and quality.
Endosperm is a biologically and economically important storage tissue within the angiosperm seed that provides nutrients and signals to the embryo during seed development. In cereal grains such as maize, endosperm comprises a large proportion of the mature seed and contains large amounts of carbohydrates and proteins that are metabolized during germination to provide nutrients for the developing seedling. These carbohydrates and proteins are also an important source of food for humankind. Approximately 50% of human calories are derived from cereal endosperm, either directly or indirectly, through animal feed. Cereal endosperm is also used as a raw material for numerous industrial products, including ethanol (1–4).
Maize endosperm development is initiated following double fertilization of the female gametophyte’s haploid egg cell and diploid central cell, which gives rise to the seed’s diploid embryo and triploid endosperm, respectively (2, 3). During 0–3 days after pollination (DAP), endosperm nuclei undergo mitosis without cytokinesis, producing a multinucleate cell, a coenocyte. At the end of this phase, the endosperm consists of 256–512 nuclei that surround a large central vacuole (1–4). At 3–4 DAP, the endosperm cellularizes and undergoes an intense period of cell division that lasts until 8–12 DAP in the central region (early mitotic phase), but continues until ∼20–25 DAP in the outer two cell layers that will become the aleurone and the subaleurone (3–5).
During 4–6 DAP, the endosperm differentiates into four major cell types: the starchy endosperm (SE), the basal endosperm transfer layer (BETL), the aleurone (AL), and the embryo-surrounding region (ESR) (3). The SE is the cell type that accumulates starch and proteins. AL cells are activated during germination and produce hydrolases that degrade the starch and proteins in SE cells. The BETL transports nutrients from the maternal tissue to the endosperm. ESR function is unclear, but this cell type appears to be involved in signaling to and/or in pathogenic defense of the developing embryo (3).
Over the past several decades, much progress has been made in understanding the physiological processes occurring during maize endosperm maturation (e.g., storage product synthesis and accumulation), which initiates at ∼10 DAP. By contrast, far less research has been devoted to understanding early endosperm development (0–10 DAP). Key developmental processes occur during this period, including coenocytic development, cellularization, cellular differentiation, and the early mitotic phase. These phases are important steps in endosperm development in cereals and most other species, and understanding these processes is central to explaining endosperm structure and function.
Comprehension of the regulation of early endosperm development will ultimately require a full understanding of the gene networks operating during this time. To this end, we have used next-generation sequencing technology to profile all of the genes expressed during early development in maize kernels and hand-dissected endosperm. As a first step toward gene network analysis, we subdivided these sequences into clusters of coexpressed genes based on temporal expression patterns. The major temporal programs identified were correlated with specific spatial programs of expression in one or more compartments or tissue types of the developing endosperm. Many of these spatiotemporal programs of gene expression correlate with expression of 399 transcription-factor genes expressed during early maize kernel and endosperm development identified in our assays. The results provide a preliminary atlas of temporal and spatial gene expression patterns to begin to decipher gene networks active in endosperm development and ultimately provide fundamental insight into the underlying mechanisms that determine seed yield and quality.
Results
Transcriptome Sequencing of Early Stages of Kernel and Endosperm Development.
To obtain a global transcriptomic map of early kernel and endosperm development in maize, we selected five stages of whole kernels (0, 2, 3, 4, and 6 DAP) and three stages of isolated endosperm (8, 10, and 12 DAP) of the B73 inbred line (Fig. S1) for RNA-Seq analysis.
Using a SOLiD sequencing platform, between ∼40 and ∼90 million 50-nt reads were generated for each RNA sample (Fig. S2A). Read quality was examined by FastQC software (www.bioinformatics.babraham.ac.uk/projects/fastqc/), and, on average, ∼5.9% of low-quality reads for each sample were removed before mapping. The retained high-quality reads were mapped to the reference B73 genome (release 5b.60) by Bowtie (6); between ∼30.9% and 49.7% of the reads from each sample mapped to the genome with no more than five aligned positions, among which ∼31% of the RNA-Seq reads mapped to a unique position in the genome (Fig. S2B). Visualization of reads for known genes expressed in the kernel or endosperm confirmed RNA quality (Fig. S2C). Therefore, our RNA-Seq data appear to represent the complexity of gene expression programs within the early developing kernel and endosperm.
Early Maize Kernel and Endosperm Transcriptomes Are Complex and Overlapping.
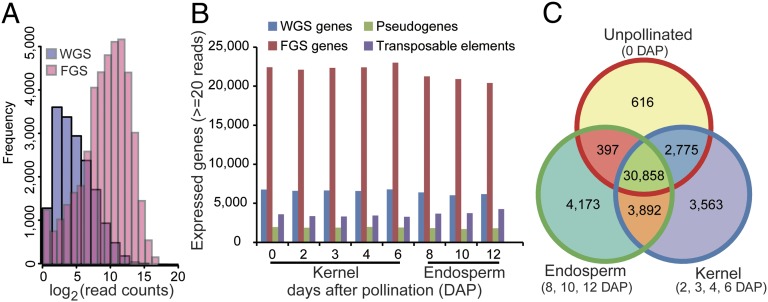
Using TopHat (7), we initially determined mRNA abundance based on the 110,028 and 39,656 gene models annotated in the working gene set (WGS) and the filtered gene set (FGS), respectively, of the maize B73 genome (Fig. 1A). Because genes in the FGS were further filtered from the WGS using in silico-based characteristics (to exclude pseudogenes, transposable elements, and partial genes), the WGS likely contains bona fide genes that were not included in the FGS. Therefore, we determined the distribution of read counts from genes in the two sets and found that the FGS genes produce, on average, ∼2,000 reads, whereas only a small number of genes (8,701) in the WGS were associated with reads higher than 100 (Fig. 1A). In order not to overlook any bona fide non-FGS genes in our analysis, we added the additional 8,701 expressed WGS genes with read counts greater than 100 to the 39,656 genes in the FGS to produce a combined gene set of 48,357 genes that was used for normalization by applying edgeR (Fig. S3) (8).
Fig. 1.
Numbers of genes expressed during early maize kernel and endosperm development. (A) Frequency of the genes from the WGS and the FGS based on mRNA levels. (B) Numbers of expressed WGS genes, FGS genes, pseudogenes, and TE-related genes in the eight developmental stages. (C) Venn diagram of the numbers of expressed genes in unpollinated kernels, early developing kernels (2–6 DAP), and endosperm stages (8–12 DAP).
Using quantitative RT-PCR (qRT-PCR) analysis of a group of candidate genes, we established 20 reads as a cutoff to determine the number of expressed genes across the eight developmental samples (SI Materials and Methods). By this criterion, 29,124 genes, on average, were classified as expressed in the five kernel developmental stages, and 27,050 genes were classified as expressed in the three endosperm developmental stages (Fig. 1B). We detected a small fraction of transcripts derived from transposable elements (TEs) annotated in release 5b.60, with an average of 3,392 expressed TEs in kernel samples and an average of 3,891 expressed TEs in the three endosperm samples (Fig. 1B). This observation is consistent with previous reports showing the endosperm genome is hypomethylated and that many TE-encoding loci are transcriptionally active (9, 10).
We found that 46,274 genes were expressed in at least one sample (Dataset S1). Of these, 30,858 genes (66.7%) produced mRNAs detected in all samples (Fig. 1C), whereas 15,416 genes (33.3%) were expressed only during specific stages of endosperm/kernel development analyzed in this study. Among the nine investigated stages, the unpollinated kernel (0 DAP) contained the smallest number of mRNAs (616) specifically detected in this stage, whereas 3,563 and 4,173 mRNAs were detected only in the developing kernel and endosperm samples, respectively (Fig. 1C). Our data indicate that the early developing kernel and endosperm express a highly complex transcriptome.
Identification of Temporally Up- and Down-Regulated Genes in Early Kernel and Endosperm Development.
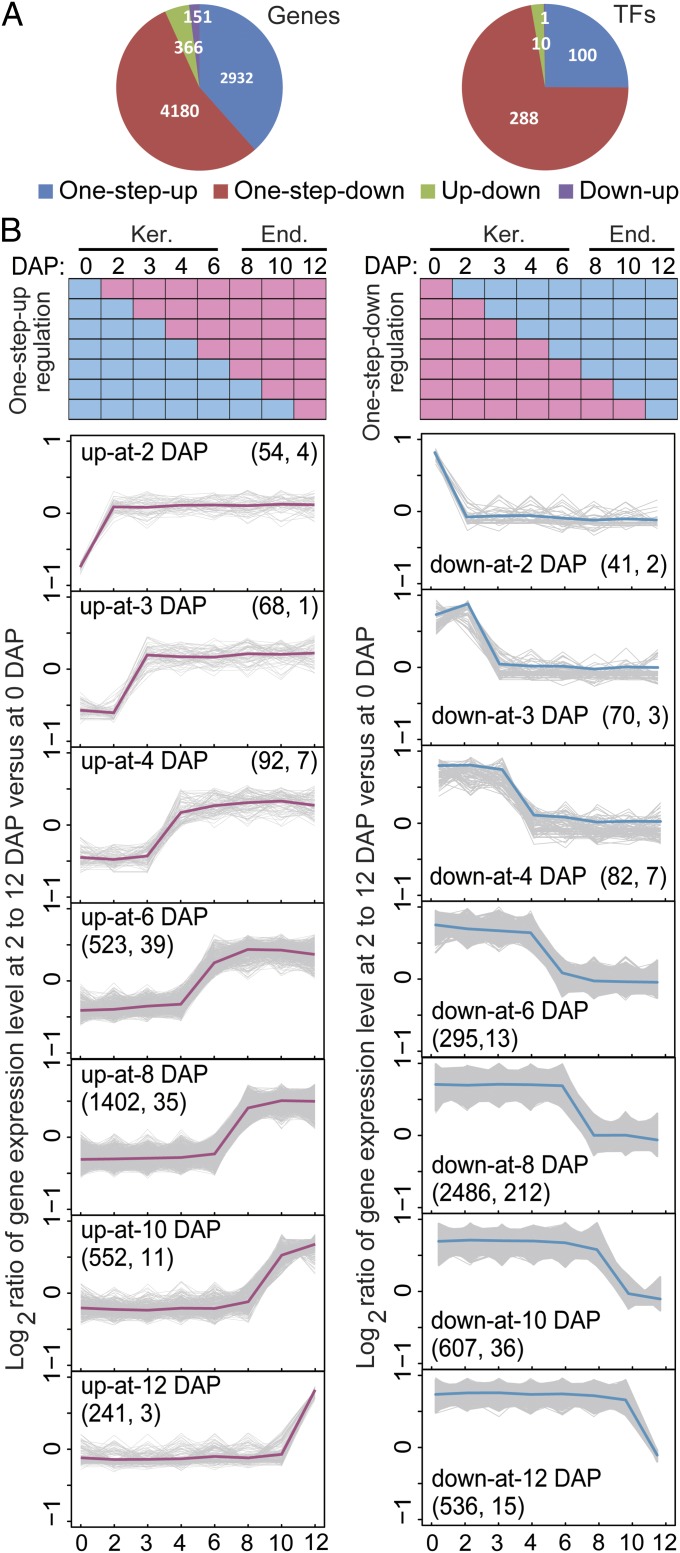
We used StepMiner analysis (11) to characterize the temporal expression patterns of endosperm/kernel-expressed genes. This algorithm is relatively more robust compared with other tools in identifying one-step transitions from a time course with limited data points (11). This analysis identified 7,629 genes (Dataset S2, Table S1) with one or two transition points in expression during the 0–12 DAP window (Fig. 2 A and B and Fig. S4A).
Fig. 2.
Identification of up- and down-regulated gene sets and the coexpressed TFs. (A) Numbers of mRNAs for all genes and the coexpressed TFs showing transitions from low to high (one-step-up) or high to low (one-step-down) in two consecutive developmental stages and two-step-up-down (up-down) or two-step-down-up (down-up) in our series of developmental stages. (B) Identification of the one-step-up (Left) and one-step-down (Right) transitions in mRNA levels for all genes and the coexpressed TFs during early kernel development. Seven expression patterns were identified to represent genes up- or down-regulated at 2, 3, 4, 6, 8, 10, and 12 DAP (Top). Gene expression profiles of individual genes are depicted in gray lines, and average expression profiles for each pattern are depicted in pink (one-step-up) or blue (one-step-down) lines. The total number of all genes (Left) and the coexpressed transcription-factor genes (Right) are indicated in parentheses for each expression pattern. End., endosperm; Ker., kernel.
StepMiner analysis identified four general temporal gene-expression patterns that we refer to as one-step-up or one-step-down (mRNA level transitions from low to high or high to low, respectively, in two consecutive developmental stages) and two-step-up-down or two-step-down-up (mRNA level transitions from low to high and back down or from high to low and back up, respectively, in a series of developmental stages). The majority of the identified genes [93.2%, including 97.2% of transcription factor (TF) genes] showed a single transition point; of these, 38.4% exhibited the one-step-up pattern, whereas 54.8% showed the one-step-down pattern. By contrast, only 6.8% of genes exhibited two transition points (two-step-up-down and two-step-down-up genes) (Fig. 2A).
We further classified the genes based on the time point during which the major expression transition occurs (Fig. 2B). For example, “up-at-6 DAP” genes showed lower expression at 0–4 DAP and higher expression during 6–12 DAP, and “up-at-8 DAP” genes showed lower expression at 0–6 DAP and higher expression during 8–12 DAP. With genes showing a single transition point (i.e., one-step-up and one-step-down genes), the transition in expression level occurred more frequently at 6–12 DAP compared with earlier time points, indicating a dramatic reprogramming of the kernel transcriptome occurs after 4 DAP (Fig. 2B).
Endosperm cell differentiation occurs during 4–6 DAP (12–14), suggesting that genes in the up-at-4-DAP and up-at-6-DAP groups may be associated with development of the AL, BETL, ESR, and SE. Accordingly, the up-at-4-DAP and up-at-6-DAP groups contain a number of previously described genes that are expressed in specific endosperm cell types, including 14 BETL-expressed genes [MRP-1 and 13 genes encoding maternally expressed genes (MEGs), BETLs, and basal layer antifungal peptides (BAPs)] (15–18) and two ESR-expressed genes (ESR1 and ESR2) (19, 20). Storage protein and carbohydrate accumulation in the SE begins during 8–12 DAP (21, 22). Consistent with this, genes up-regulated at 8 DAP include those previously shown to be involved in storage protein synthesis in the SE, including the transcription-factor genes Opaque-2/ZmbZIP1 and Prolamin-box Binding Factor (PBF)/ZmDOF3 (23, 24) and multiple genes encoding storage proteins including 11 α-, three γ-, one β-, and one δ-zein (25–28). These results indicated that our data set provides sufficient temporal resolution to distinguish genes that function at different stages of maize endosperm development.
A stringent Gene Ontology (GO) term enrichment analysis (29) of these unique expression patterns identified key biological processes showing significant enrichment at three developmental stages. These included defense response and translation processes among genes up-regulated at 6 DAP, amino acid biosynthesis and translation among genes up-regulated at 8 DAP, and carbohydrate biosynthesis and energy reserve metabolic processes among genes up-regulated at 10 DAP (Fig. S4B and Dataset S3). These data indicate that gene expression programs in early developing kernel and endosperm change dramatically starting at 6 DAP in preparation for the onset of storage protein and starch production.
Identification of Temporally Up-Regulated Transcription-Factor Genes During Early Endosperm Development.
To begin to understand the nature of the regulatory processes during early kernel and endosperm development, we focused on TF genes exhibiting a one-step-up pattern (Fig. 2B and Fig. S4). We selected this group because they are definitively expressed in the endosperm based on their relatively high expression levels in one or more endosperm stages (8–12 DAP). Furthermore, genes associated with specific developmental processes (e.g., cell differentiation) would be expected to be activated at specific time points during the developmental progression.
Collectively, 100 TF genes fell into the one-step-up groups (Fig. 2B; Dataset S2, Tables S1 and S2). Of these, most were in the up-at-6-DAP (39%) and up-at-8-DAP (35%) groups (Fig. 2B; Dataset S2, Table S2). These results suggest that the specific patterns of gene expression detected during early kernel development are associated with TF gene up-regulation.
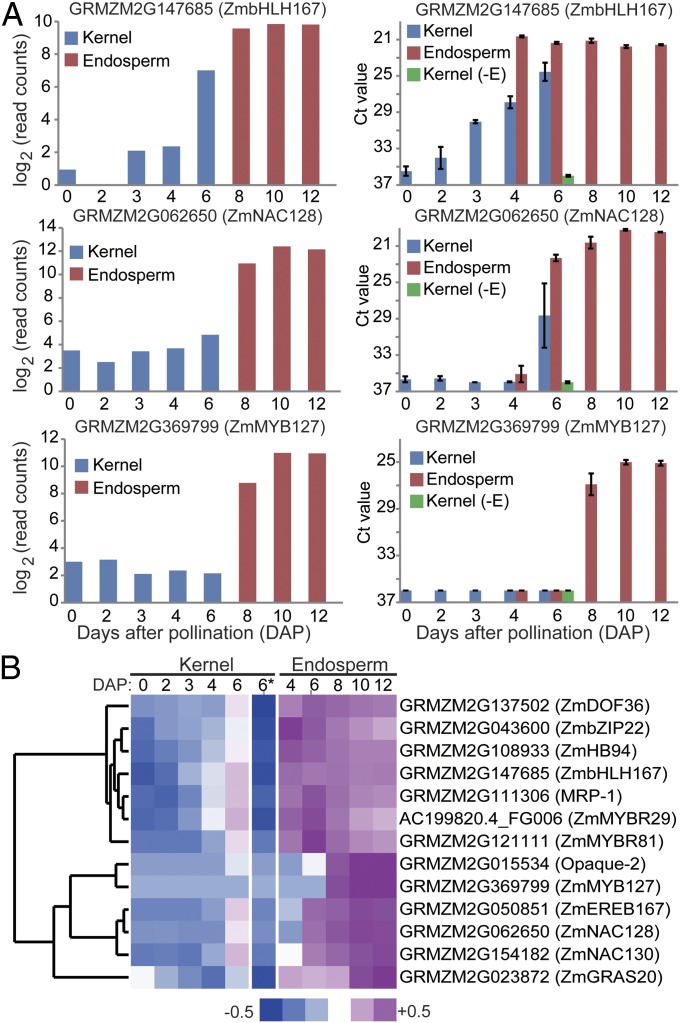
To confirm the dynamics of TF gene up-regulation within developing endosperm, we performed qRT-PCR analysis of 13 TF genes (Fig. 3; Dataset S4, Table S1). For these experiments, we also assayed mRNA levels in endosperm isolated from 4- and 6-DAP kernels and from 6-DAP kernels with the endosperm removed. With all 13 genes, the qRT-PCR patterns matched the RNA-Seq patterns. Moreover, these genes showed high specificity of expression in the endosperm (Fig. 3A). The qRT-PCR results suggest that most, if not all, of the TF genes exhibit the temporal patterns identified by StepMiner analysis. Clustering of the qRT-PCR data identified two temporal programs: one gene set up-regulated in 4-DAP endosperm and subsequently down-regulated (7 genes) and a second set exhibiting delayed up-regulation starting in 6- or 8-DAP endosperm and maintaining a persistently high level of mRNA through 12 DAP (Fig. 3B). This suggested a finely controlled temporal program of TF gene expression that may underlie key developmental programs during 4–8 DAP. In fact, the MRP-1 TF gene involved in regulation of BETL differentiation (15) clustered with the former set, whereas Opaque-2, which is known to control storage-protein gene expression in the SE, clustered in the latter set (Fig. 3B). Together, the RNA-Seq analysis of temporal stages of kernel and endosperm development helped identify coexpressed gene sets with associated up-regulated TF genes that could represent regulatory nodes for spatiotemporal control of endosperm development.
Fig. 3.
Quantitive RT-PCR validation of TFs preferentially expressed in endosperm. (A) The mRNA levels of three TFs based on RNA-Seq (Left) and qRT-PCR (Right) data. Kernel (−E) denotes the kernel at 6 DAP with the endosperm removed. (B) Heatmap of qRT-PCR data for 13 TF genes (including 11 randomly chosen showing an up-at-6/8-DAP pattern and for two known endosperm-expressed TF genes, MRP-1 and Opaque-2) showing preferential expression in the endosperm clustered based on the Ct values of qRT-PCR of three bio-reps of 0, 2, 3, 4, and 6 DAP kernels; 4, 6, 8, 10, and 12 DAP hand-dissected endosperm; and 6-DAP kernel with the endosperm removed (−E, denoted by 6*).
Correlation of Temporal and Spatial Programs of Gene Expression in Early Kernel and Endosperm.
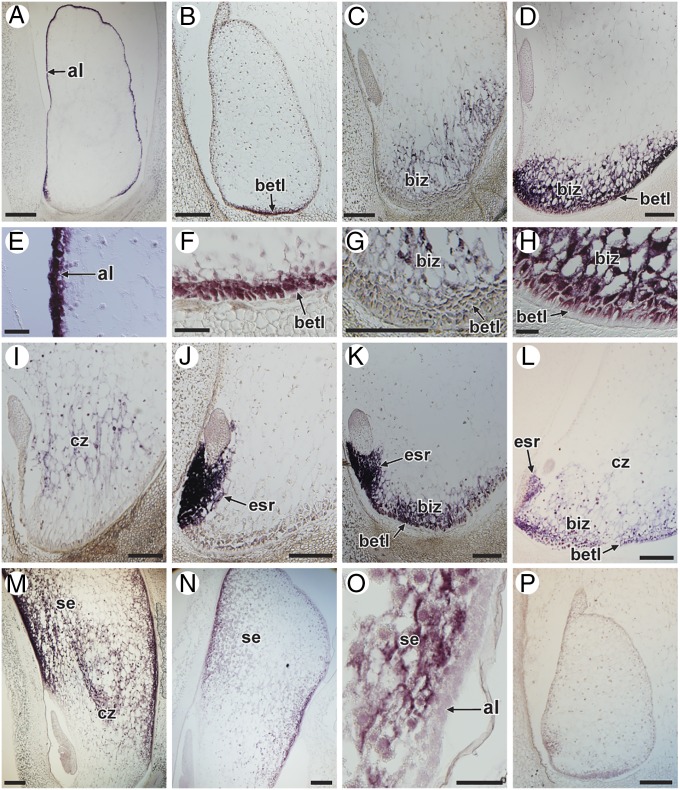
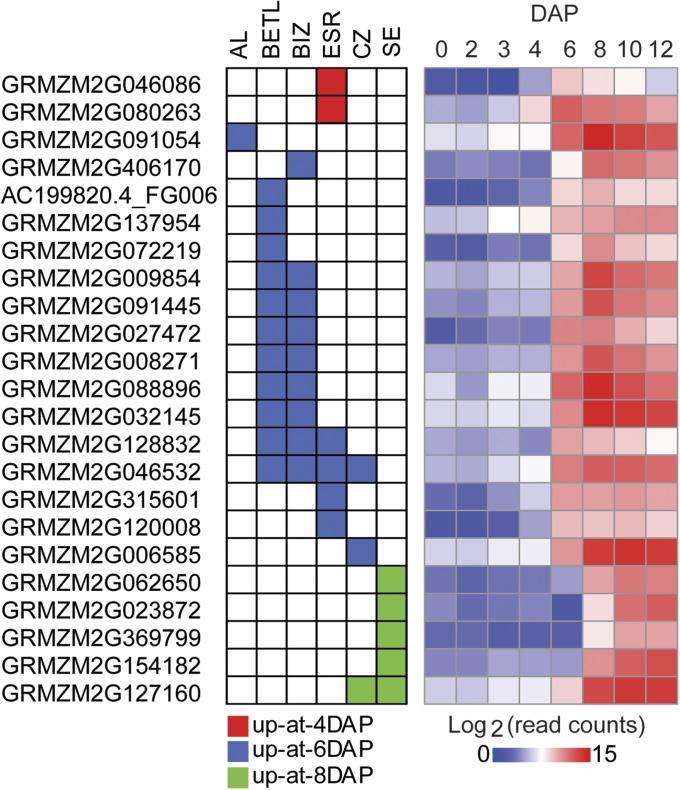
To further validate endosperm expression, we performed in situ localization studies with 23 genes, including 6 TF genes (ZmGRAS20, ZmMYBR29, ZmMYBR127, ZmNAC128, ZmNAC130, and ZmPLATZ12). All of the selected genes showed a distinct mRNA localization pattern. Most were expressed in the endosperm in just one (14 genes) or two (7 genes) cell types (Figs. 4 and 5 and Fig. S5). Most of these genes were expressed in the previously described endosperm cell types (i.e., AL, BETL, ESR, and SE) including the conducting zone (CZ), a region of elongated cells overlying the basal cells, which presumably act as a conducting tissue for the developing endosperm (30–32). However, one gene (GRMZM2G406170) was expressed specifically in an endosperm cell type not previously described (Figs. 4C and 5). This cell type lies between the BETL and the CZ, and we refer to it as the basal-intermediate zone (BIZ). Nine additional genes showed expression in the BIZ, in combination with other cell types, particularly the BETL (Fig. 4 D and K; Fig. 5; Fig. S5).
Fig. 4.
Cell-specific expression of endosperm genes as determined by in situ hybridization. (A) Expression in the AL only. Section of an 8-DAP kernel hybridized with a probe for GRMZM2G091054 (AL9). (B) Expression in the BETL only. Section of a 6-DAP kernel hybridized with a probe for AC199820.4_FG006 (ZmMYBR29). (C) Expression in the BIZ only. Section of an 8-DAP kernel hybridized with a probe for GRMZM2G406170 (BURP domain-containing). (D) Expression in the BETL and BIZ. Section of an 8-DAP kernel hybridized with a probe for GRMZM2G088896 (uncharacterized). (E) Close-up of the image in A showing expression localized to the AL. (F) Close-up of the image in B showing expression localized to the BETL. (G) Close-up of the image in C showing expression localized to the BIZ. (H) Close-up of the image in D showing expression localized in the BETL and BIZ. (I) Expression in the CZ only. Section of a 7-DAP kernel hybridized with a probe for GRMZM2G006585 (ZmPLATZ12). (J) Expression in the ESR only. Section of a 7-DAP kernel hybridized with a probe for GRMZM2G120008 (uncharacterized). (K) Expression in the ESR, BETL, and BIZ. Section of a 7-DAP kernel hybridized with a probe for GRMZM2G128832 (uncharacterized). (L) Expression in the ESR, BETL, BIZ, and CZ. Section of a 7-DAP kernel hybridized with a probe for GRMZM2G046532 (uncharacterized). (M) Expression in the SE and CZ. Section of a 10-DAP kernel hybridized with a probe for GRMZM2G127160 (tryptophan aminotransferase). (N) Expression in the SE only. Section of a 10-DAP kernel hybridized with a probe for GRMZM2G023872 (ZmGRAS20). (O) Close-up of the image in N. (P) Negative control. Section of a 6-DAP kernel hybridized with a probe for GRMZM2G453555 (transposable element), which does not produce a signal. (Scale bars: 2 mm for A, B, C, D, G, I, J, K, L, M, N, and P; 500 µm for E, F, H, O). al, aleurone; betl, basal endosperm transfer layer; biz, basal intermediate zone; cz, conducting zone; emb, embryo; esr, embryo surrounding region; se, starchy endosperm.
Fig. 5.
Correlation of the temporal and spatial patterns of individual genes expressed during early kernel and endosperm development. Spatial patterns of mRNA accumulation were identified based on in situ hybridizations in this study (graph on Left) compared with the temporal accumulation of the same mRNAs based on our RNA-Seq data (heat map on Right; 0–6 DAP from kernel and 8–12 DAP from hand-dissected endosperm). Individual gene IDs are shown on the left.
Overall, the genes analyzed exhibited 10 different expression patterns: SE-specific (four genes); ESR-specific (four genes); BETL-specific (three genes); AL-specific (one gene); BIZ-specific (one gene); CZ-specific (one gene); BETL and BIZ (6 genes); SE and CZ (one gene); BETL, BIZ, and ESR (one gene); and BETL, BIZ, CZ, and ESR (one gene) (Fig. 5 and Fig. S5). The two genes in the up-at-4-DAP group were expressed in the ESR. All of the genes in the up-at-8-DAP group were expressed in the SE specifically (four genes) or the SE plus CZ (one gene) (Fig. 5 and Fig. S5). Therefore, our findings indicate that the specific temporal programs of gene expression identified from analysis of the RNA-Seq data correspond essentially to a series of spatial programs of gene expression occurring during the differentiation of endosperm cell types or compartments.
Discussion
We carried out a transcriptome analysis of early kernel and endosperm in maize. Related analyses of grain endosperm have been performed previously in wheat, rice, oat, and maize (33–37). However, unlike the prior studies, our analysis focused on early developmental processes, which impact important kernel agronomic traits, including seed size and grain weight (38–40). In addition, our analysis used successive stages of development to determine the temporal expression pattern of each identified gene.
Our analysis has identified nearly 34,000 mRNAs (reads ≥20) in the 0- to 6-DAP period of kernel development and nearly an equivalent number of mRNAs (∼33,000, reads ≥20) in endosperm samples collected from kernels at 8–12 DAP (Fig. 1 and Dataset S1). These numbers suggest that the sequencing of kernel and endosperm RNA samples was sufficiently thorough to capture nearly all of the genes expressed in these seed structures. These numbers are similar to those in a recent profiling of maize organ and tissue mRNAs using RNA-Seq analysis (41) (Dataset S4, Table S3). Our measured number of expressed maize genes is significantly higher than the ∼9,000–14,000 mRNAs reported for early Arabidopsis seeds (42). This is likely due to the differences in profiling platforms (GeneChip vs. RNA-Seq). We also detected >97.5% of the endosperm proteins at 8, 10, and 12 DAP (Dataset S4, Table S3) in a recent study of maize seed proteotypes (43). However, this study found a poor correlation between protein and mRNA abundances for a large proportion of detectable proteins (43). Therefore, it is expected that many kernel/endosperm-expressed genes are regulated by posttranscriptional regulatory mechanisms that may positively or negatively impact the relative levels of the encoded proteins.
The maize endosperm/kernel-expressed genes that we identified include 7,629 temporally regulated genes. The temporal patterns fall into four general groups (Fig. 2), but the majority (93.2%) of genes show a one-step-up or one-step-down pattern. Furthermore, most of the one-step-up and one-step-down genes exhibit an expression transition at 6–12 DAP. These results suggest that a dramatic reprogramming of the kernel transcriptome occurs after 4 DAP (Fig. 2 A and B and Fig. S4).
Several of the temporal patterns that we identified correlate with key developmental transitions associated with kernel and endosperm development (4). The up-at-4-DAP and up-at-6-DAP gene sets likely play a role in differentiation of the AL, BETL, ESR, and SE as they include many previously described endosperm cell-specific genes including MRP-1 (15), as well as the ESRs (19), MEGs, BETLs, and BAPs (16–18). Furthermore, our in situ hybridization analysis of 18 genes within these gene sets demonstrated that all were expressed in specific endosperm cell types (Figs. 4 and 5).
The up-at-8-DAP and up-at-10-DAP gene sets correlate with the onset of storage product accumulation, suggesting that these gene sets play a role in preparation for deposition of storage protein and carbohydrate storage products, which occurs primarily in SE. Consistent with this, the up-at-8-DAP gene set is highly represented by genes associated with translation, amino acid metabolism, amine metabolism, etc., and that the up-at-10-DAP gene set is over-represented in genes associated with energy reserve and carbohydrate biosynthesis (Fig. 2 and Dataset S3). Prior work indicates that zeins, the major storage proteins of maize endosperm, are synthesized between 10 and 45 DAP (21, 44), and our analysis indicates up-regulation of zein genes at 8 DAP (Fig. S6). This program follows temporally the up-regulation of Opaque-2 (GRMZM2G015534), which falls within the up-at-6-DAP gene set (Dataset S2, Table S1; Fig. 3B; Dataset S2, Table S2). The PBF transcription-factor gene (GRMZM2G146283) was also identified among the up-at-8-DAP group. Opaque-2 activates the expression of 22-kDa α-zein genes (45), and PBF binds the prolamin box in zein gene promoters and interacts with Opaque-2 (24) to presumably control activation of a more extended set of storage-function genes in the starchy endosperm. In support of this, we show a number of transcription-factor genes that are up-regulated at 8 DAP and expressed in SE (Figs. 4 and 5; and Fig. S5). Therefore, we find that as early as 6 DAP a cascade of gene-regulatory processes culminate in activation of the major storage-protein genes involved in seed germination.
This analysis makes a significant step toward identifying spatiotemporal programs of gene expression that operate during early endosperm development and regulate key developmental transitions, including the coenocytic-cellular transition, cell differentiation, and mitotic proliferation of the endosperm, which are essential to support postgerminative development. However, the nature of the relationships between the various structural genes and their regulatory factors remains to be determined. Understanding these interactions will require more detailed understanding of cell-specific patterns of gene expression for both the transcription factors and their gene targets. Efforts to identify spatial patterns of gene expression in endosperm are currently in progress, and together with the temporal programs described in this report, these data will help determine the full extent of the spatiotemporal programs of gene activity during endosperm development.
Materials and Methods
Profiling Kernel/Endosperm mRNAs and Determination of Temporal Expression Patterns.
Detailed information about the collection of kernel and endosperm samples, RNA extraction and purification, and library construction, sequencing, quality control of RNA-Seq reads, alignment to the maize reference genome, data normalization, identification of up/down-regulated genes using StepMiner, and identification of enriched GO categories are provided in SI Materials and Methods.
Confirmation of Temporal Patterns and mRNA Localization Studies.
Standard procedures for validation of the levels of specific mRNAs using qRT-PCR and localization of mRNA patterns using in situ hybridization are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation Grant IOS-0923880 (to J.M.D., G.N.D., B.A.L., and R. Yadegari).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE54131).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406383111/-/DCSupplemental.
References
- 1.Lopes MA, Larkins BA. Endosperm origin, development, and function. Plant Cell. 1993;5(10):1383–1399. doi: 10.1105/tpc.5.10.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsen OA. Endosperm development: Cellularization and cell fate specification. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:233–267. doi: 10.1146/annurev.arplant.52.1.233. [DOI] [PubMed] [Google Scholar]
- 3.Olsen OA. Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell. 2004;16(Suppl):S214–S227. doi: 10.1105/tpc.017111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabelli PA, Larkins BA. The development of endosperm in grasses. Plant Physiol. 2009;149(1):14–26. doi: 10.1104/pp.108.129437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lending CR, Larkins BA. Changes in the zein composition of protein bodies during maize endosperm development. Plant Cell. 1989;1(10):1011–1023. doi: 10.1105/tpc.1.10.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodrigues JA, et al. Imprinted expression of genes and small RNA is associated with localized hypomethylation of the maternal genome in rice endosperm. Proc Natl Acad Sci USA. 2013;110(19):7934–7939. doi: 10.1073/pnas.1306164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh TF, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324(5933):1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahoo D, Dill DL, Tibshirani R, Plevritis SK. Extracting binary signals from microarray time-course data. Nucleic Acids Res. 2007;35(11):3705–3712. doi: 10.1093/nar/gkm284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiesselbach TA, Walker ER. Structure of certain specialized tissues in the kernel of corn. Am J Bot. 1952;39(8):561–569. [Google Scholar]
- 13.Schel JHN, Kieft H, Vanlammeren AAM. Interactions between embryo and endosperm during early developmental stages of maize caryopses (Zea mays) Can J Bot. 1984;62(12):2842–2853. [Google Scholar]
- 14.Kiesselbach TA. 1949. The Structure and Reproduction of Corn (University of Nebraska College of Agriculture, Agricultural Experiment Station, Lincoln, NE), 96 pp.
- 15.Gómez E, Royo J, Guo Y, Thompson R, Hueros G. Establishment of cereal endosperm expression domains: Identification and properties of a maize transfer cell-specific transcription factor, ZmMRP-1. Plant Cell. 2002;14(3):599–610. doi: 10.1105/tpc.010365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutiérrez-Marcos JF, et al. maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell. 2004;16(5):1288–1301. doi: 10.1105/tpc.019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hueros G, Varotto S, Salamini F, Thompson RD. Molecular characterization of BET1, a gene expressed in the endosperm transfer cells of maize. Plant Cell. 1995;7(6):747–757. doi: 10.1105/tpc.7.6.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serna A, et al. Maize endosperm secretes a novel antifungal protein into adjacent maternal tissue. Plant J. 2001;25(6):687–698. doi: 10.1046/j.1365-313x.2001.01004.x. [DOI] [PubMed] [Google Scholar]
- 19.Opsahl-Ferstad HG, Le Deunff E, Dumas C, Rogowsky PM. ZmEsr, a novel endosperm-specific gene expressed in a restricted region around the maize embryo. Plant J. 1997;12(1):235–246. doi: 10.1046/j.1365-313x.1997.12010235.x. [DOI] [PubMed] [Google Scholar]
- 20.Bonello JF, et al. Esr proteins are secreted by the cells of the embryo surrounding region. J Exp Bot. 2002;53(374):1559–1568. doi: 10.1093/jxb/erf010. [DOI] [PubMed] [Google Scholar]
- 21.Woo YM, Hu DWN, Larkins BA, Jung R. Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell. 2001;13(10):2297–2317. doi: 10.1105/tpc.010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wittich PE, Vreugdenhil D. Localization of sucrose synthase activity in developing maize kernels by in situ enzyme histochemistry. J Exp Bot. 1998;49(324):1163–1171. [Google Scholar]
- 23.Schmidt RJ, Burr FA, Aukerman MJ, Burr B. Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc Natl Acad Sci USA. 1990;87(1):46–50. doi: 10.1073/pnas.87.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci USA. 1997;94(14):7685–7690. doi: 10.1073/pnas.94.14.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedersen K, Devereux J, Wilson DR, Sheldon E, Larkins BA. Cloning and sequence analysis reveal structural variation among related zein genes in maize. Cell. 1982;29(3):1015–1026. doi: 10.1016/0092-8674(82)90465-2. [DOI] [PubMed] [Google Scholar]
- 26.Pedersen K, Argos P, Naravana SVL, Larkins BA. Sequence analysis and characterization of a maize gene encoding a high-sulfur zein protein of Mr 15,000. J Biol Chem. 1986;261(14):6279–6284. [PubMed] [Google Scholar]
- 27.Prat S, Pérez-Grau L, Puigdomènech P. Multiple variability in the sequence of a family of maize endosperm proteins. Gene. 1987;52(1):41–49. doi: 10.1016/0378-1119(87)90393-3. [DOI] [PubMed] [Google Scholar]
- 28.Kirihara JA, Petri JB, Messing J. Isolation and sequence of a gene encoding a methionine-rich 10-kDa zein protein from maize. Gene. 1988;71(2):359–370. doi: 10.1016/0378-1119(88)90053-4. [DOI] [PubMed] [Google Scholar]
- 29.Du Z, Zhou X, Ling Y, Zhang ZH, Su Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38(Web Server issue):W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becraft PW. Cell fate specification in the cereal endosperm. Semin Cell Dev Biol. 2001;12(5):387–394. doi: 10.1006/scdb.2001.0268. [DOI] [PubMed] [Google Scholar]
- 31.Charlton WL, et al. Endosperm development in Zea mays: Implication of gametic imprinting and paternal excess in regulation of transfer layer development. Development. 1995;121(9):3089–3097. [Google Scholar]
- 32.Cooper DC. Caryopsis development following matings between diploid and tetraploid strains of Zea Mays. Am J Bot. 1951;38(9):702–708. [Google Scholar]
- 33.Gao Y, Xu H, Shen YY, Wang JB. Transcriptomic analysis of rice (Oryza sativa) endosperm using the RNA-Seq technique. Plant Mol Biol. 2013;81(4-5):363–378. doi: 10.1007/s11103-013-0009-4. [DOI] [PubMed] [Google Scholar]
- 34.Gillies SA, Futardo A, Henry RJ. Gene expression in the developing aleurone and starchy endosperm of wheat. Plant Biotechnol J. 2012;10(6):668–679. doi: 10.1111/j.1467-7652.2012.00705.x. [DOI] [PubMed] [Google Scholar]
- 35.Gutierrez-Gonzalez JJ, Tu ZJ, Garvin DF. Analysis and annotation of the hexaploid oat seed transcriptome. BMC Genomics. 2013;14:471. doi: 10.1186/1471-2164-14-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu XD, et al. The differential transcription network between embryo and endosperm in the early developing maize seed. Plant Physiol. 2013;162(1):440–455. doi: 10.1104/pp.113.214874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pellny TK, et al. Cell walls of developing wheat starchy endosperm: Comparison of composition and RNA-Seq transcriptome. Plant Physiol. 2012;158(2):612–627. doi: 10.1104/pp.111.189191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones RJ, Schreiber BMN, Roessler JA. Kernel sink capacity in maize: Genotypic and maternal regulation. Crop Sci. 1996;36(2):301–306. [Google Scholar]
- 39.Reddy VM, Daynard TB. Endosperm characteristics associated with rate of grain filling and kernel size in corn. Maydica. 1983;28(4):339–355. [Google Scholar]
- 40.Capitanio R, Gentinetta E, Motto M. Grain weight and its components in maize inbred lines. Maydica. 1983;28(4):365–379. [Google Scholar]
- 41.Sekhon RS, et al. Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS ONE. 2013;8(4):e61005. doi: 10.1371/journal.pone.0061005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le BH, et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA. 2010;107(18):8063–8070. doi: 10.1073/pnas.1003530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walley JW, et al. Reconstruction of protein networks from an atlas of maize seed proteotypes. Proc Natl Acad Sci USA. 2013;110(49):E4808–E4817. doi: 10.1073/pnas.1319113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kodrzycki R, Boston RS, Larkins BA. The opaque-2 mutation of maize differentially reduces zein gene transcription. Plant Cell. 1989;1(1):105–114. doi: 10.1105/tpc.1.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt RJ, Ketudat M, Aukerman MJ, Hoschek G. Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell. 1992;4(6):689–700. doi: 10.1105/tpc.4.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.