Significance
Gibberellin (GA)-dependent degradation of DELLA protein, a key negative regulator, is essential for GA action. However, it is unclear how DELLA regulates downstream gene expression. Although possessing strong transactivation activity, DELLA lacks a DNA-binding domain. Therefore, a model has been proposed in which DELLA acts as a transcriptional coactivator with another transcription factor or factors containing a DNA-binding domain. Here, we show that some members of the INDETERMINATE DOMAIN (IDD) protein family are such intermediate proteins. The DELLA/IDD complex up-regulates the expression of the GA-positive regulator SCARECROW-LIKE 3 (SCL3). Meanwhile, SCL3 protein also interacts with IDD proteins to suppress its own expression. We propose that a coregulator exchange system between DELLA (as coactivator) and SCL3 (as corepressor) regulates the expression of SCL3.
Keywords: transcription factor, gibberellin feedback regulation, coactivator/corepressor exchange regulation system
Abstract
DELLA protein is a key negative regulator of gibberellin (GA) signaling. Although how DELLA regulates downstream gene expression remains unclear, DELLA has been proposed to function as a transcriptional activator. However, because DELLA lacks a DNA-binding domain, intermediate protein(s) mediating the DELLA/DNA interaction are believed to be necessary for activating DELLA target genes. Here, using yeast hybrid screenings, we identified five members of INDETERMINATE DOMAIN (IDD) protein family which bind physically to both DELLA and the promoter sequence of the GA-positive regulator SCARECROW-LIKE 3 (SCL3), which previously was characterized as a DELLA direct target gene. Transient assays using Arabidopsis protoplasts demonstrated that a luciferase reporter controlled by the SCL3 promoter was additively transactivated by REPRESSOR of ga1-3 (RGA) and IDDs. Phenotypic analysis of transgenic plants expressing AtIDD3 (one of the 16 IDDs in the Arabidopsis genome) fused with the plant-specific repression domain (SRDX) supported the possibility that AtIDD3 is positively involved in GA signaling. In addition, we found that SCL3 protein also interacts with IDDs, resulting in the suppression of its target gene expression. In this context, DELLA and SCL3 interact competitively with IDD proteins to regulate downstream gene expression. These results suggest that the coregulators DELLA and SCL3, using IDDs as transcriptional scaffolds for DNA binding, antagonistically regulate the expression of their downstream targets to control the GA signaling pathway.
Gibberellins (GAs) are diterpene phytohormones that regulate many cellular and developmental events such as cell elongation, leaf expansion, flowering, pollen maturation, and the transition from vegetative growth to flowering (1–4). Several protein factors involved in GA signaling have been identified. Among these, DELLA protein is a key player in the regulation of GA responses. DELLA proteins are characterized by a DELLA/TVHYNP motif at the N terminus and a GRAS domain [named after its first three members: GA INSENSITIVE (GAI), REPRESSOR of ga1-3 (RGA), and SCARECROW (SCR)] at the C terminus, placing DELLAs within the GRAS family of transcriptional regulators. GRAS-domain transcription factors have diverse functions in growth and development. Recent intensive studies revealed how GA is perceived by the GA receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) and how the perceived GA signal is transmitted to DELLA. By binding to active GAs, GID1 acquires the ability to interact with DELLA, allowing further interaction with an F box protein, SLEEPY1/GID2. DELLA is polyubiquitinated by E3 ubiquitin-ligase SCFSLY1/GID2 and finally is degraded through the 26S proteasome. However, how DELLA regulates downstream gene expression in GA signaling has remained unclear.
In Arabidopsis, five DELLA genes have been identified; GAI, RGA, and three RGA-LIKE proteins (RGL1, RGL2, and RGL3) (1–4). Rice and barley each have a single DELLA protein, SLENDER1 (SLR1) and Slender1 (Sln1), respectively (1–4). Previous studies have predicted dual functions for DELLAs in regulating downstream genes and allowing GA to regulate various developmental processes (3). One possible mode of function is that DELLA interacts with other transcription factors to inhibit their DNA-binding and transcriptional activities. For example, Arabidopsis DELLA interacts with PHYTOCHROME INTERACTING FACTOR 3 and 4 (PIF3 and PIF4) and blocks their DNA-binding abilities to their target gene promoters, resulting in short hypocotyls in light-grown plants (5, 6). Likewise, DELLA is reported to interfere with several components of hormonal and developmental signaling pathways through protein–protein interaction (3, 4).
On the other hand, Zentella et al. (7) and Gallego-Bartolomé et al. (8) have used transcriptome analyses in transgenic plants expressing gain-of-function versions of DELLA to identify DELLA target genes. More recent work has demonstrated that DELLA interacts with the core subunit of chromatin remodeling factor SWI/SNF to increase the transcription of DELLA target genes (9). Furthermore, Hirano et al. (10) have demonstrated that the N-terminal DELLA/TVHYNP motif of the rice DELLA, SLR1, possesses transactivation activity. The transactivation activity of mutated SLR1s observed in yeast corresponds well to the severity of dwarfism in rice plants overexpressing mutated SLR1, indicating that SLR1 suppresses plant growth through its transactivation activity. Thus, DELLA also functions as a transactivator in planta. Because DELLA is thought not to possess a DNA-binding domain (DBD), one or more other transcription factors may serve as an intermediate protein between DELLA and DNA to up-regulate their downstream genes. However, such intermediate proteins have not yet been identified.
To identify intermediate proteins mediating DELLA/DNA interaction, we conducted yeast two-hybrid (Y2H) screening using RGA, an Arabidopsis DELLA, as bait against a transcription factor library. We also conducted yeast one-hybrid (Y1H) screening using the promoter of a GA-positive regulator, SCARECROW-LIKE 3 (SCL3), which is a putative direct target of RGA (7, 11). From these screenings we identified five transcription factors belonging to the C2H2 zinc finger family that interact with both the RGA protein and SCL3 promoter. Interestingly, all the candidates belong to a single subfamily, the INDETERMINATE DOMAIN (IDD) family. Subsequent experiments confirmed that RGA uses these IDD proteins as transcriptional scaffolds to up-regulate SCL3 expression for GA signaling regulation. Furthermore, we found that the SCL3 protein interacts competitively with IDD against RGA and interferes with transactivation by the RGA/IDD complex. In conclusion, we propose a coactivator/corepressor exchange regulation system in which DELLA and SCL3 are used as a transcriptional coactivator and a corepressor, respectively, and IDDs are used as transcriptional scaffolds to regulate the expression of SCL3 and other genes in GA signaling.
Results
Members of the IDD Family Interact with both DELLA and the Promoter Region of SCL3.
One possible reason for the failure to identify DELLA/DNA-mediating factors to date may be that DELLA has strong self-transactivation activity in the N-terminal DELLA/TVHYNP motif, complicating screening for its interacting proteins by a Y2H approach. However, we previously identified the GRAS domain, which is separated from the transactivation domain in the N-terminal region of DELLA, as the region through which DELLA interacts with other proteins for DNA binding (10). Therefore, a Y2H screen using the GRAS domain lacking self-transactivating activity is likely to return fewer false-positive clones but retain the ability to identify genuine partners of DELLA that are important for DNA interaction and transactivation of DELLA targets. Thus, we used a truncated version of RGA (RGA-GRAS) lacking the N-terminal region (amino acids 186–587) as bait construct for Y2H screening. Another considerable problem is that the available cDNA libraries often do not include the transcription factors of interest because of their low expression levels. To overcome this drawback, we used a cDNA library composed only of transcription factors, which covered ∼75% of all Arabidopsis transcription factors (12). By this approach, using Y2H and Y1H screenings, we identified five clones encoding proteins belonging to the C2H2 type zinc finger superfamily.
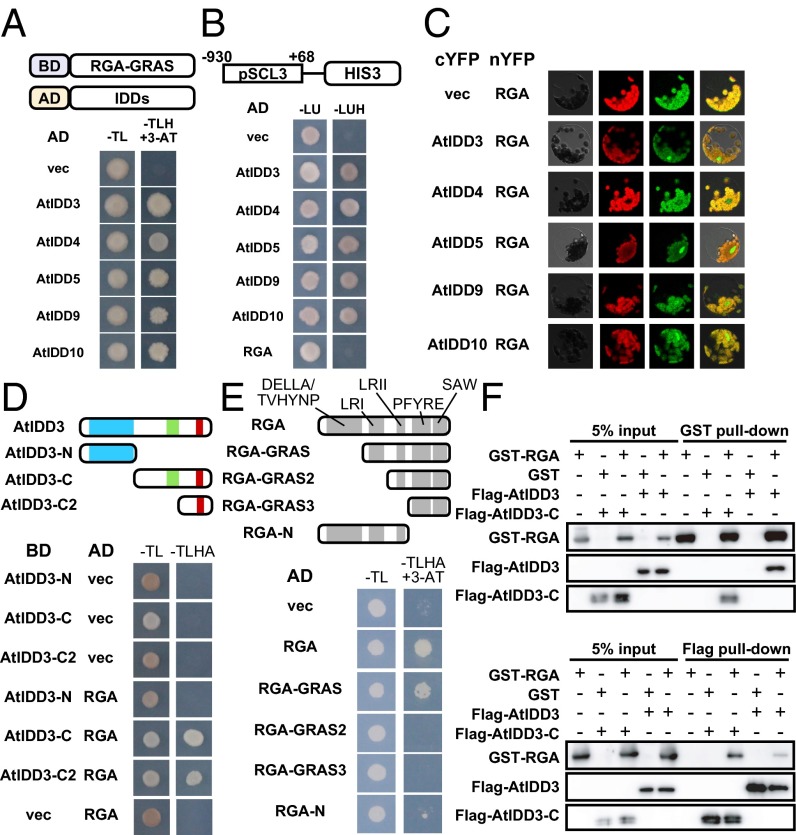
Interestingly, all five C2H2 type zinc finger superfamily proteins identified by the Y2H and Y1H screens also belong to the IDD subfamily. The Arabidopsis genome encodes 16 IDD proteins, collectively designated AtIDDs (13). The proteins of this subfamily have a characteristic domain, the IDD, which includes traditional and irregular zinc finger motifs at the N terminus that possess DNA-binding ability, as well as two other conserved motifs at the C terminus (Fig. S1) (13, 14). In Y2H screens, the identified IDD proteins AtIDD3 (AT1G03840), AtIDD4 (AT2G02080), AtIDD5 (AT2G02070), AtIDD9 (AT3G45260), and AtIDD10 (AT5G03150) interacted with RGA-GRAS (Fig. 1A). Furthermore, we confirmed that AtIDD3 interacts with intact RGA as well as with other DELLA members (Fig. S2). These IDD proteins also bound to the promoter region of SCL3 (Fig. 1B). SCL3, which also is a GRAS family member, is thought to act as a tissue-specific integrator of the GA pathway in the root endodermis by antagonizing DELLA, and its expression is positively regulated by RGA through indirect interaction between RGA and its promoter sequence (11, 15). The interaction of these IDD proteins with RGA in vivo was confirmed by bimolecular fluorescence complementation (BiFC) assays (Fig. 1C). A strong fluorescence signal was observed in the nucleus of Arabidopsis mesophyll protoplasts that were cotransfected with RGA-YFPN and IDD-YFPC but not in protoplasts transfected with the vector control.
Fig. 1.
Some members of the IDD protein family interact with RGA protein and the promoter sequence of SCL3. (A) Interactions between RGA-GRAS and IDDs in a Y2H assay. Schematic representations of the construct, BD-RGA-GRAS, and AD-IDDs are shown in the upper panel. AD, GAL4-activating domain; BD, GAL4-binding domain; −TL, synthetic complete medium lacking Trp and Leu; −TLH, synthetic complete medium lacking Trp, Leu, and His; 3-AT, 3-aminotriazole, which is a competitive inhibitor of HIS3 enzyme. The −TLH +3-AT medium contained 10 mM 3-AT. (B) Interaction between IDDs and the promoter of SCL3 (pSCL3) in a Y1H assay. Schematic representations of the construction, pSCL3::HIS3 is shown in the upper panel in which the numbers correspond to the distance from the transcription start site. −LU, synthetic complete medium lacking Leu and Ura; −LUH, synthetic complete medium lacking Leu, Ura, and His. (C) BiFC analyses testing the interaction between IDDs and RGA in Arabidopsis mesophyll protoplasts. Individual and merged images of YFP (green) and chlorophyll autofluorescence (red) as well as differential interference contrast images of protoplasts are shown. (D) Domain analyses of AtIDD3 in Y2H assays. The upper panel indicates AtIDD3 and its derivatives showing the conserved domain/motifs by color: N-terminal ID domain (light blue box) and two C-terminal motifs, MSATALLQKAA (light green box) and TR/LDFLG (red box). −TLHA, synthetic complete medium lacking Trp, Leu, His, and adenine. (E) Domain analyses of RGA in Y2H assays. The upper panel indicates RGA and its derivatives. The selection medium contained 50 mM 3-AT. (F) In vitro pull-down assays assessing physical interaction between RGA and AtIDD3. AtIDD3-C indicates the truncated AtIDD3, as used in D. GST–RGA fusion protein or GST (Upper) and Flag-AtIDD3 or Flag-AtIDD3-C (Lower) were used as bait.
Next, we analyzed a domain of AtIDD3 necessary for interaction with RGA (Fig. 1D). AtIDD3 containing only the N-terminal region, including the DBD (AtIDD3-N, amino acids 1–229) did not interact with DELLA, whereas AtIDD3 variants containing only C-terminal regions (AtIDD3-C, amino acids 230–506, and AtIDD3-C2, amino acids 401–506) were able to interact with DELLA. These results suggest that IDD proteins can interact simultaneously with DNA and DELLA, unlike other reported DELLA-interaction partners whose interaction with DELLA abolishes their DNA-binding activities (3–6).
We also performed domain analysis of RGA for interaction with AtIDD3 (Fig. 1E). The GRAS domain of RGA can be subdivided into five distinct motifs: leucine-rich region I (LRI), VHIID, leucine-rich region II (LRII), PFYRE, and SAW (16). The entire RGA and RGA-GRAS containing these five motifs interacted with AtIDD3, whereas RGA-GRAS2, which lacks only the LRI domain, could not. RGA-N, which contains the DELLA/TVHYNP, LRI, and LRII domains, also was unable to interact with AtIDD3. These results indicate that the LRI domain of RGA is essential but not sufficient for the interaction between RGA and AtIDD3 and that at least one other C-terminal region is necessary. In vitro pull-down assays also confirmed the interaction between GST-tagged full-length RGA and Flag-tagged AtIDD3 regardless of the DBD of AtIDD3 (Fig. 1F). Taken together, these results strongly suggest that the AtIDD3, -4, -5, -9, and -10 members of the IDD family are able to mediate the regulatory function of RGA for the expression of SCL3.
The DELLA/IDD Complex Promotes the Expression of SCL3.
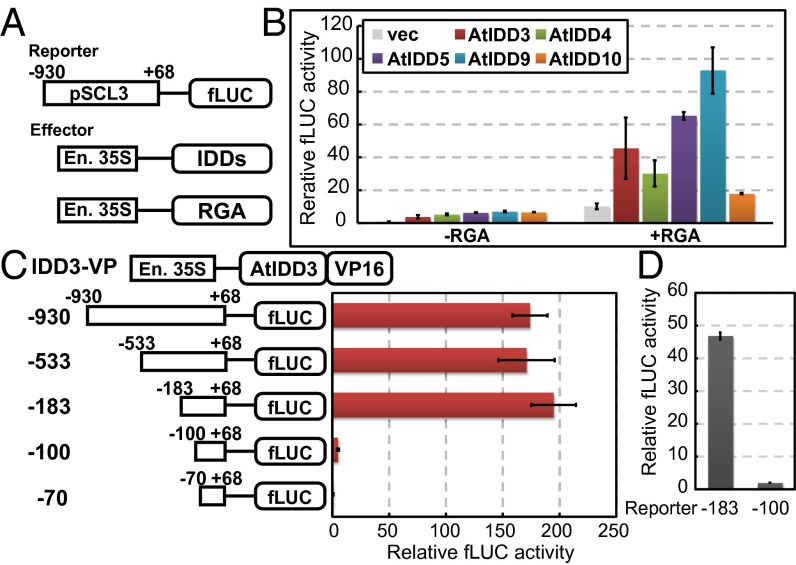
A transient reporter assay was used to study the transcriptional activity of the DELLA/IDD complex, in which a firefly luciferase (fLUC) reporter under the control of the 1-kb 5′ upstream promoter region of SCL3, with or without effector plasmids (enhanced 35S::AtIDD3, -4, -5, -9, -10 and RGA), was introduced into Arabidopsis protoplasts (Fig. 2A). When protoplasts were transfected with the fLUC reporter alone, only trace fLUC activity was detected (Fig. 2B), whereas cotransfection of the reporter and IDDs (−RGA), resulted in a significantly increased fLUC activity. Cotransfection of the reporter and both IDD and RGA plasmids (+RGA) again was able to produce substantially higher fLUC activity, which was dependent on the IDD protein variant used. These results demonstrate that IDDs function as intermediate proteins for the transcriptional activation of SCL3 by RGA, whereas IDDs alone possess only a small transcriptional activation activity. To localize target sites of the DELLA/IDD complex in the SCL3 promoter, we conducted a transient reporter assay using a series of truncated SCL3 promoters together with an AtIDD3 protein fused with a transcriptional activation domain from viral protein 16 (VP16) replacing RGA (IDD3-VP) (Fig. 2C). High fLUC activity was observed when the promoter was −183 bp (or more) upstream of the transcription start site, but promoters less than −100 bp upstream of the transcription start site displayed no fLUC activity, indicating that the region from −183 to −100 bp upstream from the transcription site is essential for transcriptional activation of SCL3 by AtIDD3. In addition, because VP16 can substitute for RGA, RGA likely acts as a transcriptional activator rather than by strengthening the binding between IDD and DNA. We performed the same experiment using AtIDD3 and RGA (instead of IDD3-VP) as effectors (Fig. 2D) and confirmed that the region from −183 to −100 is essential for the transcription of SCL3 by the DELLA/IDD complex.
Fig. 2.
The effects of IDDs and RGA on the expression of SCL3. (A and B) A transient reporter assay was used to examine the transactivation effects of IDDs and RGA on the expression of SCL3 in protoplasts from Arabidopsis T87 suspension-cultured cells. (A) Schematic representations of the effector and reporter constructs. (B) Bars marked −RGA represent experiments in which the reporter, IDD effector plasmid, and empty RGA effector plasmids were cotransfected. Bars marked +RGA correspond to experiments in which each of the IDDs, RGA, and reporter plasmids were cotransfected. As a negative control, an empty effector vector (vec) was used in place of the IDD plasmids. The relative activity caused by vector control (far left bar) was set as 1. Results represent the means of three experiments; error bars represent SD. En. 35S, enhanced 35S promoter whose details are described in SI Materials and Methods. (C) Transient reporter assay in cultured cell protoplast to determine where AtIDD3 exerts its effect on the SCL3 promoter. Schematic representations of the effector construct, IDD3-VP16, and SCL3 promoter deletion are shown on the left. (D) Transient reporter assays using AtIDD3, RGA, and the −183 and −100 SCL3 promoter constructs. The AtIDD3 and RGA constructs were cotransfected as effectors. The relative activity caused by vector control was set as 1.
To determine which DNA sequences within the SCL3 promoter are targeted by the DELLA/IDD complex, we searched for sequences homologous to the consensus target motifs of IDD homologs ZmID1, AtIDD8, and OsIDD10 (14, 17, 18) and found one candidate carrying AGACAA from −111 to −106 in the SCL3 promoter (Fig. S3). The physical interaction between AtIDD3 and the biotinylated DNA fragments containing the region from −123 to −94 was confirmed by an EMSA, whereas the same DNA fragment in which the target sequence (AGACAA) is replaced by CTCAGG lost interacting activity (Fig. S3). These data indicate that RGA positively regulates the expression of SCL3 through interaction with AtIDD3, which binds to specific DNA sequences containing AGACAA as a core motif.
atidd10/jkd-4 and AtIDD3-SRDX Plants Induced Characteristic Phenotypes Related to GA.
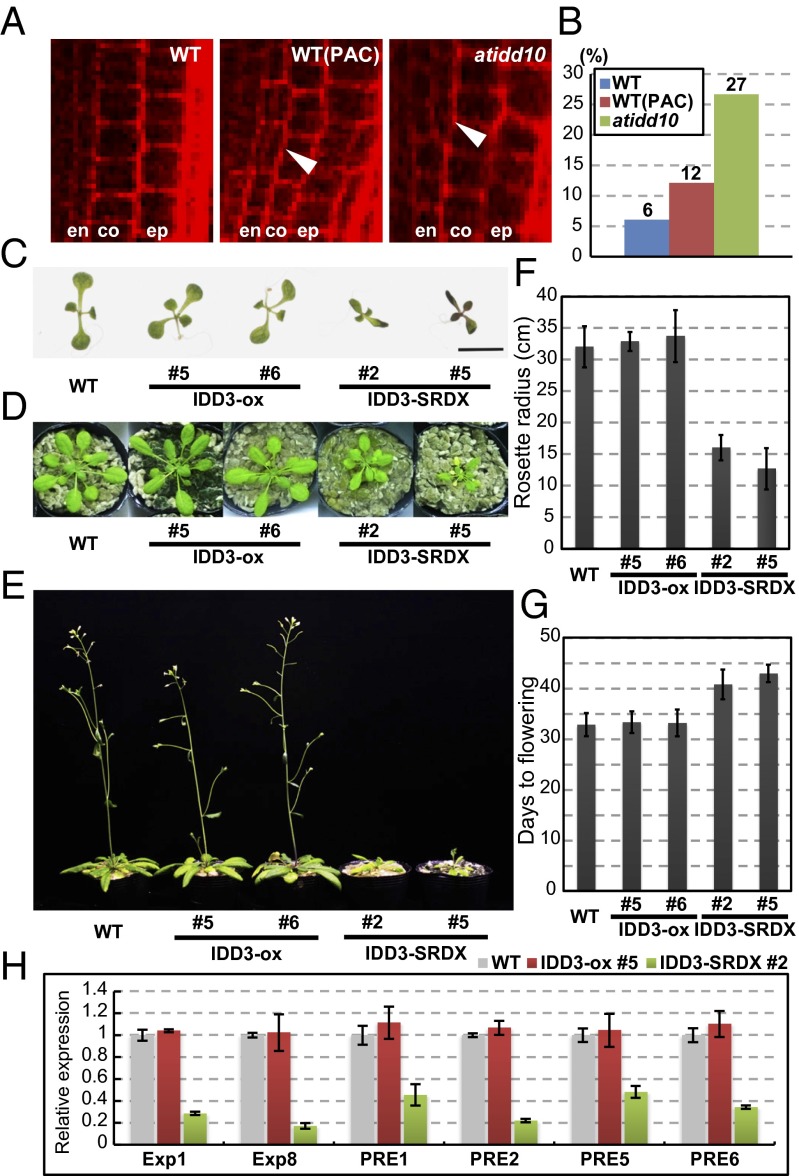
Welch et al. (19) reported that AtIDD3 (MAGPIE; MGP) and AtIDD10 (JACKDAW; JKD) are involved in the root development, which also is controlled by DELLA and SCL3 (15). Therefore, we compared the root phenotype of atidd10/jkd-4 (SALK_054242C) with WT and GA-deficient plants. We observed an unusually high frequency of periclinal cell division in the root endodermis of atidd10 plants, which also was observed in the WT plants under GA-deficient conditions (Fig. 3 A and B and Fig. S4) and in GA-deficient and scl3 mutants, as previously reported (15). However, we were unable to observe additional atidd10 phenotypes typical of GA-related defects or any such phenotypes in plants with down-regulated AtIDD3 expression as previously reported (17, 19), perhaps because of the partially redundant activities of IDDs. Therefore, we generated transgenic plants overexpressing AtIDD3 (IDD3-ox) and plants in which the IDD function was suppressed by the introduction of AtIDD3 fused with the plant-specific repression domain (IDD3-SRDX) (20). IDD3-SRDX plants should be phenocopies of mutants with defects in which multiple IDDs (including AtIDD3, -4, -5, -9, and -10) share the common target sequence; therefore it is considered useful for investigating the physiological functions of the redundant transcription factors (20). IDD3-ox plants were rarely different from WT; however, the IDD3-SRDX plants had smaller leaves, shorter roots, and delayed flowering as compared with WT plants (Fig. 3 C–G and Fig. S5A), mimicking the previously reported phenotypes of GA-signaling and -deficient mutants (1–4, 21, 22). Furthermore, cell numbers in the first true leaves were reduced in both IDD3-SRDX plants and paclobutrazol-treated WT plants as compared with untreated WT plants (Fig. S5B), a finding that is consistent with GA promoting cell proliferation in young leaves (23). Taken together, these results suggest that IDDs are related to plant GA signaling. IDD3-SRDX also exhibited an abnormal leaf phenotype at 1 wk but not at 4 wk after germination; this phenotype did not seem to be related to GA signaling (Fig. 3 C and D). To confirm that these GA-related phenotypes in IDD3-SRDX were caused by repression of GA signaling, we analyzed the gene expressions of the Expansin1 and -8, and paclobutrazol resistance factor (PRE) 1, 2, 5, and 6, which previously were reported to be up-regulated by GA (24, 25). Quantitative RT-PCR (qRT-PCR) analyses showed that the expression of the GA-regulated genes was reduced significantly in IDD3-SRDX plants but was not obviously changed in IDD3-ox plants (Fig. 3H). The SCL3 expression pattern was similar in WT, IDD3-ox, and IDD3-SRDX plants (Fig. S6).
Fig. 3.
The mutation of AtIDD10 and the expression of AtIDD3 fused with SRDX reduce GA signaling. (A) Higher-magnification view of periclinal cell division in the endodermis. Whole images are shown in Fig. S4. Arrowheads indicate periclinal cell division in the endodermis. en, endodermis; ep, epidermis; co, cortex; WT(PAC), WT plants treated with the GA biosynthesis inhibitor paclobutrazol. (B) The frequency of periclinal cell division in the endodermis at 4 d after germination. (C and D) Leaf phenotype in 1-wk-old (C) and 4-wk-old (D) plants. AtIDD3 in the IDD3-ox plants and AtIDD3 fused with SRDX in the IDD3-SRDX plants were both driven by the 35S promoter. (E) Late flowering of IDD3-SRDX in 6-wk-old plants. (F and G) Quantification of data in D and E. Results represent the means of more than three samples; error bars indicate SD. (H) Relative transcript levels in the leaves from 1-wk-old seedlings. The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase C subunit (GAPC), whose expression is not responsive to GA, was used to normalize different samples. Biological triplicate samples were averaged. Error bars indicate SD of the means.
Given these results, we conclude that IDD proteins regulate the GA-signaling pathway by serving as an intermediate protein between DELLA and the region of the promoters of downstream genes containing the sequence AGACAA.
RGA and SCL3 Antagonize Each Other Through the Competition to Interact with IDD Proteins.
Previous studies demonstrated that SCL3 protein functions antagonistically against RGA as a GA-positive regulator (11). Additionally, DELLA and SCL3 are both members of the GRAS family, although SCL3 lacks the DELLA domain, and DELLA interacts with IDDs via their GRAS domain, possibly suggesting that SCL3 interacts with its target DNA in a manner similar to that of the DELLA protein (11). This circumstantial evidence led us to speculate that SCL3 may interfere with the interaction between DELLA and IDDs by interacting directly with IDDs. Indeed, a physical interaction between SCL3 and IDD proteins was confirmed by Y2H (Fig. 4A) and BiFC assays (Fig. S7). Moreover, Y2H results have shown that the interaction of AtIDD3 with SCL3 occurs through their C-terminal domains, as is the case for DELLA (Fig. S8). We next performed a yeast three-hybrid (Y3H) assay to examine the competitive relationship between the RGA/IDD and SCL3/IDD interactions (Fig. 4B). The activity of a reporter gene HIS3, which indicates the interaction between SCL3 and AtIDD3, apparently was reduced in the presence of RGA. An in vitro pull-down assay demonstrated that an increasing abundance of RGA causes a decrease in the SCL3/AtIDD3 complex (Fig. 4C). These results indicated a competitive relationship between RGA and SCL3 for interaction with AtIDD3 rather than the formation of a triple RGA/SCL3/AtIDD3 complex. Such a competitive relationship also was demonstrated in vivo, in which protoplast fLUC activity promoted by the RGA/AtIDD3 complex was diminished by SCL3 expression (Fig. 4D). Taken together, these results suggest the SCL3 corepressor also is able to interact with IDDs, thus competing with the coactivator DELLA; this competition may result in a feedback loop mechanism for transcriptional activity of DELLA and SCL3 (Fig. 4E and Discussion).
Fig. 4.
SCL3 competes with DELLA and prevents its transactivation activity. (A) Interactions between IDDs and SCL3 in the Y2H assay. Schematic representations of the construct, BD-SCL3, and AD-IDDs are shown in the upper panel. (B) Y3H assay to determine the physical relationship between RGA, SCL3, and AtIDD3. The two lines on the right indicate yeasts on selection medium containing 5 and 10 mM 3-AT, respectively. (C) In vitro pull-down assays for determining the physical association of SCL3 and AtIDD3 and competition between RGA and SCL3 for AtIDD3. Flag-AtIDD3-C was used as bait. Numbers indicate the concentrations (in mM) of each protein in the reaction solution. (D) Transient reporter assay for identifying the effect of SCL3 on the transcriptional regulation of the DELLA/IDD complex. The −183 construct shown in Fig. 2C was used as the reporter. Results represent the means of three experiments; error bars indicate SD. The relative activity caused by vector control was set as 1. (E) A model for GA feedback regulation by the interaction among DELLA, SCL3, and IDD.
Discussion
It has been suggested that GA-regulated growth is governed by the GA-dependent de-repression of a growth-repressive factor, DELLA, and that GA-dependent degradation of DELLA is a core mechanism in the GA-signaling pathway (1–4). In this context, Davière and Achard (3) proposed two possible functions of DELLA. The first is that DELLA functions to block the transcriptional activity of transcription factors by physical interaction, as for the PIF proteins. The other possibility is that DELLA functions as a transcriptional coactivator through interaction with another transcription factor(s) for DNA binding. We conducted Y1/2H screening from a library covering almost all Arabidopsis transcription factors to identify proteins that contain a DBD and that interact with both DELLA and the promoter sequence of SCL3, one of the known target genes of DELLA, both properties being consistent with a role as a DELLA transcriptional coactivator (7, 11). We identified five such proteins, all of which were members of the IDD family (Fig. 1 A and B). Further analyses confirmed that DELLA acts as a transcriptional coactivator through the IDD proteins (Fig. 2 B–D and Fig. S3).
The IDD family is highly conserved protein family in angiosperms (13). Because the first IDD family gene identified (ZmID1) was isolated as a causal gene for a late-flowering mutant of maize (26), the function of this family has been discussed in the context of flowering time regulation. For example, a rice ZmID1 homolog, OsID1, promotes flowering, as does AtIDD8 (NUTCRACKER; NUC) (17, 27–29). Later, a functional role in gravitropism was reported for AtIDD15 (SHOOT GRAVITROPISM 5; SGR5) and OsIDD14 (LOOSE PLANT ARCHITECTURE1; LPA1) (30–32). To date, only one report has described physical interactions of AtIDD1 (ENHYDROUS; ENY) with each of the five DELLA members (33). Feurtado et al. (33) also demonstrated that AtIDD1 represses the expression of GA biosynthesis and signaling genes, including SCL3, and discussed the possibility that AtIDD1 may disrupt the interaction between DELLA and other proteins, including PIFs. This proposed function of AtIDD1 is different from our description that DELLA and IDDs cooperate to up-regulate their downstream genes. It is noteworthy that AtIDD1 possesses a putative repression domain, whereas AtIDD3, -4, - 5, -9, and -10 do not (34), suggesting that there are functional and physical differences between AtIDD1 and the other IDD family members. In addition, although the library we used contains AtIDD1 (12), we failed to isolate it in our Y2H screening; this result suggests that the binding affinity of AtIDD1 for RGA is likely to be lower than that of the other IDDs. Here, we propose the novel function of IDDs as intermediate proteins between DELLA and target gene promoters, acting to enhance gene expression. That at least five of the 16 Arabidopsis IDDs have been demonstrated to have such a function suggests this is a principal function of the IDD family proteins. However, because some of IDD3-SRDX phenotypes [e.g., the abnormal leaf phenotype at 1 wk after germination (Fig. 3C)] were not the same as those of GA-deficient mutants, IDDs also may work in other signaling pathways, in addition to GA signaling.
GA deficiency and AtIDD10 knockout mutation increased the frequency of periclinal cell division in the root endodermis (Fig. 3 A and B and Fig. S4) (15, 35). Recent work has demonstrated that SCL3 also is involved in this division in the context of GA signaling (15). The IDD family members are expressed in the endodermal cell, where RGA and SCL3 are expressed also (19, 36–39). Such overlapping expression of IDDs, RGA, and SCL3 strongly supports our model in which DELLA, SCL3, and IDD proteins cooperate to control GA signaling in the endodermis by regulating downstream gene expression. Consistent with previous studies (11, 15), we considered the relationship of the three factors in this study: DELLA promotes the periclinal cell division in the root endodermis by repressing GA signaling; on the contrary, SCL3, a positive regulator of GA signaling, suppresses the periclinal cell division; and lack of AtIDD10 causes a defect in DELLA-mediated feedback, thus resulting in increased cell division.
We found that RGA up-regulates the expression of SCL3 by collaborating with IDDs (Fig. 2 B–D and Fig. S3). Like DELLA, SCL3 is a putative transcription factor of the GRAS family, but, unlike DELLA, SCL3 acts as a positive regulator of GA signaling (11, 40). A previous study also has shown that SCL3 interferes with the function of DELLA to repress transcription of GA biosynthesis genes as well as its own (SCL3) transcription (11). In this study, we identified an interaction between SCL3 and IDDs (Fig. 4A and Fig. S7). We also found that RGA and SCL3 interact competitively with IDDs to regulate the expression of SCL3 (Fig. 4 B–D). According to these results, we propose a novel feedback-loop system to regulate GA signaling (Fig. 4E). DELLA activates the expression of the downstream genes, including SCL3, by IDD-mediated interaction with their promoters. The subsequent increase in the abundance of SCL3 protein results in an increase of the SCL3/IDD complex, decreasing the formation of the DELLA/IDD complex and consequent suppression of SCL3 expression (Fig. 4E). This DELLA-SCL3–mediated feedback loop explains homeostatic regulation of protein levels of the downstream genes, including the positive regulator SCL3, resulting in homeostatic GA signaling. Therefore, the transcription activity of AtIDD3 may depend on which coregulator it binds to. This model can explain the observations that the IDD3-ox plants were rarely different from WT (Fig. 3 C–E). In fact, although the increase in IDD protein does not determine the level of SCL3, the balance between DELLA and SCL3 protein level does. On the other hand, IDD3-SRDX plants exhibited a GA-deficient phenotype (Fig. 3 C–E), which can be explained by the IDD3-SRDX fusion protein functioning as a repressor of the downstream gene expression to negatively regulate GA signaling regardless of the presence of coregulators.
To our knowledge, such a coactivator/corepressor exchange regulation system (Fig. 4E) has not been described previously for any plant signaling pathway. However, similar regulation systems have been well studied in animals (41). For example, estrogen nuclear receptor can act as a transcriptional activator or repressor by interacting with coactivators such as histone acetyltransferase p300 and a mediator recruiting RNA polymerase complex or with corepressors such as HDAC and NCoR (42). A similar system also has been observed in the regulation of cortisol biosynthesis (43) and the function of Liver X receptor, one of the nuclear oxysterol receptors (44). Rosenfeld et al. (41) reported that this regulation system plays important roles in integrating signal-dependent programs of transcriptional responses at the molecular level in animal systems. Our findings demonstrated that the coactivator/corepressor exchange system should explain, at least in part, the intricate regulation of GA signaling in plants. Moreover, such a system may be involved in transcriptional regulation of other signaling pathway.
Materials and Methods
Yeast One-Hybrid/Two-Hybrid Screening.
Yeast one-hybrid/two-hybrid (Y1/2H) screening was performed as previously described (12) with yeast strains AH109 harboring BD-RGA-GRAS and YM4271 harboring pSCL3::HIS3. Selection medium lacking histidine but containing 5 mM 3-amino-1,2,4-triazole (3-AT; Wako) or 10 mM 3-AT were used for Y1H and Y2H screening, respectively.
Transient Reporter Assays.
Plasmids were extracted using the NucleoBond Xtra Midi Plus according to the manufacture’s instruction (Takara). Transient reporter assays using protoplasts prepared from Arabidopsis T87 suspension-cultured cells were performed and calculated as previously described (45). The fLUC activity was normalized according to the humanized renilla luciferase activity in each assay, and the relative ratio was determined by comparing this ratio with that obtained with the empty vector. The mean relative ratios were calculated from three or four independent experiments.
Primers used in this study are listed in Table S1.
Supplementary Material
Acknowledgments
We thank Taisuke Nishimura, Akiko Yamamoto, Takashi Kuroha, and Chiharu Ueguchi (Nagoya University) for helpful advice about Arabidopsis experiments; Hironaka Tsukagoshi (Nagoya University) for advice about Arabidopsis root assays; Kazuko Yamaguchi-Shinozaki and Satoshi Kidokoro (The University of Tokyo) for advice about transient expression assays using Arabidopsis mesophyll protoplast; Fumie Tobe (National Institute of Advanced Industrial Science, Japan) for technical assistance in Y1/2H screening; Tsuyoshi Nakagawa (Shimane University) for providing pGWB502, pGWnY, and pGWc; Yasuaki Kagaya (Mie University) and Akiko Yamamoto (Nagoya University) for providing pE2113_GUS; and Peter McCourt (University of Toronto) for critical reading of the manuscript and insightful comments. This work was supported by the Japan Society for the Promotion of Science (JSPS) through the World-Leading Researchers (M.U.-T.), JSPS Grant-in-Aid for Scientific Research 23113005 (to M.M.), and the Japanese Ministry of Education, Culture, Sports, Science, and Technology Program for Leading Graduate Schools “Integrative Graduate Education and Research in Green Natural Sciences” (H.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321669111/-/DCSupplemental.
References
- 1.Itoh H, Ueguchi-Tanaka M, Matsuoka M. Molecular biology of gibberellins signaling in higher plants. Int Rev Cell Mol Biol. 2008;268:191–221. doi: 10.1016/S1937-6448(08)00806-X. [DOI] [PubMed] [Google Scholar]
- 2.Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol. 2011;21(9):R338–R345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 3.Davière JM, Achard P. Gibberellin signaling in plants. Development. 2013;140(6):1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- 4.Locascio A, Blázquez MA, Alabadí D. Genomic analysis of DELLA protein activity. Plant Cell Physiol. 2013;54(8):1229–1237. doi: 10.1093/pcp/pct082. [DOI] [PubMed] [Google Scholar]
- 5.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451(7177):480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 6.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451(7177):475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zentella R, et al. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19(10):3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallego-Bartolomé J, Alabadí D, Blázquez MA. DELLA-induced early transcriptional changes during etiolated development in Arabidopsis thaliana. PLoS ONE. 2011;6(8):e23918. doi: 10.1371/journal.pone.0023918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarnowska EA, et al. DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiol. 2013;163(1):305–317. doi: 10.1104/pp.113.223933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirano K, et al. The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J. 2012;71(3):443–453. doi: 10.1111/j.1365-313X.2012.05000.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang ZL, et al. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci USA. 2011;108(5):2160–2165. doi: 10.1073/pnas.1012232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitsuda N, et al. Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 2010;51(12):2145–2151. doi: 10.1093/pcp/pcq161. [DOI] [PubMed] [Google Scholar]
- 13.Colasanti J, et al. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics. 2006;7:158. doi: 10.1186/1471-2164-7-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozaki A, Hake S, Colasanti J. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 2004;32(5):1710–1720. doi: 10.1093/nar/gkh337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heo JO, et al. Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc Natl Acad Sci USA. 2011;108(5):2166–2171. doi: 10.1073/pnas.1012215108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun X, Jones WT, Rikkerink EH. GRAS proteins: The versatile roles of intrinsically disordered proteins in plant signalling. Biochem J. 2012;442(1):1–12. doi: 10.1042/BJ20111766. [DOI] [PubMed] [Google Scholar]
- 17.Seo PJ, Ryu J, Kang SK, Park CM. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J. 2011;65(3):418–429. doi: 10.1111/j.1365-313X.2010.04432.x. [DOI] [PubMed] [Google Scholar]
- 18.Xuan YH, et al. Indeterminate domain 10 regulates ammonium-mediated gene expression in rice roots. New Phytol. 2013;197(3):791–804. doi: 10.1111/nph.12075. [DOI] [PubMed] [Google Scholar]
- 19.Welch D, et al. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 2007;21(17):2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oshima Y, et al. Novel vector systems to accelerate functional analysis of transcription factors using chimeric repressor gene-silencing technology (CRES-T) Plant Biotechnol. 2011;28(2):201–210. [Google Scholar]
- 21.Hedden P, Phillips AL. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000;5(12):523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- 22.Griffiths J, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18(12):3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Achard P, et al. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol. 2009;19(14):1188–1193. doi: 10.1016/j.cub.2009.05.059. [DOI] [PubMed] [Google Scholar]
- 24.Bai MY, et al. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol. 2012;14(8):810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogawa M, et al. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell. 2003;15(7):1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colasanti J, Yuan Z, Sundaresan V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell. 1998;93(4):593–603. doi: 10.1016/s0092-8674(00)81188-5. [DOI] [PubMed] [Google Scholar]
- 27.Wu C, et al. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc Natl Acad Sci USA. 2008;105(35):12915–12920. doi: 10.1073/pnas.0806019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsubara K, et al. Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol. 2008;148(3):1425–1435. doi: 10.1104/pp.108.125542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park SJ, et al. Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J. 2008;56(6):1018–1029. doi: 10.1111/j.1365-313X.2008.03667.x. [DOI] [PubMed] [Google Scholar]
- 30.Morita MT, et al. A C2H2-type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems. Plant J. 2006;47(4):619–628. doi: 10.1111/j.1365-313X.2006.02807.x. [DOI] [PubMed] [Google Scholar]
- 31.Tanimoto M, Tremblay R, Colasanti J. Altered gravitropic response, amyloplast sedimentation and circumnutation in the Arabidopsis shoot gravitropism 5 mutant are associated with reduced starch levels. Plant Mol Biol. 2008;67(1-2):57–69. doi: 10.1007/s11103-008-9301-0. [DOI] [PubMed] [Google Scholar]
- 32.Wu X, Tang D, Li M, Wang K, Cheng Z. Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 2013;161(1):317–329. doi: 10.1104/pp.112.208496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feurtado JA, et al. The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell. 2011;23(5):1772–1794. doi: 10.1105/tpc.111.085134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsuda N, et al. The new FioreDB database provides comprehensive information on plant transcription factors and phenotypes induced by CRES-T in ornamental and model plants. Plant Biotechnol. 2011;28(2):123–130. [Google Scholar]
- 35.Paquette AJ, Benfey PN. Maturation of the ground tissue of the root is regulated by gibberellin and SCARECROW and requires SHORT-ROOT. Plant Physiol. 2005;138(2):636–640. doi: 10.1104/pp.104.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birnbaum K, et al. A gene expression map of the Arabidopsis root. Science. 2003;302(5652):1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 37.Hassan H, Scheres B, Blilou I. JACKDAW controls epidermal patterning in the Arabidopsis root meristem through a non-cell-autonomous mechanism. Development. 2010;137(9):1523–1529. doi: 10.1242/dev.048777. [DOI] [PubMed] [Google Scholar]
- 38.Cruz-Ramírez A, et al. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell. 2012;150(5):1002–1015. doi: 10.1016/j.cell.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shani E, et al. Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc Natl Acad Sci USA. 2013;110(12):4834–4839. doi: 10.1073/pnas.1300436110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18(1):111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- 41.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20(11):1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 42.Métivier R, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115(6):751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 43.Dammer EB, Leon A, Sewer MB. Coregulator exchange and sphingosine-sensitive cooperativity of steroidogenic factor-1, general control nonderepressed 5, p54, and p160 coactivators regulate cyclic adenosine 3′,5′-monophosphate-dependent cytochrome P450c17 transcription rate. Mol Endocrinol. 2007;21(2):415–438. doi: 10.1210/me.2006-0361. [DOI] [PubMed] [Google Scholar]
- 44.Jakobsson T, Treuter E, Gustafsson JÅ, Steffensen KR. Liver X receptor biology and pharmacology: New pathways, challenges and opportunities. Trends Pharmacol Sci. 2012;33(7):394–404. doi: 10.1016/j.tips.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Maeo K, et al. An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 2009;60(3):476–487. doi: 10.1111/j.1365-313X.2009.03967.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.