Significance
Viruses that are seemingly unrelated in genomic studies, and which infect hosts in different domains of life, show similarities in virion structure that indicate deep evolutionary relationships. We report the cryo-EM structure, at near-atomic resolution, of the fungal dsRNA Penicillium chrysogenum virus. Its capsid protein is a duplication of a single primordial α-helical domain. This domain has a fold that is conserved among dsRNA viruses; it has increased its complexity through an early gene duplication event, followed by insertion of distinct segments in preferential “hotspots.” We show evidence that this preserved hallmark indicates an ancestral fold, and we suggest a relationship among current viral lineages.
Keywords: 3D cryo-EM, chrysovirus, structural homology, virus evolution
Abstract
Viruses evolve so rapidly that sequence-based comparison is not suitable for detecting relatedness among distant viruses. Structure-based comparisons suggest that evolution led to a small number of viral classes or lineages that can be grouped by capsid protein (CP) folds. Here, we report that the CP structure of the fungal dsRNA Penicillium chrysogenum virus (PcV) shows the progenitor fold of the dsRNA virus lineage and suggests a relationship between lineages. Cryo-EM structure at near-atomic resolution showed that the 982-aa PcV CP is formed by a repeated α-helical core, indicative of gene duplication despite lack of sequence similarity between the two halves. Superimposition of secondary structure elements identified a single “hotspot” at which variation is introduced by insertion of peptide segments. Structural comparison of PcV and other distantly related dsRNA viruses detected preferential insertion sites at which the complexity of the conserved α-helical core, made up of ancestral structural motifs that have acted as a skeleton, might have increased, leading to evolution of the highly varied current structures. Analyses of structural motifs only apparent after systematic structural comparisons indicated that the hallmark fold preserved in the dsRNA virus lineage shares a long (spinal) α-helix tangential to the capsid surface with the head-tailed phage and herpesvirus viral lineage.
High-resolution X-ray crystallography and near-atomic 3D cryo-EM structural studies of the capsid, the virus hallmark (1), have shown deep evolutionary relationships among distantly related viruses that infect hosts of different domains of life (2). Structure-based comparisons suggest that, based on capsid protein (CP) folds, evolution has resulted in a small number of viral lineages, probably due to capsid assembly constraints (3). The question nonetheless remains as to whether the last universal common ancestor was infected by a progenitor of current viral lineages before host and virus diverged. Icosahedral viruses are grouped in four lineages: the dsDNA viruses with an upright double β-barrel CP (prototypes are phage PRD1 and adenovirus), the head-tailed phages and herpesviruses that share the Hong Kong 97 (HK97)-like CP fold (4), the picornavirus-like superfamily with a single β-barrel as the CP fold (5), and the dsRNA or bluetongue virus (BTV)-like viruses. PRD1- and HK97-like lineages include archaeal, bacterial, and eukaryotic viruses, suggesting that the last universal ancestral host was infected by the progenitors of current viral lineages before the host organism diverged (6–9). Even poxviruses have been linked to icosahedral viruses; the vaccinia virus is related to PRD1-like viruses because the D13 scaffolding protein has two fused β-barrels, suggesting evolutionary descent from an icosahedral ancestor (10, 11).
The dsRNA viruses are found in bacteria, as well as in simple (fungi and protozoa) and complex (animals and plants) eukaryotes (12), but no archaea-infecting viruses are reported. Their genomes are isolated within a specialized icosahedral capsid or cell microcompartment that remains structurally undisturbed throughout the viral cycle, thus avoiding induction of host cell defense mechanisms (13, 14). The capsid also participates in genome metabolism by organizing packaged dsRNA molecules and viral polymerase complex(es) for transcription and replication reactions, and in the extrusion of transcripts for protein synthesis or virion progeny (15). Probably due to these stringent functional requirements, dsRNA virus capsids have a similar architecture; they are built from 60 asymmetrical dimers of a single CP in a T = 1 icosahedral lattice (i.e., 120 copies of a major CP instead of 60 copies as in standard T = 1 capsids). T = 1 capsids with 60 dimers are described only in members of the Reoviridae and Picobirnaviridae (which mainly infect higher eukaryotes), in bacteriophages of the Cystoviridae, and in Totiviridae and Partitiviridae (which infect fungi and protozoa). Eleven T = 1 layer CPs, which show negligible sequence similarity, have been resolved at atomic resolution; they include six reoviruses and a picobirnavirus (PBV), two phages (ϕ6 and ϕ8), a yeast L-A virus (a totivirus), and Penicillium stoloniferum virus F (a partitivirus) (Fig. S1). Additional capsids can be added to this inner core, as in the case of the T = 13 icosahedral capsid observed in cystovirus and reoviruses.
The Penicillium chrysogenum virus (PcV) of the Chrysoviridae has an authentic T = 1 capsid, formed by 60 copies of a 109-kDa polypeptide (16). PcV is a mycovirus with a multipartite genome consisting of four monocistronic dsRNA segments, each of which is encapsidated separately in a similar particle (17). Here, we report the 3D cryo-EM structure of the PcV capsid at near-atomic resolution; the PcV CP is a duplication of a single domain. This basic domain provides information regarding the primordial fold of the dsRNA virus lineage and its evolutionary mechanisms.
Results and Discussion
Cryo-EM Structure of PcV at Near-Atomic Resolution.
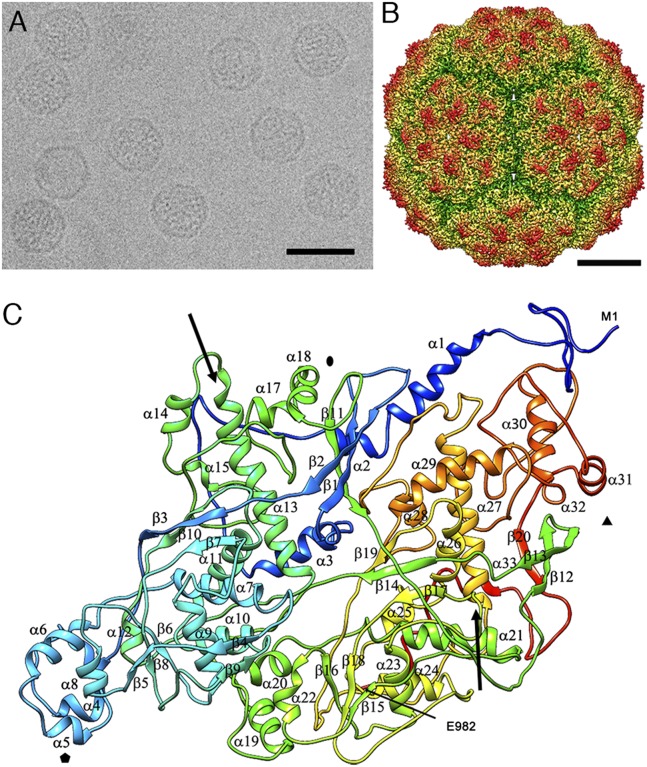
Purified PcV particles were imaged at −170 °C in a 300-kV Tecnai F30 cryoelectron microscope (Fig. 1A). A total of 27,566 particle images from 489 micrographs were merged to calculate an ∼4.1-Å resolution map, as estimated by the criterion of a 0.143 Fourier shell correlation (FSC) coefficient (Fig. S2). The 400-Å diameter T = 1 capsid shows 12 outwardly protruding pentons (Fig. 1B), each containing five copies of the CP, as described (18). Structural features, such as deep grooves of helices, separated β-strands in β-sheets, and turns, were clearly resolved, as well as 73% of the densities of voluminous side chains (Fig. S3A). The Cα model was built in parallel with the Coot and Gorgon programs. A full atomic model of the CP was built manually with Coot, initially based on consensus structural predictions for some amino acid sequence segments (Fig. S3B).
Fig. 1.
Three-dimensional cryo-EM reconstruction of PcV virions at a resolution of 4.1 Å. (A) Cryo-EM image of PcV. (Scale bar: 500 Å.) (B) Radially color-coded, surface-shaded virion capsid viewed along a twofold axis. Protruding pentamers (orange) are visible in the T = 1 lattice. (Scale bar: 100 Å.) (C) Ribbon diagram of the PcV capsid protein (top view), rainbow-colored from blue (N terminus) to red (C terminus). The first (Met1) and last (Glu982) residues are indicated. Symbols indicate icosahedral symmetry axes, and arrows indicate the longest helices: α13 and α27.
The 982-residue PcV CP (Fig. 1C), built from 33 α-helices and 20 β-strands, had two domains with similar morphology, the N-terminal A domain (residues 1–498) and the C-terminal B domain (residues 516–982). Both PcV CP halves had a long α-helix tangential to the capsid surface (Fig. 1C, arrows), the α13 helix in domain A (36 Å long, 24 residues), and the α27 helix in domain B (31.5 Å long, 21 residues).
Structural Duplication of the PcV CP.
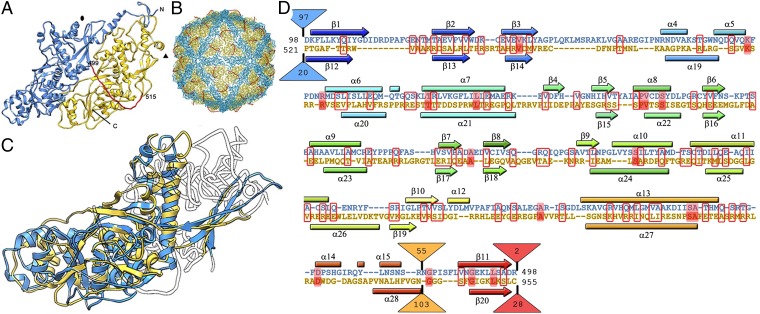
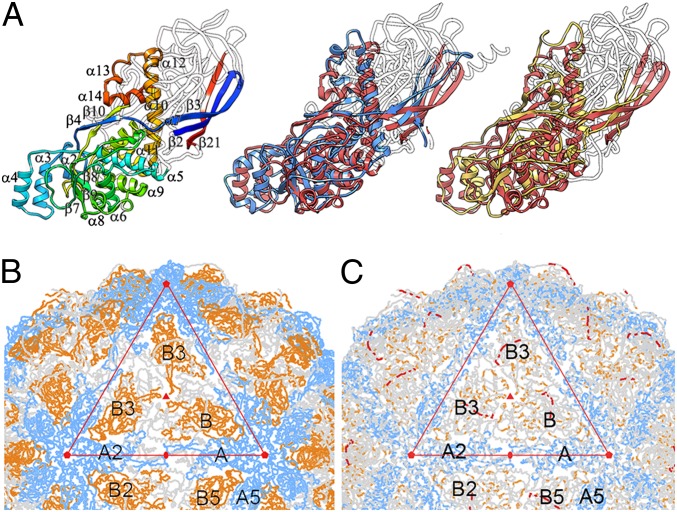
Domains A and B are connected by a 16-residue linker (Ala499-Ile515) accessible from the outer capsid surface (Fig. 2A, red). These domains are arranged in two sets of five; five A domains directly surround the icosahedral fivefold axis, and five B domains are intercalated between them, forming a pseudodecamer (Fig. 2B). This organization, shared among chrysoviruses (19), generates an architecture similar to that of the 120-subunit T = 1 capsid in reovirus, cystovirus, and totivirus, in which the asymmetrical dimer is approximately parallel. Partitivirus and PBV have a distinct quaternary organization, because their CP dimer has almost perfect local twofold symmetry (Fig. S4).
Fig. 2.
PcV capsid protein is a structural duplication. (A) Atomic model of a PcV CP showing the N-terminal domain A (1–498, blue), the linker segment (499–515, red), and the C-terminal domain B (516–982, yellow). (B) Atomic model of the PcV capsid viewed along a twofold axis. (C) Superimposed A and B domains (white regions indicate nonsuperimposed regions for both domains). (D) Sequence alignment of domains A (blue) and B (yellow) resulting from the Dali structural alignment. The α-helices (rectangles) and β-strands (arrows) are rainbow-colored from blue (N terminus) to red (C terminus) for each domain. Triangles represent nonaligned segments (sizes indicated). Strictly conserved residues are on a red background, and partially conserved residues are in a red rectangle.
The A and B α-helical domains were compact and showed substantial similarity. Structural alignment between these two domains using the Dali server showed an rmsd of 4.4 Å for 316 superposed Cα (z-score = 12.2), indicating a common fold in both CP halves; nine α-helices and nine β-strands matched very well and required only minor adjustment (Fig. 2C and Movie S1). Equivalent secondary structure elements (SSEs) had the same polarity, and β-sheets were formed by the same number of β-strands. Despite the lack of sequence similarity between the two halves (∼5%), the CP is an almost perfect structural duplication of a single domain with two continuous sequence segments, Asp98-Arg425 (domain A) and Pro521-Asn838 (domain B) (Fig. 2D). Gene duplication is a recurrent evolutionary event in other virus lineages and in bacterial carboxysome structural proteins (20), whose outer shell is reminiscent of a viral capsid (21).
In addition to the N and C termini, we detected a single “hotspot” on the outer capsid surface into which variations had been introduced by insertion of 50–100 residue segments preceding β-strands 11 and 20 of the A and B halves, respectively (Fig. 2D). A preferential insertion site would have a dual role, allowing acquisition of new functions while preserving the basic CP fold. Structural and functional requirements for capsid assembly and dsRNA metabolism have thus acted as evolutionary constraints.
Protein–Protein Interactions That Stabilize the PcV Capsid.
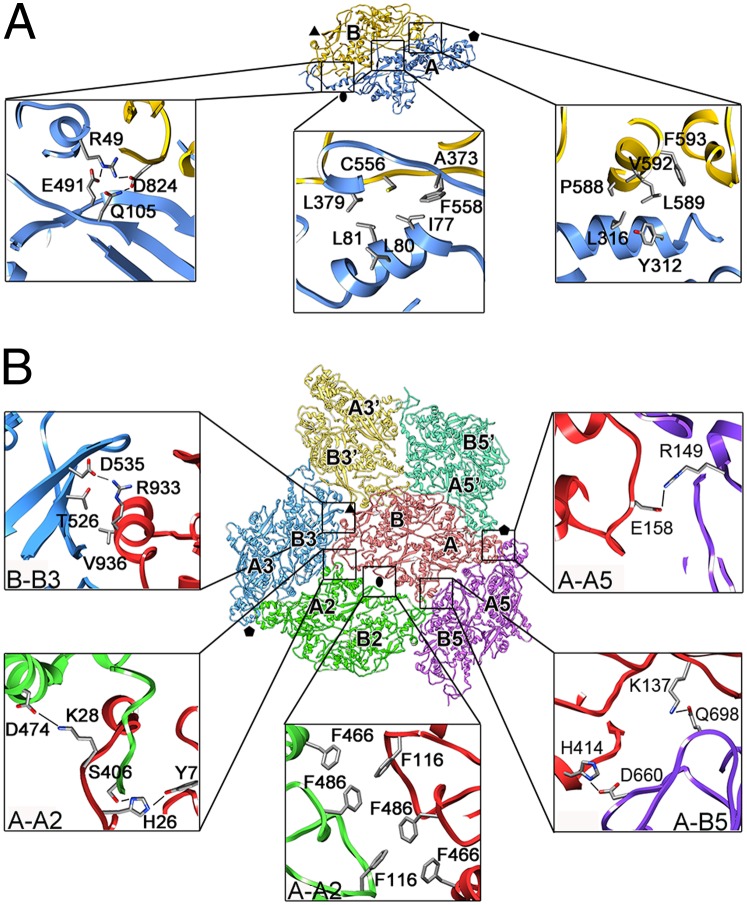
The interfaces between domains A and B had a buried area of ∼2,400 Å2, in which two distinct regions were seen; that closer to the fivefold axis had mostly hydrophobic interactions, whereas polar interactions dominated in the region closer to the twofold axis (Fig. 3A and Table S1).
Fig. 3.
Intra- and intermolecular interactions in the PcV CP. (A) Intrasubunit interactions between PcV CP domains A (blue) and B (yellow) (top view). Close-up views show three regions of the interface between domains A and B. The interface near the twofold axis (oval) has mostly polar interactions (Left), whereas hydrophobic interactions dominate in the region closer to the fivefold axis (Center and Right) (Table S1). (B) Intersubunit interactions. A PcV CP subunit (red) with surrounding subunits (green, yellow, blue, light green, and violet) is shown; each CP interacts with seven domains of adjacent CP subunits, mediated by 110 residues. The symmetry relationships of domains A and B relative to the subunit labeled A-B (red) are indicated (e.g., domains A5 are symmetrically related by a fivefold axis to domain A). Close-up views show representative interfaces. A comprehensive list of interactions is given in Table S2.
Intermolecular interactions (8,500-Å2 area) were also consistent with our CP atomic model (Fig. 3B, Table S2, and Movie S2). Each CP interacts with seven domains of adjacent CP subunits; many interactions correspond to intrapentameric contacts (64 of 110). Domain A establishes polar and ionic interactions with fivefold-related A domains (referred to as A-A5′ and A-A5), and domain B interacts with A domains of adjacent subunits mediated by hydrophobic and hydrophilic residues (B-A5′ and A-B5). The numerous contacts suggest that the pentamer constitutes the basic capsid assembly unit. Interpentameric contacts were mediated by hydrophobic interactions of A and B domains with threefold-related B domains (A-B3, B-A3′, B-B3, and B-B3′) and interactions between twofold-related A domains (A-A2). N termini also have interpentameric contacts with two asymmetrical subunits (A-A2 and A-B3).
RNA–Protein Interactions and Pores in the PcV Capsid.
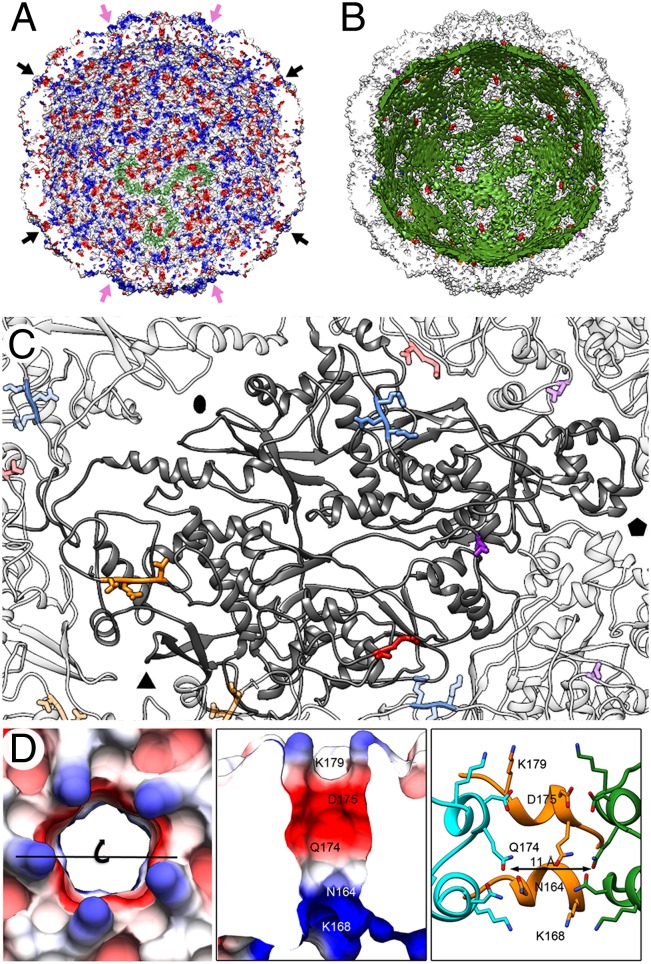
The duplicated structural signature for PcV CP reconciles the structural details observed in the PcV capsid with the concept that a 120-subunit T = 1 layer is an ubiquitous architecture for management of dsRNA metabolism. The PcV capsid has retained the capacity to organize the packaged genome, and for transcript passage to host cytoplasm (Fig. 4). The electrostatic potential of the capsid inner surface showed highly positively charged triskelion-shaped areas (Fig. 4A) that maintain RNA density in close contact with the inner capsid surface (Fig. 4B, green). In addition, there were 180 direct interactions mediated by three regions (Fig. 4C and Fig. S5A): the Arg67-Lys69 segment (Fig. 4C, blue) and the Leu981-Glu982 segment (Fig. 4C, red) of an adjacent CP, Leu564 (Fig. 4C, violet), and the Leu884-Leu887 segment (Fig. 4C, orange). The dsRNA internal ordering is a general feature in chrysoviruses, such as Cryphonectria nitschkei chrysovirus 1 (19), and might facilitate dsRNA genome mobility within the capsid for its transcription and replication. Fungal viruses, such as L-A and PcV, share a spacious capsid (18), and average genome density is looser than that of replicative cores of the Reoviridae. Reovirus T = 1 cores with a multipartite 10- to 12-segment genome are highly organized, probably to avoid tangling during segment transport through the active sites of the RNA-dependent/RNA polymerase (RdRp) complexes, which is simultaneous in independent transcription/replication reactions. In PcV, ∼50% of the genome volume is located in the ordered RNA layer in contact with the inner capsid surface, leaving the central region with disordered dsRNA at low density. The associated increase in dsRNA mobility might be necessary for optimal PcV RdRp activity that, compared with the L-A–RdRP complex, is not expressed as a fusion protein with CP.
Fig. 4.
PcV CP–dsRNA interactions and pore structure. (A) PcV capsid inner surface represented with electrostatic potentials, showing the distribution of negative (red) and positive (blue) charges. The dsRNA densities have been removed computationally. Arrows indicate the capsid pores at the fivefold (black) and threefold (pink) axes. Note the triskelion-shaped electropositive areas (one is indicated in green). (B) View as for A with the PcV capsid (white) and the dsRNA outer layer (green) in close contact with the capsid inner surface (the innermost RNA shells have been stripped away). Most of three-blade–like dsRNA densities colocalize with the triskelion-shaped areas. (C) Location of the dsRNA-interacting residues in a CP subunit (dark gray): Arg67-Lys69 (blue), Leu564 (violet), Leu884-Leu887 (orange), and Leu981-Glu982 (red). Neighboring subunits and contacting residues are similarly coded in paler colors. (D) Pores at the fivefold axes showing charge distribution on the outer surface (top view, Left) and on the channel walls (slabbed side view, Center). (Right) Side view of the channel is shown, with CP subunits (green, orange and blue) represented as ribbons and the side chains exposed at the outer surface and channel wall as sticks. Note an electronegative ring toward the outer exit (built of Asp175 and Gln174) and an electropositive ring toward the inner exit (built of Asn164 and Lys168). The channel is surrounded on the outer surface by five Lys179 residues.
The Arg67-Lys69 and Leu884-Leu887 segments were solved in the density map calculated with full particles only, suggesting a dual function: The CP would organize the genome, and the genome would act as a scaffold for some CP regions.
The ∼11-Å diameter, ∼26-Å long pores at the fivefold axis would allow the exit of ssRNA viral transcripts by a simple rearrangement of Asn164 side chains that face the channel wall (Fig. 4D). Pores at the threefold axis left an ∼5-Å diameter hole between Ala531 residues, which would allow nucleotide diffusion (Fig. 4A, pink arrows, and Fig. S5B).
Structural Comparison of PcV and L-A CPs.
The conserved fold between domains A and B (∼350 residues) constitutes the smallest polypeptide of the dsRNA T = 1 CP, and could have evolved from an ancestral domain to the structures we observed. We compared the PcV conserved domain with the Gag CP of L-A virus, a fungal virus with a single-genome dsRNA segment. Although they belong to different dsRNA families, both of these mycoviruses are transmitted by cytoplasm interchange (i.e., they lack an extracellular cycle) and have no outer shells on the rough outer surface of the T = 1 capsids, which differs from the smooth outer surface of reovirus and cystovirus capsids.
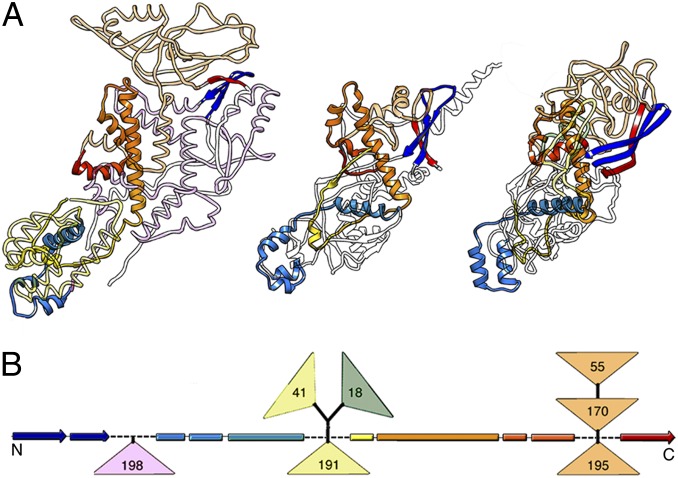
Dali structural alignment between Gag (680 residues) and the PcV A domain superimposed 288 Cα (rmsd = 4.4 Å, z-score = 14.8), and that between Gag and the PcV B domain superimposed 276 Cα (rmsd = 4.8 Å, z-score = 12.5) (Fig. 5A, Fig. S6, and Movie S3). The fold common to all three domains included eight α-helices and eight β-chains. The most similar segments between Gag and the A and B domains were near the fivefold axis, at the lateral interface between A and B subunits, and at the two- and threefold axes (Fig. 5B).
Fig. 5.
Structural homology of the PcV fold with L-A virus Gag CP. (A) Gag structure, rainbow-colored, indicating each conserved SSE relative to the PcV basic CP domain (Left) and structural alignments of domains A (blue, Center) and B (yellow, Right) with Gag (red). White segments are nonsuperimposed regions (the 36-residue N-terminal segment of domain A is omitted). (B) PcV capsid showing A (blue) and B (yellow) domain segments structurally aligned with Gag A and B monomers (variable regions, gray). Interactions at the fivefold axis (pentagons) are fully conserved, as are those at the twofold (oval) and threefold (solid triangle) axes. The large triangle defines an icosahedral face. A and B domains (or monomers for L-A) are related to neighboring domains by icosahedral symmetry (A to A5 by fivefold rotation). (C) PcV capsid showing A (blue) and B (yellow) domain segments and the linker (red) conserved among several chrysovirus CP sequences (variable regions, gray).
We found an unambiguous relationship between chrysovirus CP sequences; whereas domain A, which participates in five- and twofold axis contacts, is relatively well-conserved (43% similarity), domain B is highly variable, although most residues that interact at the threefold axis are conserved (Fig. 5C and Fig. S7). These results indicated evolutionary constraints on the CP to preserve the 120-domain or subunit capsid assembly.
Two evolutionary trends can be envisaged for dsRNA virus CP, depending on whether the basic helical structural subunit is a joined fold or not. In chrysoviruses, which have fused folds, domain A would be a relatively invariable scaffold for capsid assembly, whereas domain B would be more flexible, a region for “trial-and-error changes,” to incorporate additional functions after variation in sequence (but not in structure). In dsRNA viruses with a genuine 120-subunit capsid, however, sequence changes are restricted in those CPs that assemble as unfused identical A and B structural subunits.
The preserved fold in Gag had three peptide insertion sites facing the outer capsid surface. The peptides are segments Gln139-Ser182 (41 residues), responsible for cellular RNA decapping activity of His154 (22, 23), as well as Met301-Ala318 (18 residues) and the C-terminal 170-residue Asp417-Ser586 segment (170 residues), which colocalized with the single-insertion hotspots of the PcV CP domains (Fig. S6). This colocalization suggests that these preferential insertion sites are ancient, and could constitute a mechanism for acquisition of new functions without altering the structural and functional motifs of the dsRNA virus CP. Insertions in Gag and PcV CP, in addition to a structural role, could be responsible for enzymatic activity; this prediction could be tested by generating chimeric CPs and their assembly products.
Structural Motifs Shared in the dsRNA Virus Lineage and Similarities with HK97 CP.
Structural comparison of PcV and L-A virus CP showed a conserved domain of ∼300 residues that is probably ancestral, the basic fold of dsRNA viruses. Despite the size difference, the ∼300-residue fold can now be compared more precisely with the 1,000–1,300-residue Reoviridae T = 1 CP, which must have much longer insertions than the basic fold in fungal viruses (Fig. 6 and Fig. S8). The Reoviridae T = 1 CP can be envisaged as a large N-terminal insertion (Fig. 6 A and B, pink) followed by three α-helices that participate in the fivefold axis contacts, another large central insertion (yellow), an α-helical region (including the long spine α-helix), and another large C-terminal insertion (orange). The N- and C-terminal ends form a conserved β-sheet that takes part in the interactions at the three- and twofold axes (Fig. 6B). These three motifs are strategically placed to form a structural skeleton.
Fig. 6.
Structural comparison of PcV, L-A, and consensus reovirus CP. (A) Structural motifs common to the reovirus CP consensus model (Left), PcV A domain (Center), and L-A Gag (Right). Conserved SSEs are rainbow-colored (bright), and nonconserved segments or insertions at the N-terminal (pale pink), middle (pale yellow), or C-terminal (pale orange) region are also shown. (B) Scheme showing the conserved SSEs among dsRNA virus CPs; triangles indicate insertions (sizes are given), rectangles indicate α-helices, and arrows indicate β-strands.
Although these virus CP folds differ between dsRNA lineage members, software tools like the Homologous Structure Finder classify reovirus with cystovirus CP and group PBV, partitivirus, and totivirus in another distant branch (24). Given that dsRNA viruses originated from a common ancestor (2), our data suggest a succession of divergent evolutionary events in the ancestral PcV-like fold, first by an early gene duplication that led to two separate or tandem folds, followed by insertion of α-helical domains in (at least) three preferential sites. Alternatively, some of these α-helical domains comprised the ancestral dsRNA virus fold that diverged to the present structures, by early incorporation of the PcV-like fold followed by other N- and C-terminal insertions in some cases.
Due to the relatively large insertions in three sites, the α-helical core would be broken into basic structural motifs or subdomains only apparent after a systematic structural comparison, which could identify ancient subdomains shared by distinct viral lineages. The major CP of the HK97 phage has a long (spinal) α-helix that is also tangential to the capsid surface (25) and, with its adjacent α-helices, resembles PcV CP helices α13 and α27.
In conclusion, dsRNA viruses share a protein nanocompartment derived from the fold of the PcV CP domains, optimal for their replication strategy and genome organization. We suggest a structural relationship between the CPs of dsRNA viruses and HK97-like viruses.
Materials and Methods
Virion Preparation.
PcV virions were purified from P. chrysogenum strain American Type Culture Collection 9480 by ultracentrifugation in a sucrose density gradient as described (16).
Cryo-EM.
Purified virions (2 μL) were applied to glow-discharged, C-flat holey carbon grids (CF-1.2/1.3 400 mesh copper grids; Protochips) and vitrified using a Gatan CP3 vitrobot. Data were collected with SerialEM as described (26). Briefly, samples were imaged on a Tecnai FEG F30 electron microscope operating at 300 kV and a Gatan 626 cryo holder. Before data acquisition, the microscope was coma-free aligned and constant sample height was maintained by adjusting the eucentric height of the stage in each imaging area. A total of 489 micrographs were recorded on Kodak ISO163 film at a nominal calibrated magnification of 58,333×, a 20-e−/Å2 electron dose, and an underfocus ranging from 1.1 to 3.8 μm.
Image Processing.
General image processing operations were performed using Xmipp [http://xmipp.cnb.csic.es/ (27)], Bsoft [http://lsbr.niams.nih.gov/bsoft/ (28, 29)], and Spider [www.wadsworth.org/spider_doc/spider/docs/ (30)]. Graphics were produced by UCSF Chimera (University of California, San Francisco) (31). Selected micrographs were digitized using a Zeiss SCAI scanner at a 7-μm step size to yield a 1.20-Å pixel size on the specimen. X3d (32) was used to select 22,835 and 7,793 individual particle images of PcV full and empty particles, respectively, manually, which were further extracted and normalized with Xmipp routines. Defocus was determined twice for each micrograph with Bshow and ctffind3 (33), and phase reversals due to the contrast transfer function were inverted as required. The published PcV structure (18), low-pass filtered to 30 Å, was used as an initial model, and an Xmipp iterative projection matching routine was used to determine the origin and orientation of each particle. Full and empty particles were processed independently and joined. After each refinement iteration, a reconstruction was computed using interpolation in Fourier space. In addition, the resolution was assessed by FSC between independent half-dataset maps. After refinement, 20,552, 7,014, and 27,566 particles were included in the final three-dimensional reconstruction (3DR) for full, empty, and joined datasets, respectively. Additionally, resolution was estimated after random separation of the particle images into two datasets that were processed independently; FSC was calculated between 3DR obtained in each independent iterative processing. Amplitude decay was calculated and corrected with Embfactor (34). A B-factor of −318 A2 was calculated and applied to the entire map, although B-factors between −310 Å2 and −340 Å2 were estimated through a trial-and-error procedure for some local density areas (Fig. S9). Frequencies higher than the maximum achieved resolution were soft low-pass filtered with a soft-edged mask.
Spherically averaged radial density profiles of full and empty maps at a resolution of ∼6 Å were calculated with Spider. For difference map calculations to establish PcV CP–dsRNA interactions, radial density profiles were normalized in intensity to match the fit between the maps, and the difference was calculated by arithmetic subtraction of the empty map from the full PcV map.
The asymmetrical unit of the capsid was segmented with UCSF Chimera by evaluating the molecular boundaries between densities at different thresholds. To determine whether the segmented density corresponds to a single asymmetrical unit without loss or overlap, the full capsid was recovered by applying icosahedral symmetry to the segmented asymmetrical unit.
Atomic Model Building and Model Refinement.
The segmented asymmetrical unit was evaluated with Gorgon software (35) to identify the SSEs in the density and their possible connectivity. Coot crystallographic modeling (36, 37) was used to replace these SSEs by ideal geometry polyalanine α-helices and β-chains that were linked by evaluating the density map boundaries at different threshold levels. To determine the Cα chain fully, we locally evaluated the map at different B-factors for amplitude decay correction. The amino acid sequence was registered by identifying clear densities of bulky side chains used as landmarks for the sequence. In this process, three regions were relevant: helix α13 [residues 380–403, which matched a previous sequence-predicted 27-residue helix (residues 376–403)]; the His517-His518-Lys519 segment; and three consecutive helices, α24, α25, and α26. Once the sequence was registered, the positions of the main chain and side chains were adjusted manually and the fit of the atomic model to the density map was improved by iterative cycles of model rebuilding using Coot. During this process, the density map scale was refined to 1.23 Å per pixel (corresponding to a calibrated magnification of 56,910×), using the averaged spacing between β-strands as standard.
To evaluate and improve the accuracy of the model further, we used Refmac5 (38) to eliminate clashes and inconsistencies in geometry. Comparison of coordinates before and after Refmac5 processing was used to locate regions that required further refinement. In addition, the geometry of the model was checked iteratively with the Procheck program (39) and Coot validation tools. This process helped detection and correction of residues outside the allowed regions in the Ramachandran plot. The final modeled coordinates were refined in 10 additional cycles of Refmac5, PROCHECK, and Coot. The quality of the final model was confirmed visually by analyzing the match between map densities and coordinates and calculation of their correlation coefficient (82.4%) and R-factor (R = 39.8) with URO software (40). The final model of the icosahedral asymmetrical unit was further refined by energy minimization with Phenix (41), using the structural information “Fobs” calculated from the final icosahedral map. This automatic refinement resulted in an overall R-factor of 0.34 (0.48 at 4.15 Å). The Ramachandran plot showed 7.5% of residues in disallowed regions.
Model Analysis.
The electrostatic potential for the PcV capsid was calculated using DelPhi software (42) and surface-colored with UCFS Chimera. Dali software (43) was used for structural alignment between PcV CP A and B domains and their overlap with L-A Gag CP.
To generate a consensus structure for Reoviridae T = 1 CP, BTV VP3 nucleocapsid protein [Protein Data Bank (PDB) ID code 2btv] was used as an initial template and aligned with RDV P3 protein (PDB ID code 1uf2) using Dali (z-score = 8.4, rmsd = 4.4 Å) to define a common core. This core was then aligned with rotavirus VP2 (PDB ID code 3kz4) to obtain a new common core (z-score = 8.5, rmsd = 5.3 Å). This approach was used iteratively to include the atomic structures for the reovirus GCRV (PDB ID code 3k1q), orthoreovirus (PDB ID code 1ej6), and CPV (PDB ID code 3cnf).
Multiple Sequence Alignment.
CP sequences of PcV (UniProt accession no. Q8JVC1), CnCV1 (C. nitschkei chrysovirus 1, UniProt accession no. C7EUC3), AfuCV (Aspergillus fumigatus chrysovirus, UniProt accession no. D8L7M0), VdCV1 (Verticillium dahliae chrysovirus 1, UniProt accession no. D6QSQ4), HvV145S (Helminthosporium victoriae 145S virus, UniProt accession no. Q8JVB6) and ACDACV (Amasya cherry disease-associated chrysovirus, UniProt accession no. Q65A73) were aligned with several programs with similar results: MUSCLE [http://toolkit.tuebingen.mpg.de/muscle (44)], Mafft [http://mafft.cbrc.jp/alignment/server/ (45)], EXPRESSO (T-Coffee) [http://igs-server.cnrs-mrs.fr/Tcoffee (46)], and PROMALS3D [http://prodata.swmed.edu/promals3d/ (47)].
Data Deposition.
The PcV cryo-EM map is deposited in the Electron Microscopy Data Bank (accession no. emd-5600), and the atomic coordinates of the PcV CP are deposited in the PDB (ID code 3j3i).
Supplementary Material
Acknowledgments
We thank N. Grigorieff for continuous technical and intellectual support, stimulating discussions, and critical reading of the manuscript; C. Xu for maintaining the Brandeis EM facility and help with data collection; and C. Mark for editorial assistance. A.F.B. was supported by a grant from the Canadian National Science and Engineering Research Council. The Brandeis EM facility is supported by National Institutes of Health Grant P01 GM62580. This work was supported by Spanish Ministry of Economy and Competitivity Grant BFU 2011-29038 (to J.L.C.), Grant BIO2011-24333 (to N.V.), and Grant BIO BFU2011-25902 (to J.R.C.), and by a grant from the National Institutes of Health Intramural Research Program and the Center for Information Technology (to B.L.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.E.J. is a guest editor invited by the Editorial Board.
Data deposition: The Penicillium chrysogenum virus (PcV) cryo-EM map is deposited in the Electron Microscopy Data Bank, www.ebi.ac.uk/pdbe/emdb (EMDB accession no. emd-5600), and the atomic coordinates of the PcV capsid protein are deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3j3i).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404330111/-/DCSupplemental.
References
- 1.Krupovič M, Bamford DH. Order to the viral universe. J Virol. 2010;84(24):12476–12479. doi: 10.1128/JVI.01489-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrescia NG, Bamford DH, Grimes JM, Stuart DI. Structure unifies the viral universe. Annu Rev Biochem. 2012;81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- 3.Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15(6):655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79(23):14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58(1):533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 6.Rissanen I, et al. Bacteriophage P23-77 capsid protein structures reveal the archetype of an ancient branch from a major virus lineage. Structure. 2013;21(5):718–726. doi: 10.1016/j.str.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pietilä MK, et al. Structure of the archaeal head-tailed virus HSTV-1 completes the HK97 fold story. Proc Natl Acad Sci USA. 2013;110(26):10604–10609. doi: 10.1073/pnas.1303047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krupovic M, Bamford DH. Virus evolution: How far does the double beta-barrel viral lineage extend? Nat Rev Microbiol. 2008;6(12):941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- 9.Benson SD, Bamford JK, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell. 2004;16(5):673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 10.Bahar MW, Graham SC, Stuart DI, Grimes JM. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure. 2011;19(7):1011–1020. doi: 10.1016/j.str.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyun JK, et al. Membrane remodeling by the double-barrel scaffolding protein of poxvirus. PLoS Pathog. 2011;7(9):e1002239. doi: 10.1371/journal.ppat.1002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patton JT. Segmented Double-Stranded RNA Viruses: Structure and Molecular Biology. Norfolk, UK: Caister Academic; 2008. [Google Scholar]
- 13.Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9(1):22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 14.Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR Activation by dsRNA. J Mol Biol. 2008;381(2):351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison SC. Principles of virus structure. In: Knipe DM, et al., editors. Fields Virology. 5th Ed. Vol 1. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 59–98. [Google Scholar]
- 16.Castón JR, et al. Three-dimensional structure of penicillium chrysogenum virus: A double-stranded RNA virus with a genuine T=1 capsid. J Mol Biol. 2003;331(2):417–431. doi: 10.1016/s0022-2836(03)00695-8. [DOI] [PubMed] [Google Scholar]
- 17.Jiang D, Ghabrial SA. Molecular characterization of Penicillium chrysogenum virus: Reconsideration of the taxonomy of the genus Chrysovirus. J Gen Virol. 2004;85(Pt 7):2111–2121. doi: 10.1099/vir.0.79842-0. [DOI] [PubMed] [Google Scholar]
- 18.Luque D, et al. The T=1 capsid protein of Penicillium chrysogenum virus is formed by a repeated helix-rich core indicative of gene duplication. J Virol. 2010;84(14):7256–7266. doi: 10.1128/JVI.00432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gómez-Blanco J, et al. Cryphonectria nitschkei virus 1 structure shows that the capsid protein of chrysoviruses is a duplicated helix-rich fold conserved in fungal double-stranded RNA viruses. J Virol. 2012;86(15):8314–8318. doi: 10.1128/JVI.00802-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein MG, et al. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol. 2009;392(2):319–333. doi: 10.1016/j.jmb.2009.03.056. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka S, et al. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319(5866):1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 22.Naitow H, Tang J, Canady M, Wickner RB, Johnson JE. L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism. Nat Struct Biol. 2002;9(10):725–728. doi: 10.1038/nsb844. [DOI] [PubMed] [Google Scholar]
- 23.Tang J, et al. The structural basis of recognition and removal of cellular mRNA 7-methyl G ‘caps’ by a viral capsid protein: A unique viral response to host defense. J Mol Recognit. 2005;18(2):158–168. doi: 10.1002/jmr.724. [DOI] [PubMed] [Google Scholar]
- 24.El Omari K, et al. Plate tectonics of virus shell assembly and reorganization in phage φ8, a distant relative of mammalian reoviruses. Structure. 2013;21(8):1384–1395. doi: 10.1016/j.str.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wikoff WR, et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289(5487):2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci USA. 2008;105(6):1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marabini R, et al. Xmipp: An Image Processing Package for Electron Microscopy. J Struct Biol. 1996;116(1):237–240. doi: 10.1006/jsbi.1996.0036. [DOI] [PubMed] [Google Scholar]
- 28.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157(1):3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Heymann JB. Bsoft: Image and molecular processing in electron microscopy. J Struct Biol. 2001;133(2-3):156–169. doi: 10.1006/jsbi.2001.4339. [DOI] [PubMed] [Google Scholar]
- 30.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 31.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 32.Conway JF, et al. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J Struct Biol. 1993;111(3):222–233. doi: 10.1006/jsbi.1993.1052. [DOI] [PubMed] [Google Scholar]
- 33.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 34.Fernández JJ, Luque D, Castón JR, Carrascosa JL. Sharpening high resolution information in single particle electron cryomicroscopy. J Struct Biol. 2008;164(1):170–175. doi: 10.1016/j.jsb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 35.Baker ML, et al. Modeling protein structure at near atomic resolutions with Gorgon. J Struct Biol. 2011;174(2):360–373. doi: 10.1016/j.jsb.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vagin AA, et al. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231(4):1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 40.Navaza J, Lepault J, Rey FA, Alvarez-Rua C, Borge J. On the fitting of model electron densities into EM reconstructions: A reciprocal-space formulation. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 10 Pt 2):1820–1825. doi: 10.1107/s0907444902013707. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rocchia W, et al. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: Applications to the molecular systems and geometric objects. J Comput Chem. 2002;23(1):128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 43.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16(6):566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 44.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Armougom F, et al. Expresso: Automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34(Web Server issue):W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pei J, Kim BH, Grishin NV. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36(7):2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]