Significance
DNA damage can be a potent block to replication. One pathway to bypass damage is translesion synthesis (TLS) by specialized DNA polymerases. These conserved TLS polymerases have higher error rates than replicative polymerases, requiring careful regulation of polymerase exchange. By reconstituting the full polymerase exchange reaction at the single-molecule level, we show how distinct sets of binding sites on the β processivity clamp regulate exchange between the Escherichia coli replicative polymerase (Pol) III and the TLS Pol IV. At low concentrations, Pol IV binds β in an inactive binding mode, promoting rapid bypass of a cognate DNA lesion, whereas at high concentrations (corresponding to SOS damage response levels), association at a low-affinity sites facilitates Pol III displacement.
Keywords: single-molecule techniques, DNA replication, DNA repair, lesion bypass, DinB
Abstract
Translesion synthesis (TLS) by Y-family DNA polymerases alleviates replication stalling at DNA damage. Ring-shaped processivity clamps play a critical but ill-defined role in mediating exchange between Y-family and replicative polymerases during TLS. By reconstituting TLS at the single-molecule level, we show that the Escherichia coli β clamp can simultaneously bind the replicative polymerase (Pol) III and the conserved Y-family Pol IV, enabling exchange of the two polymerases and rapid bypass of a Pol IV cognate lesion. Furthermore, we find that a secondary contact between Pol IV and β limits Pol IV synthesis under normal conditions but facilitates Pol III displacement from the primer terminus following Pol IV induction during the SOS DNA damage response. These results support a role for secondary polymerase clamp interactions in regulating exchange and establishing a polymerase hierarchy.
Despite the action of several DNA repair pathways, unrepaired damage is encountered by replicative DNA polymerases, which stall at DNA-distorting lesions. Translesion synthesis (TLS), most notably by Y-family polymerases, is one pathway that alleviates such roadblocks. In TLS, a Y-family polymerase switches with a stalled replicative polymerase, synthesizes across from and past the lesion, and then, switches back to allow resumption of normal synthesis (1). The ability of Y-family polymerases to bypass damaged DNA comes at the cost of lower fidelity, requiring careful regulation of polymerase exchange (2).
Processive synthesis by DNA polymerases requires their tethering to the protein-binding cleft of a ring-shaped processivity clamp by a conserved clamp-binding motif (CBM). Canonical clamps, such as the bacterial β and eukaryotic proliferating cell nuclear antigen (PCNA), are multimeric, with a binding cleft on each protomer. Biochemical experiments with bacterial (3, 4) and eukaryotic (5) proteins have suggested that clamps can simultaneously bind multiple DNA polymerases during active DNA synthesis, serving as a molecular toolbelt (6). This multivalency may facilitate rapid polymerase exchange and lesion bypass. However, it remains unclear if and when large multisubunit replicative polymerases can accommodate Y-family polymerases on the clamp. Furthermore, most organisms have multiple Y-family polymerases and many other clamp-binding proteins—at least 10 in Escherichia coli (7) and over 50 in humans (8). It is consequently an open question how the correct polymerase is selected at a DNA lesion (1).
To further elucidate the role of processivity clamps in polymerase trafficking, we studied the E. coli replicative polymerase, the polymerase (Pol) III heterotrimer αεθ, and the Y-family Pol IV, which individually bind the dimeric β clamp. Pol IV, encoded by the gene dinB, is widely conserved across the three domains of life, and it is the homolog of human Pol κ (9). In addition to its function in lesion bypass (10), E. coli Pol IV is required for the mechanistically controversial phenomenon of stress-induced mutagenesis (11, 12), which is proposed to occur by its preferential synthesis at double-strand break intermediates (13, 14), and involved in reactive oxygen species-mediated antibiotic lethality by its incorporation of oxidized nucleotides into the genome (15).
Interactions of Pol IV with β and their implications for the toolbelt model have generated widespread interest (4, 16). The structure of the C-terminal little finger domain of Pol IV bound to β revealed that Pol IV can simultaneously interact with the cleft and the rim, a secondary site of β near its dimer interface, which positions the Pol IV catalytic domain well away from the DNA running through the center of the clamp (17). Although this potential inactive binding mode for Pol IV has been interpreted as evidence for the toolbelt model, Pol III was shown to contain a second weak CBM in its ε exonuclease subunit (18) in addition to the CBM in its α catalytic subunit that has a strong affinity for the β clamp. A recent study proposed that Pol III would, therefore, occlude Pol IV from clamp binding during replication, only accommodating simultaneous binding after a lesion-induced stall (19).
Previous efforts to reconstitute this model system for polymerase exchange have involved stalling Pol III on β at a primer terminus by nucleotide omission to synchronize a population of molecules and simulate a lesion-induced block (3, 4). These studies were not able to resolve an exchange back to Pol III after Pol IV synthesis. To bypass these limitations and elucidate the molecular mechanism of exchange between Pol III and Pol IV, we developed a single-molecule assay to observe the whole TLS reaction, quantifying polymerase exchange and bypass at site-specific DNA lesions. Here, we show that Pol III and Pol IV can simultaneously bind β during active synthesis, enabling rapid lesion bypass, and report a previously unidentified inactive binding mode for Pol III. We also observe that, at high concentrations (corresponding to up-regulated levels during the SOS DNA damage response), Pol IV occupies a secondary contact on β, promoting dissociation of Pol III. These results support a model in which secondary contacts between processivity clamps and Y-family polymerases establish a hierarchy for polymerase selection.
Results
Single-Molecule Assay to Measure DNA Polymerase Activity.
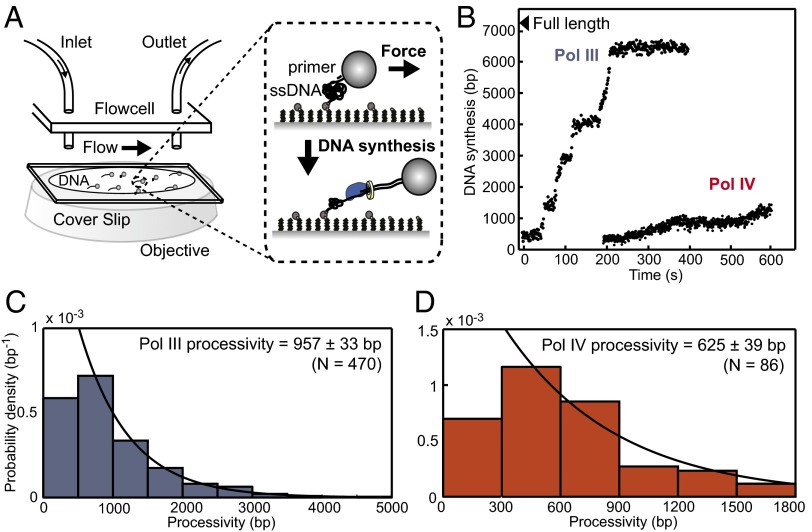
We used an assay that exploits the differential elasticity of ssDNA and dsDNA to observe primer extension on individual DNA molecules within a microfluidic flow cell (20, 21). Each primed ssDNA substrate, constructed from a 7.2-kb phage M13 genome (Fig. S1A), was coupled to a micrometer-scale bead. Laminar flow of buffer through the flow cell exerts a constant force of ∼3 pN on the bead and by extension, uniformly throughout the DNA tether. At this low force, ssDNA is entropically coiled, whereas dsDNA is stretched to nearly its crystallographic length (Fig. S1C); conversion of ssDNA to dsDNA causes motion of the bead in the direction of buffer flow and can be tracked with high accuracy in space (σ ∼ 70 bp) and time (0.5 s) to determine the amount of DNA synthesized (Fig. 1A). DNA synthesis began after the introduction of the components required to reconstitute processive synthesis—polymerase(s), β, the clamp loader complex τ3δδ′χψ, and nucleotides.
Fig. 1.
A single-molecule primer extension assay. (A) Under an applied force of ∼3 pN, ssDNA is entropically collapsed, whereas dsDNA is extended to nearly its crystallographic length. Primer extension results in motion of tethered beads in the direction of flow. (B) Representative trajectories of synthesis by Pol III and Pol IV on individual DNA molecules (Fig. S1 E and F). (C and D) Processivity distributions for (C) Pol III (5 nM) and (D) Pol IV (30 nM); values represent means ± SEMs.
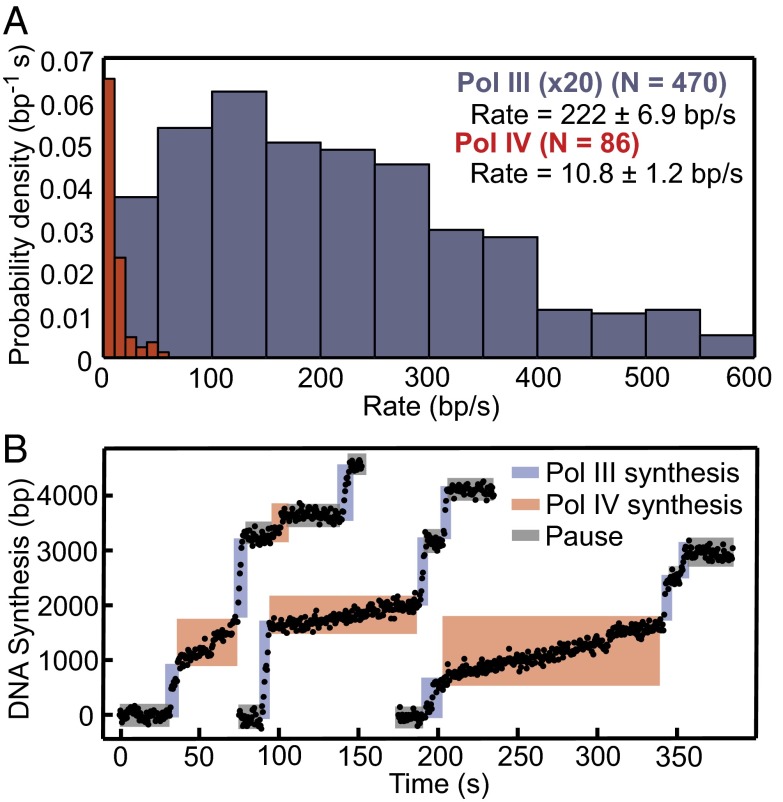
Using this technique, we characterized primer extension by Pol III and Pol IV. Synthesis by either polymerase occurred in discrete steps of processive synthesis interspersed by pauses (Fig. 1B and Fig. S1 E and F). Distributions for the processivity (Fig. 1 C and D) and rate (Fig. 2A) of each synthesis step were generated from a large number of events; the data were in agreement with previous single-molecule experiments for Pol III (22) and bulk data for Pol IV (23). Pauses between synthesis steps were exponentially distributed, consistent with a single rate-limiting step, and we observed that increasing the concentration of Pol III from 5 to 30 nM reduced the pause length (Fig. S2 A–C, time constant τ decreases from 19.7 to 12.4 s). Biophysical and structural data suggest that only one Pol III binds the clamp dimer (4, 18, 24, 25), arguing that pauses observed during synthesis result from stochastic dissociation of Pol III from the clamp and the diffusion-limited recruitment of a new polymerase from solution (22).
Fig. 2.
Observing exchange between Pol III and Pol IV. (A) Rate distributions for Pol III (blue) and Pol IV (red); values represent means ± SEMs. Probability densities for Pol III are multiplied by 20 to facilitate comparison. (B) Sample trajectories of rapid exchange between Pol III and Pol IV.
In contrast, results from structural and biophysical experiments suggest that two Pol IV molecules may simultaneously bind to the dimeric β (4, 17). To test this model, we purified a mutant clamp with a single binding cleft, β+/βC (26). Although increasing the concentration of Pol IV from 5 to 30 nM also decreased pauses between Pol IV synthesis steps (Fig. S2 D–F, τ decreases from 58.9 to 16.8 s), pausing was not affected by the use of β+/βC (Fig. S2G). The Pol IV processivity, however, dropped almost in one-half in experiments with β+/βC (Fig. S3). Together, these data imply that two Pol IV molecules can occupy β simultaneously but that exchange between the two occurs on a timescale faster than our resolution, increasing the apparent processivity. Similar to Pol III, the concentration-dependent pauses observed result from recruitment of a Pol IV molecule to the clamp from solution.
Observation of Pol III–Pol IV Exchange and Lesion Bypass.
The dramatically different rates of the two polymerases (Fig. 2A) enable assignment of synthesis events to either Pol III or Pol IV. We, therefore, performed primer extension with a mixture of Pol III (5 nM) and Pol IV (30 nM). This ratio was chosen to approximate that found in cells during exponential growth (2), with concentrations reduced (from about 20 nM for Pol III and 300 nM for Pol IV) so that distinct synthesis events could be resolved. If the fraction of active protein differs for each one, then the molar ratio of active polymerases will be shifted by a constant factor. Under these conditions, Pol III performed 78% of DNA synthesis (Fig. S4), likely because of stronger interactions with β (2); however, one or more Pol IV events were also observed in 75% of trajectories (Fig. 2B), exchanging with Pol III.
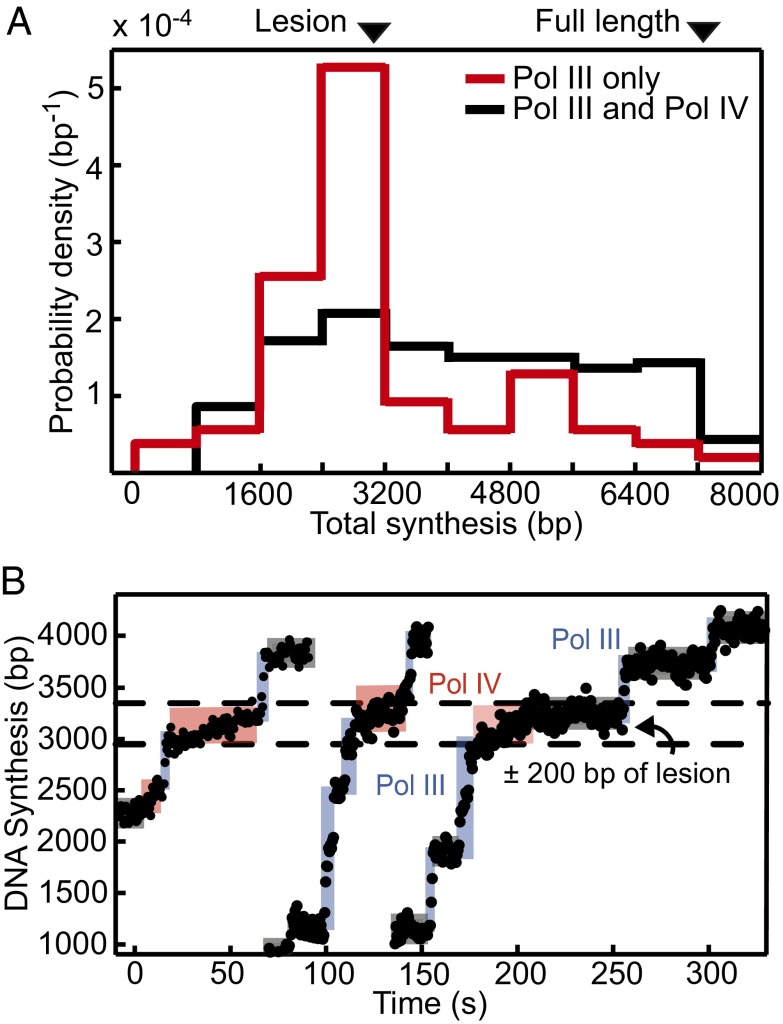
To observe polymerase exchange in the physiological context of a DNA lesion, we adapted a protocol (27) to generate single-stranded M13 substrates with a site-specific N2-furfuryl-dG adduct: a cognate lesion for Pol IV (Fig. S1B). N2-furfuryl-dG is a minor groove lesion that is efficiently and accurately bypassed by Pol IV and an analog of the primary adduct formed by the antibiotic nitrofurazone, an agent to which Pol IV KO strains are significantly sensitive (10).
Although the lesion strongly blocked Pol III in ensemble synthesis in the absence of the clamp (Fig. S5A), we found that it blocked only 65% of trajectories in single-molecule experiments (Fig. 3A and Fig. S5B). Previous studies have shown that Pol III lesion bypass efficiency is strongly promoted by β (28) and increased dNTP levels, which bias polymerase over exonuclease activity (29); we, indeed, observed that higher dNTP levels increased bypass (Fig. S5B). The addition of both polymerases to the primer extension reaction alleviated the block at the N2-furfuryl-dG position (Fig. 3A) and revealed polymerase exchange at the lesion site and bypass by Pol IV (Fig. 3B).
Fig. 3.
A single-molecule reconstitution of polymerase exchange and bypass at a DNA lesion. (A) An N2-furfuryl-dG lesion at ∼3,150 bp blocks processive synthesis by Pol III (5 nM; n = 69) but is bypassed when Pol IV (30 nM) is added (n = 175). (B) Rapid exchange from Pol III to Pol IV and back is observed at the lesion site.
Kinetics of Polymerase Exchange Support the Toolbelt Model.
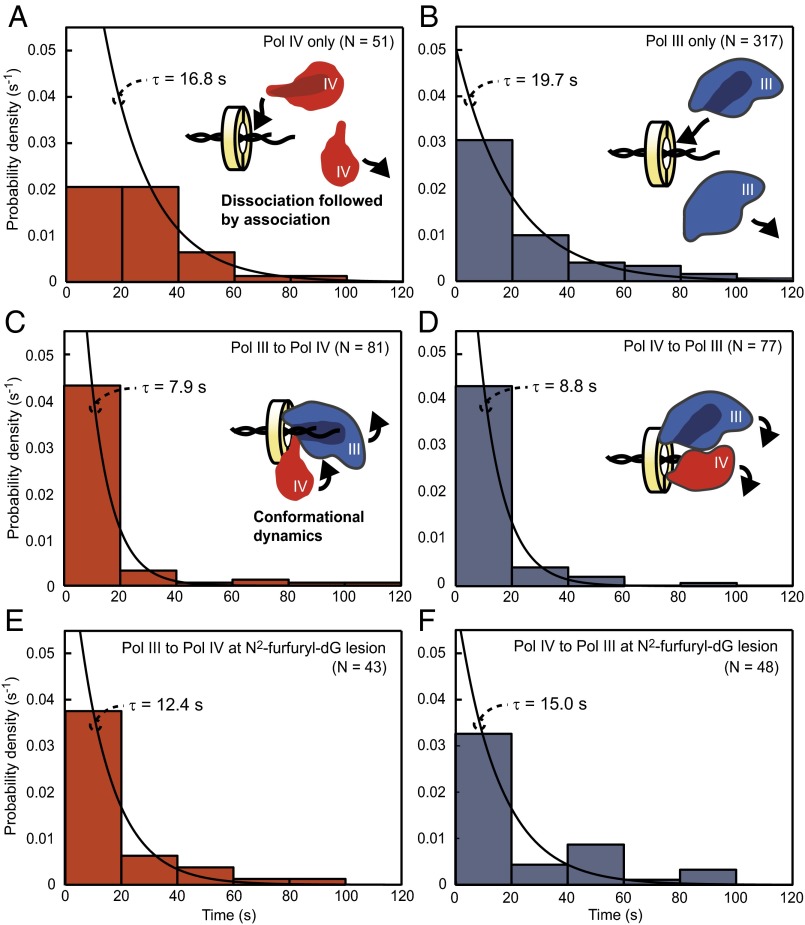
The observation of exchange from Pol III to Pol IV and back is uniquely accessible in our single-molecule reconstitution and permits us to investigate the role of β in polymerase trafficking. In the toolbelt model, polymerase exchange is only limited by the timescale of conformational changes of Pol III and Pol IV simultaneously bound to β. In an alternative model, in which steric effects prevent concurrent binding, exchange requires dissociation of the first polymerase followed by recruitment of the second from solution, which would be sensitive to protein dilution. Quantifying exchange by measuring the time between the termination of synthesis by one polymerase and the subsequent initiation of synthesis by the other allows us to distinguish between these two models.
The time for exchange from Pol III to Pol IV on undamaged DNA (Fig. 4C) was more rapid than the diffusion-limited recruitment time of Pol IV from solution (Fig. 4A and Fig. S2) seen in exponential fits and a statistical comparison of the two datasets (P < 10−5) (SI Experimental Procedures). Furthermore, reducing the concentration of Pol IV in exchange experiments from 30 to 15 nM did not affect the timescale of exchange (Fig. S6), whereas the same dilution increased pause times in experiments with Pol IV alone (Fig. 4A and Fig. S2E). These data argue that exchange during active synthesis occurs between two polymerases bound to the clamp. Our observation of β-mediated exchange implies that the second Pol III CBM in the ε subunit does not exclude Pol IV from binding the clamp in the absence of a lesion-induced stall in contrast to a previous suggestion (19); rather, Pol IV can compete with ε for a cleft, allowing Pol IV to bind β while Pol III is synthesizing DNA.
Fig. 4.
β acts as a molecular toolbelt for Pol III and Pol IV to promote lesion bypass. Each distribution is fit to an exponential with associated time constant τ. Pauses represent association of a new polymerase from solution in experiments with (A) Pol IV (30 nM) or (B) Pol III alone (5 nM). (C) Exchange from Pol III (5 nM) to Pol IV (30 nM; P < 10−5 vs. A) and (D) back to Pol III (P < 10−9 vs. B) is rapid because of simultaneous binding of both polymerases to the clamp. (E) Exchange to Pol IV at the N2-furfuryl-dG site is also rapid (P < 0.01 vs. A and NS vs. C), indicating that lesion bypass is β-mediated. (F) Exchange back to Pol III is intermediate between B (P = 0.04) and D (P = 0.02), suggesting that both types of exchange occur. SI Experimental Procedures has additional analysis.
Importantly, we observed that exchange back to Pol III after Pol IV synthesis was also more rapid than the recruitment time of Pol III from solution (Fig. 4 B and D) (P < 10−9). Furthermore, exchange from Pol IV to Pol III in the presence of the single-cleft β+/βC (Fig. S7A, τ = 27.3 s) was slower than in experiments with the WT clamp (P < 10−3), closely matching the recruitment time of Pol III from solution [not significant (NS) vs. Fig. 4B]. These data show that Pol III can bind the opposing cleft of β in an inactive conformation while Pol IV is carrying out synthesis and that eliminating the second cleft abolishes rapid exchange. The fact that the Pol IV processivity preceding exchange to Pol III matches that of Pol IV on β+/βC (Figs. S3B and S7B) shows that a single Pol IV is bound to β in the presence of Pol III and strongly implies that Pol III does not displace Pol IV during the exchange back but takes over after Pol IV stochastically releases DNA.
To test if polymerase exchange can occur in the physiological context of Pol III encountering a DNA lesion, we collected data for exchange from Pol III to Pol IV within the experimental resolution (±200 bp) of the N2-furfuryl-dG position (Fig. 3B). These exchange times (Fig. 4E) matched the times for exchange on undamaged DNA (NS vs. Fig. 4C) and were more rapid than recruitment of Pol IV from solution (P < 0.01 vs. Fig. 4A), indicating that Pol IV was bound to β when Pol III encountered the lesion. The switch back to Pol III after the lesion (Fig. 4F) was intermediate between rapid β-mediated exchange and recruitment from solution, suggesting a mixture of these two types of exchange.
Binding of Pol IV at a Secondary Site on β Reduces the Pol III Processivity.
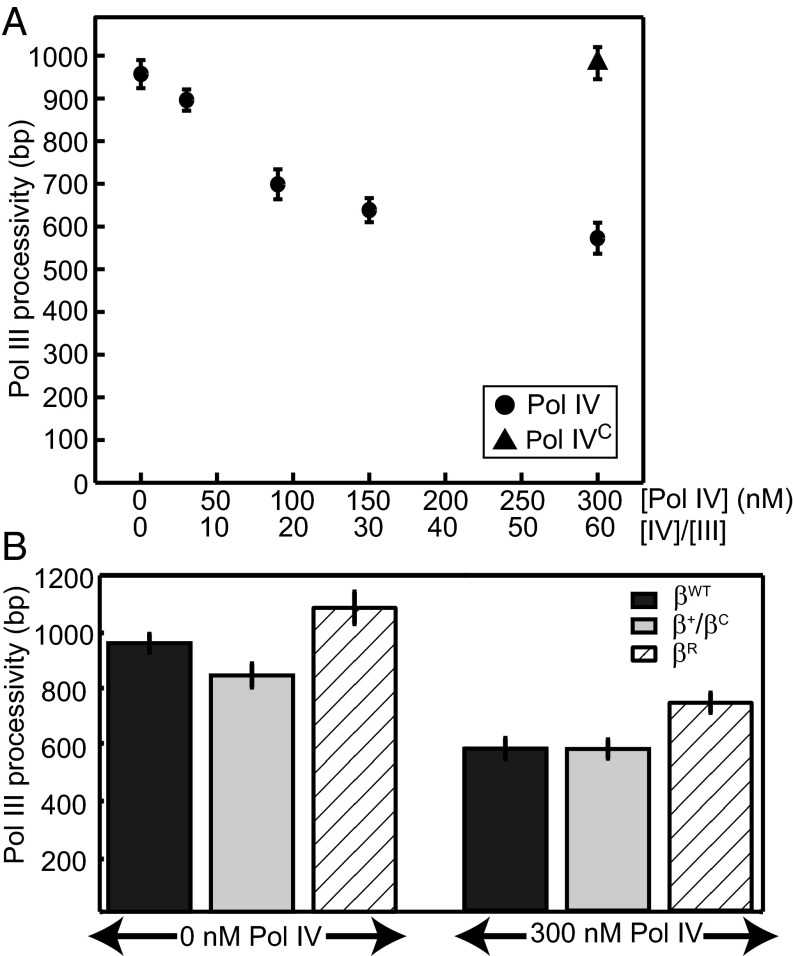
Our results support a model in which Pol IV, at a ratio to Pol III consistent with normal growth, can bind β in an inactive conformation, thereby promoting rapid bypass of DNA lesions encountered during synthesis. During the SOS DNA damage response, however, the cellular concentration of Pol IV increases roughly 10-fold, whereas Pol III levels remain constant (2). To test if an increased ratio of Pol IV to Pol III alters polymerase exchange, we performed primer extension experiments with 5 nM Pol III and 300 nM Pol IV. At these concentrations, Pol IV outcompeted Pol III, performing 78% of DNA synthesis (Fig. S4). Although the average processivity of Pol III synthesis preceding exchange was only modestly affected by Pol IV under normal conditions, possibly because of disruption of the binding of the ε subunit of Pol III to β (Fig. S8A), under SOS-like conditions, it dropped almost by half (Fig. 5A) (P < 10−5), reflecting a decrease of the lifetime of Pol III on the clamp (Fig. S8B). This reduction in processivity was dose-dependent with the Pol IV concentration and β-mediated; Pol IVC, a mutant that lacks its CBM, did not reduce the Pol III processivity (Fig. 5A).
Fig. 5.
Binding of Pol IV at a secondary site on β reduces the Pol III processivity. (A) The Pol III processivity decreases with increasing concentrations of Pol IV (●) but is unaffected by Pol IVC, a mutant lacking its CBM (▲). (B) Reduction in the Pol III processivity with βWT (black bars) also occurs with the single-cleft mutant clamp β+/βC (gray bars; NS) but is partially alleviated by βR, a clamp with a weakened Pol IV-interacting rim interface (cross-hatched bars; P < 0.01). N ranges from 71 to 470 for A and B, respectively. Values represent means ± SEMs.
To further define which interactions with β mediate this activity, we tested the effects of mutant clamps under SOS-like conditions; βR, a clamp mutant that weakens the secondary Pol IV–β-interaction at the rim site (4), did not affect the synthesis of Pol III alone but partially restored the Pol III processivity at a high Pol IV concentration (Fig. 5B) (P < 0.01). Although the processivity of Pol III was reduced slightly with the single-cleft β+/βC, consistent with the model that the ε subunit stabilizes it on the clamp (18, 19), a high concentration of Pol IV with β+/βC reduced the Pol III processivity equivalently to the WT clamp condition (Fig. 5B), further supporting a role for the noncleft rim contact in Pol III displacement.
Discussion
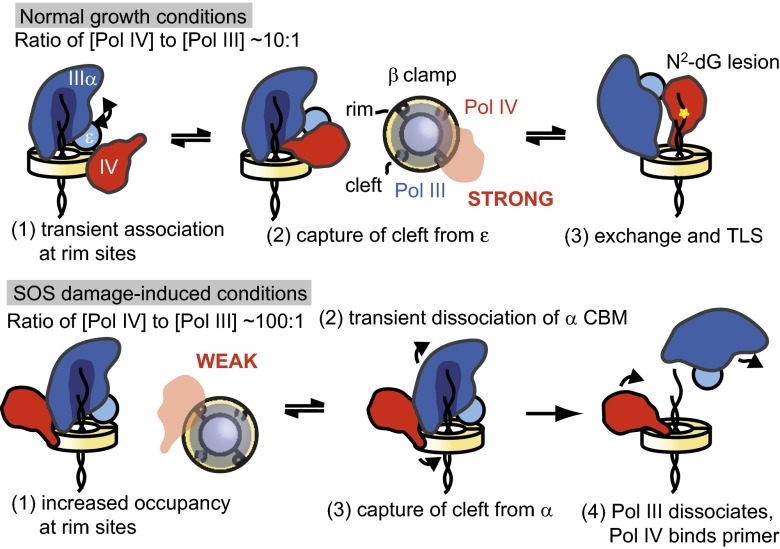
By visualizing the TLS reaction in its entirety at the single-molecule level, our data provide a comprehensive view of how Pol IV access to the replication fork is regulated through interactions with β (Fig. 6). Pol IV, at relatively low concentrations during normal growth, is able to associate with the rim site and compete with the weakly bound ε subunit of Pol III for its cleft (Fig. 4). By occupying the rim and cleft sites of β in an inactive mode during normal growth conditions, Pol IV is available for rapid exchange and translesion synthesis when Pol III stalls on encountering a lesion, which was proposed in the toolbelt model (6).
Fig. 6.
A model for how the Pol IV occupancy at rim sites and competition with Pol III subunits dictate polymerase exchange at different Pol IV concentrations. The small θ subunit of Pol III, which binds ε, is not represented for clarity.
We have also shown that a previously unidentified inactive binding mode for Pol III allows it to remain bound to the cleft of one protomer of β until the switch back (Fig. 4 and Fig. S7A), the other half of the polymerase exchange reaction that has been difficult to resolve by bulk biochemical studies (3, 4). The Pol IV processivity preceding a switch back to Pol III is not reduced from that of Pol IV alone (Fig. S7B), suggesting that Pol III does not actively displace Pol IV during translesion synthesis but relies on the lower processivity of Pol IV to minimize the mutagenic load.
At higher concentrations of Pol IV (corresponding to the SOS damage response), we observed a decrease in the Pol III processivity (Fig. 5A). We propose that this decrease is caused by an increased occupancy of Pol IV at the low-affinity rim sites of β (Fig. 6). Adjacent to the Pol III α subunit, Pol IV would be positioned to dynamically replace the strongly bound CBM of α during a transient release from the cleft, which can be seen in a molecular model of both polymerases bound to β (Fig. S9). This role for the rim contact is consistent with biochemical results that β+/βC can support polymerase exchange to Pol IV when Pol III is stalled by nucleotide omission (4). Without the critical α–β contact after cleft capture by Pol IV, Pol III would dissociate from the primer terminus, allowing Pol IV to take its place. Unless the displaced Pol III could bind an adjacent unoccupied protomer or is stabilized by additional interactions, it would likely dissociate from the clamp entirely.
The requirement of the Pol IV CBM indicates that binding at rim sites is not sufficient for a reduction of the Pol III lifetime on DNA and that Pol IV must also compete for the cleft bound by α. Shared contacts, such as a single cleft of β during competition between Pol III and Pol IV bound at additional sites on β, have been proposed to be important in facilitating dissociation and subunit exchange in multiprotein complexes upon transient contact release (30). This phenomenon has also been observed in the dynamic processivity of phage T4 and T7 replication, where additional polymerases are able to associate with moving replisomes and undergo exchange on a timescale faster than that of stochastic dissociation of the synthesizing polymerase (31–33), and the facilitated dissociation of the E. coli DNA-binding protein Fis by nucleoid proteins (34). Secondary contacts, such as the rim site for Pol IV shown here, play an important role in orienting proteins to exploit transient changes in occupancy of these shared sites, resulting in binding partner exchange.
During coordinated leading and lagging strand replication, a displaced Pol III may remain associated with the replisome through additional contacts with the clamp loader complex. These contacts, however, do not seem to prevent Pol IV from accessing β; previous biochemical experiments with a fully reconstituted replisome have shown that Pol IV can replace Pol III (35). Furthermore, overexpression of Pol IV beyond SOS levels in cells has been shown to arrest replication and induce toxicity because of unregulated access of Pol IV to the replication fork (15, 35, 36). Removing the CBM residues alleviates Pol IV toxicity, whereas mutating the rim-contacting residues partially alleviates it (16). These data are explained by our model: contacts with the rim site and subsequently, the cleft provide a molecular path for Pol IV to displace Pol III from the primer terminus after SOS induction.
A putative interaction with the rim site could also position the other E. coli Y-family polymerase, Pol V, on the clamp when it is expressed later in the SOS damage response (37). This binding activity would create a hierarchy for access to the primer terminus, a view of the toolbelt model in which clamp–polymerase interactions do more than merely increase the local polymerase concentration at the DNA template. During normal growth conditions, Pol III, which is preferentially loaded to the primer terminus (38), performs the majority of DNA synthesis. Pol IV is able to simultaneously bind β in an inactive mode, although it is currently unclear how other β-interacting proteins would influence its occupancy in vivo. After SOS induction, the rim site positions Pol IV to preferentially bind a cleft of β when it becomes available. Such a competitive advantage would ensure timely access of Y-family polymerases to the primer template and is likely to be important with several proteins competing for an open cleft.
Noncleft contacts may play a similar role in regulating access to the DNA template in other domains of life. PCNA plays a key role in coordinating the handoff of DNA intermediates between a polymerase, flap endonuclease, and ligase during both Okazaki fragment maturation and long-patch base excision repair in eukaryotes (39). Structural studies have revealed that Flap endonuclease-1, Polymerase β, and Ligase I bind overlapping but distinct regions of DNA intermediates, which may facilitate displacement during handoff (40). In the archaeon Sulfolobus solfataricus, the three enzymes each bind distinct monomers of the heterotrimeric PCNA during Okazaki fragment maturation (41). However suggestive, it remains unknown if the homotrimeric eukaryotic PCNA can simultaneously bind any combination of these three proteins.
Eukaryotic TLS is regulated, in part, by ubiquitination of PCNA; all four human Y-family polymerases have ubiquitin-binding domains (1). Structural data of monoubiquitinated PCNA and its conformations support the model that the secondary ubiquitin site allows the TLS polymerase Pol η to bind an occupied clamp and position it to compete for the cleft on transient dissociation of the replicative polymerase (42, 43). In contrast to bacteria, where the occupancy of the rim site is controlled by polymerase concentration, analogous sites in eukaryotes are introduced by posttranslational modification. We anticipate that the single-molecule approaches described here will serve as powerful tools to elucidate the role of these interactions in translesion synthesis.
Experimental Procedures
Proteins and Buffers.
Pol III core (αεθ), Pol IV, β (WT and mutants), and other E. coli replisome components were expressed and purified as described in SI Experimental Procedures. Experiments were performed in replication buffer [50 mM Hepes-KOH, pH 7.9, 12 mM Mg(OAc)2, 80 mM KCl, 0.1 mg mL−1 BSA] with 5 mM DTT, 1 mM ATP, 760 μM dNTPs, 15 nM τ3δδ′χψ, 30 nM clamp (β, βR, or β+/βC as dimers), and the indicated concentrations of Pol III and/or Pol IV; 60 μM dNTPs were used for single-molecule lesion bypass experiments. Single-stranded DNA-binding (SSB) protein was excluded from primer extension experiments, because SSB protein extends ssDNA at low force, reducing the contrast with dsDNA that is used to observe replication (44).
ssDNA Constructs.
DNA constructs were generated from circular M13 ssDNA with either M13mp18 ssDNA (New England Biolabs) or purified M13mp7(L2) containing a site-specific N2-furfuryl-dG lesion and constructed using a previously described protocol (27). M13mp7(L2) phage stock was a gift from John Essigmann (Massachusetts Institute of Technology, Cambridge, MA). Additional details on substrate construction and a list of oligonucleotides used (Table S1) can be found in SI Experimental Procedures.
Single-Molecule Experiments.
Single-molecule primer extension experiments were performed in custom microfluidic flow cells constructed with functionalized glass coverslips as described previously (45). Statistical comparisons were made between full datasets with the two-tailed Wilcoxon rank sum test using the Matlab function ranksum. A significance level of P = 0.05 was used. Additional details on the experimental approach and data analysis are described in SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Jamie Foti for providing purified Pol IV, Deyu Li for assistance with purifying the N2-furfuryl-dG oligonucleotide, and Johannes Walter and Seungwoo Chang for helpful discussions and critical reading of the manuscript. This work was funded by National Institutes of Health Grants T32 GM008313 (to J.E.K.), R01 GM066094 (to M.D.S.), R01 CA021615 (to G.C.W.), and P30ES002019 (to G.C.W.); a National Science Foundation Graduate Research Fellowship (to J.E.K.); the Smith Family Award in Biomedical Excellence (to J.J.L.); the Stuart Trust Fellows Award (to J.J.L.); and Australian Research Council Grant DP0877658, including an Australian Professorial Fellowship (to N.E.D.). G.C.W. is an American Cancer Society Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321076111/-/DCSupplemental.
References
- 1.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13(3):141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutton MD. Coordinating DNA polymerase traffic during high and low fidelity synthesis. Biochim Biophys Acta. 2010;1804(5):1167–1179. doi: 10.1016/j.bbapap.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Indiani C, McInerney P, Georgescu R, Goodman MF, O’Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell. 2005;19(6):805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Heltzel JM, Maul RW, Scouten Ponticelli SK, Sutton MD. A model for DNA polymerase switching involving a single cleft and the rim of the sliding clamp. Proc Natl Acad Sci USA. 2009;106(31):12664–12669. doi: 10.1073/pnas.0903460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuang Z, et al. Regulation of polymerase exchange between Pol eta and Pol delta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc Natl Acad Sci USA. 2008;105(14):5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagès V, Fuchs RP. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21(58):8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- 7.Wijffels G, et al. Inhibition of protein interactions with the β2 sliding clamp of Escherichia coli DNA polymerase III by peptides from β2-binding proteins. Biochemistry. 2004;43(19):5661–5671. doi: 10.1021/bi036229j. [DOI] [PubMed] [Google Scholar]
- 8.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129(4):665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Ohmori H, et al. The Y-family of DNA polymerases. Mol Cell. 2001;8(1):7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 10.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439(7073):225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 11.McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol Cell. 2001;7(3):571–579. doi: 10.1016/s1097-2765(01)00204-0. [DOI] [PubMed] [Google Scholar]
- 12.Slechta ES, et al. Adaptive mutation: General mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc Natl Acad Sci USA. 2003;100(22):12847–12852. doi: 10.1073/pnas.1735464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19(6):791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 14.Pomerantz RT, Kurth I, Goodman MF, O’Donnell ME. Preferential D-loop extension by a translesion DNA polymerase underlies error-prone recombination. Nat Struct Mol Biol. 2013;20(6):748–755. doi: 10.1038/nsmb.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science. 2012;336(6079):315–319. doi: 10.1126/science.1219192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner J, Etienne H, Fuchs RP, Cordonnier A, Burnouf D. Distinct β-clamp interactions govern the activities of the Y family PolIV DNA polymerase. Mol Microbiol. 2009;74(5):1143–1151. doi: 10.1111/j.1365-2958.2009.06920.x. [DOI] [PubMed] [Google Scholar]
- 17.Bunting KA, Roe SM, Pearl LH. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the β-clamp. EMBO J. 2003;22(21):5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jergic S, et al. A direct proofreader-clamp interaction stabilizes the Pol III replicase in the polymerization mode. EMBO J. 2013;32(9):1322–1333. doi: 10.1038/emboj.2012.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toste Rêgo A, Holding AN, Kent H, Lamers MH. Architecture of the Pol III-clamp-exonuclease complex reveals key roles of the exonuclease subunit in processive DNA synthesis and repair. EMBO J. 2013;32(9):1334–1343. doi: 10.1038/emboj.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wuite GJ, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404(6773):103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 21.Maier B, Bensimon D, Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc Natl Acad Sci USA. 2000;97(22):12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanner NA, et al. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat Struct Mol Biol. 2008;15(2):170–176. doi: 10.1038/nsmb.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner J, Fujii S, Gruz P, Nohmi T, Fuchs RP. The β clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 2000;1(6):484–488. doi: 10.1093/embo-reports/kvd109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamers MH, Georgescu RE, Lee S-G, O’Donnell M, Kuriyan J. Crystal structure of the catalytic α subunit of E. coli replicative DNA polymerase III. Cell. 2006;126(5):881–892. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 25.Ozawa K, et al. Proofreading exonuclease on a tether: The complex between the E. coli DNA polymerase III subunits α, epsilon, θ and β reveals a highly flexible arrangement of the proofreading domain. Nucleic Acids Res. 2013;41(10):5354–5367. doi: 10.1093/nar/gkt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scouten Ponticelli SK, Duzen JM, Sutton MD. Contributions of the individual hydrophobic clefts of the Escherichia coli β sliding clamp to clamp loading, DNA replication and clamp recycling. Nucleic Acids Res. 2009;37(9):2796–2809. doi: 10.1093/nar/gkp128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delaney JC, Essigmann JM. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- 28.Tomer G, Reuven NB, Livneh Z. The β subunit sliding DNA clamp is responsible for unassisted mutagenic translesion replication by DNA polymerase III holoenzyme. Proc Natl Acad Sci USA. 1998;95(24):14106–14111. doi: 10.1073/pnas.95.24.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 2011;108(48):19311–19316. doi: 10.1073/pnas.1113664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ha T. Single-molecule approaches embrace molecular cohorts. Cell. 2013;154(4):723–726. doi: 10.1016/j.cell.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Zhuang Z, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proc Natl Acad Sci USA. 2004;101(22):8289–8294. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamdan SM, et al. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27(4):539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 33.Loparo JJ, Kulczyk AW, Richardson CC, van Oijen AM. Simultaneous single-molecule measurements of phage T7 replisome composition and function reveal the mechanism of polymerase exchange. Proc Natl Acad Sci USA. 2011;108(9):3584–3589. doi: 10.1073/pnas.1018824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graham JS, Johnson RC, Marko JF. Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA. Nucleic Acids Res. 2011;39(6):2249–2259. doi: 10.1093/nar/gkq1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Indiani C, Langston LD, Yurieva O, Goodman MF, O’Donnell M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc Natl Acad Sci USA. 2009;106(15):6031–6038. doi: 10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchida K, et al. Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal. Mol Microbiol. 2008;70(3):608–622. doi: 10.1111/j.1365-2958.2008.06423.x. [DOI] [PubMed] [Google Scholar]
- 37.Beuning PJ, Sawicka D, Barsky D, Walker GC. Two processivity clamp interactions differentially alter the dual activities of UmuC. Mol Microbiol. 2006;59(2):460–474. doi: 10.1111/j.1365-2958.2005.04959.x. [DOI] [PubMed] [Google Scholar]
- 38.Downey CD, McHenry CS. Chaperoning of a replicative polymerase onto a newly assembled DNA-bound sliding clamp by the clamp loader. Mol Cell. 2010;37(4):481–491. doi: 10.1016/j.molcel.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapados BR, et al. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116(1):39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 40.Tsutakawa SE, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145(2):198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beattie TR, Bell SD. Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation. EMBO J. 2012;31(6):1556–1567. doi: 10.1038/emboj.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freudenthal BD, Gakhar L, Ramaswamy S, Washington MT. Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange. Nat Struct Mol Biol. 2010;17(4):479–484. doi: 10.1038/nsmb.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsutakawa SE, et al. Solution X-ray scattering combined with computational modeling reveals multiple conformations of covalently bound ubiquitin on PCNA. Proc Natl Acad Sci USA. 2011;108(43):17672–17677. doi: 10.1073/pnas.1110480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu H, Le S, Chen H, Muniyappa K, Yan J. Force and ATP hydrolysis dependent regulation of RecA nucleoprotein filament by single-stranded DNA binding protein. Nucleic Acids Res. 2013;41(2):924–932. doi: 10.1093/nar/gks1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanner NA, van Oijen AM. Visualizing DNA replication at the single-molecule level. Methods Enzymol. 2010;475:259–278. doi: 10.1016/S0076-6879(10)75011-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.