Significance
The nucleotide-binding domain and leucine rich repeat containing family, pyrin domain containing 3 (NLRP3) inflammasome regulates capase-1-dependent maturation of interleukin-1β during infection with Gram-negative bacterial pathogens such as enterohemorrhagic Escherichia coli. Here we identified bacterial RNA:DNA hybrids as well as RNA as critical mediators of these responses. RNA:DNA hybrids and RNA gained access to the cytosol from phagolysosomal compartments during infection, leading to the assembly of NLRP3 inflammasome complex. Delivery of synthetic RNA:DNA hybrids into the cytosol triggered NLRP3-dependent responses, whereas introduction of RNase H, which degrades hybrids, abolished inflammasome activation. Notably, an E. coli rnhA mutant, incapable of producing RNase H, induced elevated levels of NLRP3-dependent inflammasome activation. Collectively, these studies define bacterial RNA:DNA hybrids as a new microbe-associated molecular pattern with innate immune stimulatory activity during microbial infections.
Abstract
Enterohemorrhagic Escherichia coli (EHEC) is an extracellular pathogen that causes hemorrhagic colitis and hemolytic uremic syndrome. The proinflammatory cytokine, interleukin-1β, has been linked to hemolytic uremic syndrome. Here we identify the nucleotide-binding domain and leucine rich repeat containing family, pyrin domain containing 3 (NLRP3) inflammasome as an essential mediator of EHEC-induced IL-1β. Whereas EHEC-specific virulence factors were dispensable for NLRP3 activation, bacterial nucleic acids such as RNA:DNA hybrids and RNA gained cytosolic access and mediated inflammasome-dependent responses. Consistent with a direct role for RNA:DNA hybrids in inflammasome activation, delivery of synthetic EHEC RNA:DNA hybrids into the cytosol triggered NLRP3-dependent responses, and introduction of RNase H, which degrades such hybrids, into infected cells specifically inhibited inflammasome activation. Notably, an E. coli rnhA mutant, which is incapable of producing RNase H and thus harbors increased levels of RNA:DNA hybrid, induced elevated levels of NLRP3-dependent caspase-1 activation and IL-1β maturation. Collectively, these findings identify RNA:DNA hybrids of bacterial origin as a unique microbial trigger of the NLRP3 inflammasome.
Enterohemorrhagic Escherichia coli (EHEC) is an important extracellular pathogen that causes diarrheal illness. EHEC colonizes the surface of colonic epithelial cells using a type III secretion system (T3SS) to inject dozens of bacterial effectors into cells to modulate a variety of signaling pathways (1). In addition, EHEC harbors virulence factors such as shiga toxin (Stx) and a pO157 plasmid-encoded enterohemolysin, both implicated in severe manifestations of EHEC disease such as hemorrhagic colitis and hemolytic uremic syndrome (HUS), the life-threatening triad of hemolytic anemia, thrombocytopenia, and renal failure (1).
Along with Stx, inflammation is thought to contribute to both hemorrhagic colitis and HUS in EHEC infection (2). Coadministration of LPS greatly potentiates the action of Stx in a murine HUS model, indicating that innate immune activation during EHEC infection may play an important role in the development of systemic disease (3). In addition, elevated serum levels of the proinflammatory cytokine IL-1β have been observed in human EHEC infection and are a major risk factor for HUS development (4).
The enormous proinflammatory potential of IL-1β is countered by critical regulatory checkpoints that control its production in immune cells. In contrast to the majority of proinflammatory cytokines, IL-1β is regulated at both the transcriptional and posttranslational levels. It is synthesized as an inactive proform following engagement of receptors such as Toll-like receptors (5) and then processed by a cysteine protease, caspase-1 (IL-1β converting enzyme; ICE). Enzymatically active caspase-1 is itself converted from the inactive procaspase-1 form by the inflammasome complex (5). Such complexes consist of a cytosolic sensor [which can be either a nucleotide-binding domain and leucine-rich repeat containing (NLR) protein or AIM2], the adaptor apoptosis-associated speck-like protein containing CARD (ASC) and the effector molecule procaspase-1. In addition to IL-1β, active caspase-1 also regulates IL-18 maturation and a form of inflammatory cell death referred to as pyroptosis (6). During infection, microbial products engage the activation of one or more NLR and/or AIM2 inflammasomes to coordinate IL-1β production. Nucleotide-binding domain and leucine rich repeat containing family, pyrin domain containing 3 (NLRP3), the best-studied inflammasome, plays a key role in host defense and is activated by numerous bacterial, viral, and fungal pathogens, as well as products associated with noninfectious inflammatory diseases, such as uric acid and silica crystals (6). Recently, bacterial mRNA was identified as a viability-associated pathogen-associated molecular pattern (vita-PAMP) important in promoting the assembly of the NLRP3 inflammasome (7, 8).
Despite the importance of IL-1 production in the serious manifestations of EHEC infection, the microbial element(s) that triggers inflammasome activation has not been identified. In this study, we found that EHEC triggers NLRP3 inflammasome activation. Bacterial factors that have been previously implicated in inflammatory responses, such as an EHEC pore-forming toxin or its T3SS, were dispensable for EHEC-induced inflammasome activation. Rather, bacterial nucleic acids, such as RNA:DNA hybrids and RNA, are the triggers of NLRP3 activation in EHEC infection. Following phagocytosis, RNA:DNA hybrids of bacterial origin were observed as discrete foci and colocalized with cytosolic inflammasome complexes containing NLRP3, ASC, and caspase-1. Collectively, these studies identify bacterial RNA:DNA hybrids as a unique class of microbe-associated molecular pattern with innate immune stimulatory activity during microbial infection.
Results
EHEC Activates the NLRP3 Inflammasome Independent of Its Major Virulence Factors.
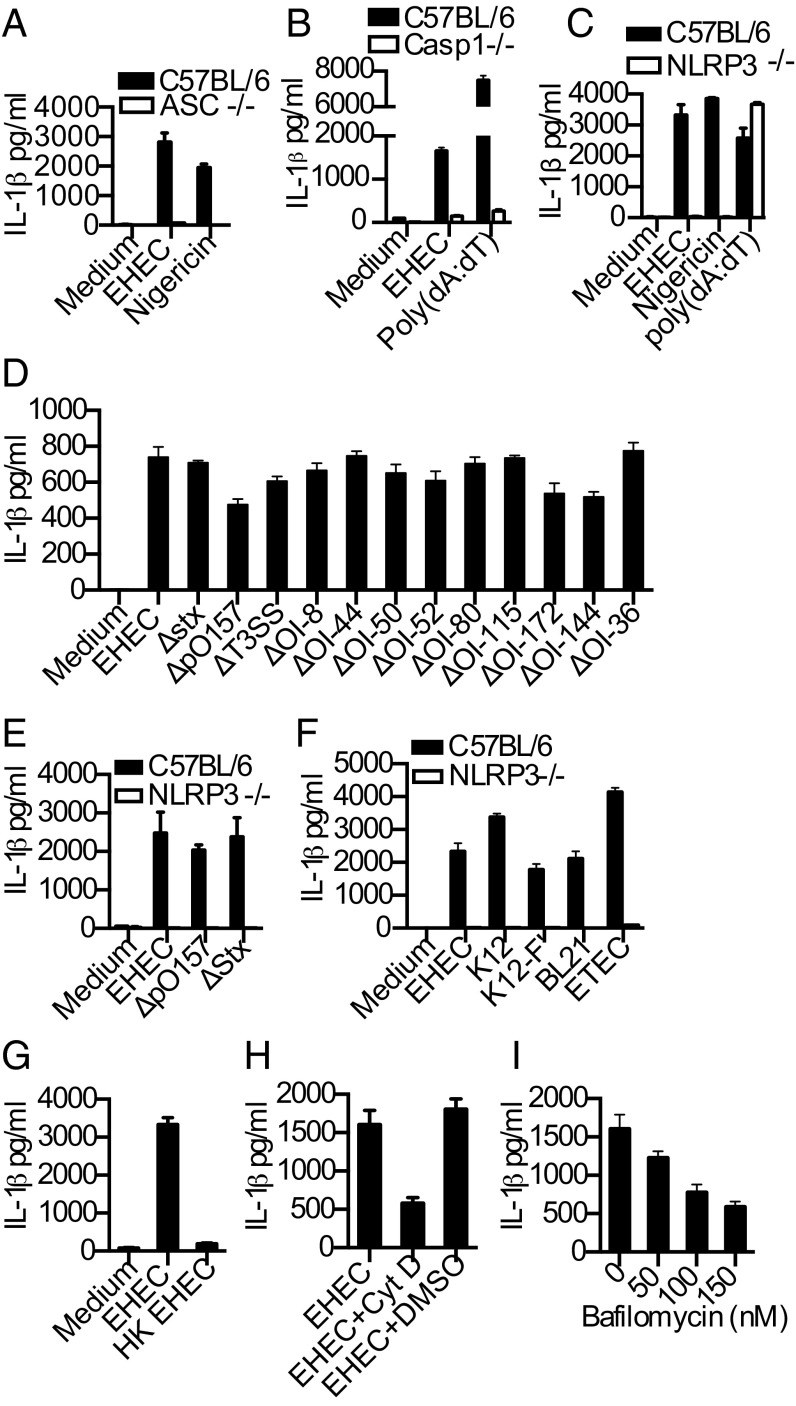
To identify host determinants required for the proteolytic maturation of IL-1β in response to EHEC, we infected bone marrow-derived macrophages (BMDMs) and dendritic cells (BMDCs) from wild-type mice or mice deficient in various inflammasome components with EHEC. EHEC-induced IL-1β production was abrogated in ASC-, caspase-1-, and NLRP3-deficient macrophages and dendritic cells (Fig. 1 A–C and Fig. S1A). Among the upstream cellular events involved in NLRP3 activation, Ca2+ mobilization was not required for EHEC-induced IL-1β production whereas reactive oxygen species production and K+ efflux were found to be required (Fig. S1 B–D). In contrast to NLRP3, the NLRC4/IPAF inflammasome, which senses T3SS, and the AIM2 inflammasome, which senses cytosolic DNA, were largely dispensable for IL-1β secretion in EHEC-infected macrophages (Fig. S1 E and F). Although recent studies identified an important role for protein kinase R (PKR), a dsRNA-activated protein kinase with well-documented antiviral activities, in the activation of inflammasomes (9), we found no role for PKR in regulating inflammasome-dependent IL-1β production in response to EHEC, nigericin, or poly(dA:dT) (Fig. S1G).
Fig. 1.
EHEC activates NLRP3 inflammasome independent of its major virulence factors. (A−G) IL-1β in supernatants of BMDMs from indicated genotypes infected with EHEC or various E. coli strains or heat-killed EHEC (HK EHEC) at a multiplicity of infection (MOI) of 25 for 8 h or treated with poly(dA:dT) for 6 h or nigericin for 1 h. (H and I) IL-1β in the supernatants of C57BL/6 BMDMs treated with cytochalasin D (CytD; 2 μM), DMSO, or bafilomycin A (50−150 nM) or left untreated for 30 min or 1 h before infecting with EHEC (MOI = 25) for 8 h. Data presented as mean SEM are from one experiment representative 2–3 experiments.
To test the role of known EHEC virulence factors in inflammasome activation, we infected macrophages with EHEC strains deficient in a variety of virulence genes. Consistent with our data from NLRC4/IPAF-deficient cells, deletion of the EHEC T3SS only marginally affected IL-1β induction (Fig. 1D). Stx, a major virulence factor required for EHEC-induced intestinal and renal disease, did not contribute to IL-1β production (Fig. 1D). EHEC lacking plasmid pO157, which encodes enterohemolysin, a pore-forming toxin previously implicated in NLRP3 activation (4), showed only a marginal defect in IL-1β production (Fig. 1D). Finally, a panel of EHEC strains lacking different O-islands (OI), i.e., segments of the EHEC genome that are lacking in the genome of the laboratory strain E. coli K12 that often encode putative or documented virulence factors (10), induced IL-1β that is comparable to wild-type EHEC-induced IL-1β (Fig. 1D). Notably, IL-1β responses elicited by pO157- and stx-deficient mutants were still entirely NLRP3 dependent (Fig. 1E), indicating the existence of a predominant, presumably virulence-independent, mechanism by which EHEC activates the NLRP3 inflammasome. Consistent with this idea, avirulent strains of E. coli such as K12, BL21, as well as different “pathotypes” such as enterotoxigenic E. coli (ETEC) harboring virulence genes distinct from those of EHEC also elicited IL-1β production in an NLRP3-dependent fashion (Fig. 1F). Heat-killing the bacteria, or blocking bacterial phagocytosis or lysosomal acidification using cytochalasin D and bafilomycin A, respectively, inhibited EHEC-induced IL-1β secretion (Fig. 1 G−I), suggesting that phagocytosis of viable bacteria followed by their degradation within the lysosomal compartment is an important event upstream of NLRP3 activation.
EHEC RNA Accesses the Cytosol and Activates the NLRP3 Inflammasome.
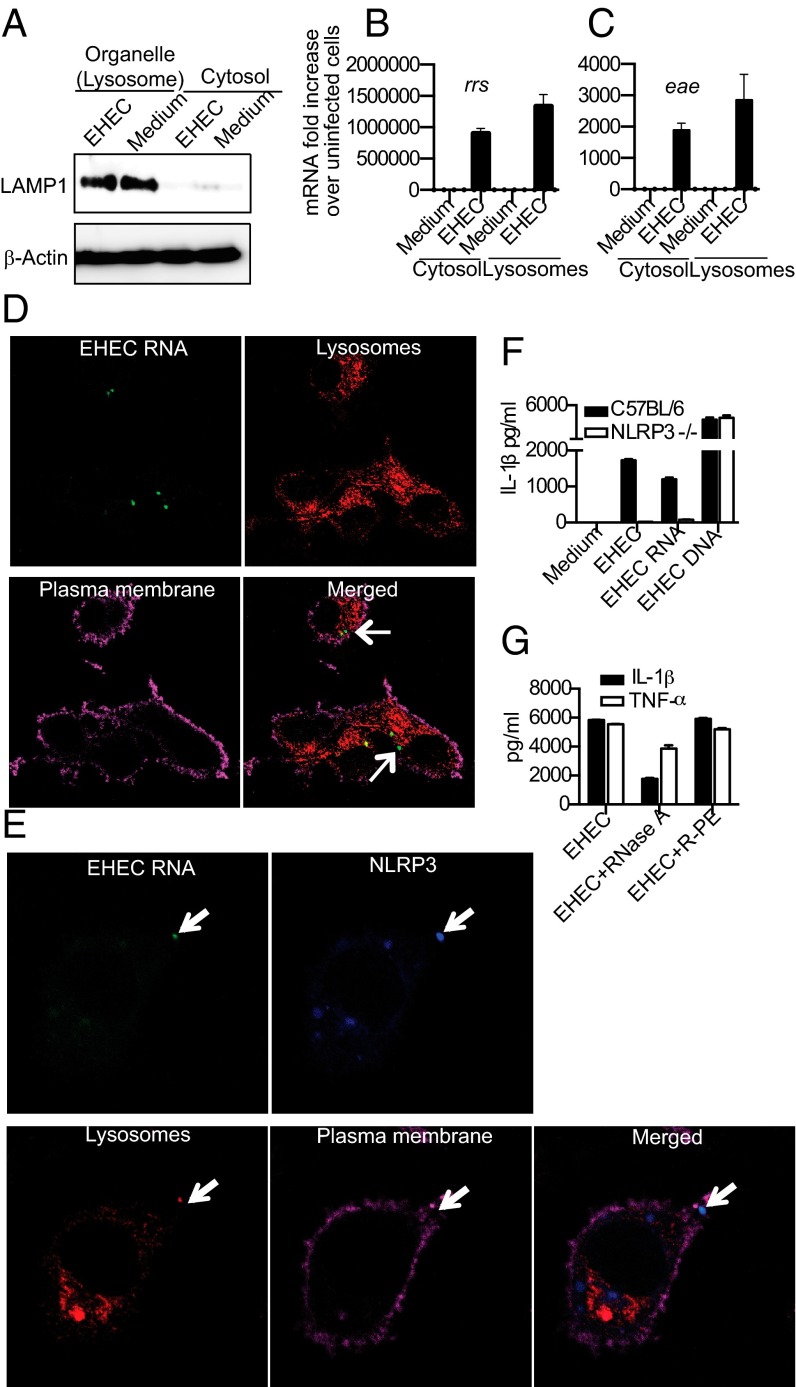
As EHEC activates the NLRP3 inflammasome, a molecular platform localized in the cytosolic compartment, we asked whether, following phagocytosis and lysosomal killing, EHEC components such as nucleic acids translocate from the lysosomes to an intracellular location to trigger inflammasome activation. Bacterial mRNA has been shown to induce NLRP3 inflammasome activation and IL-1 responses (7, 8). However, it is not clear if bacterial nucleic acids, including mRNA, are released from the lysosome into the cytosol following bacterial killing and degradation. To determine whether EHEC RNA is released into the cytosol of BMDMs during infection, we performed subcellular fractionation of infected and uninfected cells (11). The purity of the cytosolic fraction was verified by the lack of LAMP1, a lysosomal marker (Fig. 2A). RNA from cytosolic or organellar (including lysosomal) fractions was subjected to quantitative real-time PCR to measure EHEC representatives of ribosomal (rrs encoding16s rRNA) or messenger (eae, encoding intimin) RNA. As predicted, cytosolic and lysosomal fractions of uninfected cells did not contain EHEC RNA, whereas lysosomal fractions of infected cells contained both transcripts (Fig. 2 B and C). Remarkably, rrs and eae RNA of EHEC were also detected in the cytosolic fraction of infected macrophages, suggesting that a significant fraction of bacterial RNA is translocated to the cytosol following phagocytosis (Fig. 2 B and C).
Fig. 2.
EHEC RNA accesses cytosol after phagocytosis and activates the NLRP3 inflammasome. (A) Immunoblots for LAMP1 and β-actin in the cytosolic and organellar (Lysosome) fractions of EHEC-infected and uninfected BMDMs. (B and C) Quantitative real-time PCR for detection of rrs and eae mRNA in the cytosolic and organellar (Lysosome) fractions of EHEC-infected and uninfected BMDMs. (D and E) Confocal microscopy of C57BL/6 BMDMs infected with EU-labeled EHEC at MOI = 10 for 2 h. EHEC RNA is visualized using clickIT reagent (green), lysosomes using anti-LAMP1 antibody (red), NLRP3 using anti-NLRP3 antibody (blue), and plasma membrane using cholera toxin B (magenta). (F) IL-1β in the supernatants of BMDMs from C57BL/6 or NLRP3−/− mice infected with EHEC at MOI = 25 or transfected with 1 μg of EHEC RNA or EHEC DNA for 8 h. (G) IL-1β and TNFα in the supernatants of C57BL/6 BMDMs transfected with 1 μg of RNase A or R-PE (control protein) or left untreated for 2 h before infection with EHEC at MOI = 25 for 8 h. Data presented as mean SEM are from one experiment representative 2–3 experiments.
To attempt to visualize the cytosolic bacterial RNA, we labeled live EHEC with a clickIT RNA labeling system, which labels newly synthesized RNA, and analyzed the cells by confocal microscopy at various time points following infection. EHEC were phagocytosed within 30 min of infection (Fig. S2A) and, by 2 h postinfection EHEC RNA (Fig. 2D and Fig. S2A) began to translocate to the cytosol from the lysosomes. Consistent with the large fraction of the rrs and eae transcripts found in the cytosol by fractionation, ∼20–25% of cells displayed one or more foci of bacterial RNA in the cytosol by 2 h of infection. These foci of RNA were not associated with bacteria, and, in fact, intact bacteria were not found in the cytosol, consistent with the fact that EHEC is an extracellular bacterium excluded from the cytosolic compartment by the bactericidal activity of the phagosomal machinery. We also observed specks of EHEC DNA in the cytosol of infected BMDMs (Fig. S2B), but, as mentioned above, the DNA sensor AIM2 was not required for EHEC-induced IL-1β production. Consistent with the hypothesis that lysosomal acidification is required for this translocation, bafilomycin A inhibited detectable release of nucleic acids into the macrophage cytosol following phagocytosis of live bacteria (Fig. S2C).
Notably, EHEC RNA released into the cytosol colocalized with NLRP3 specks (Fig. 2E), which are indicative of the assembly of the macromolecular NLRP3 inflammasome complex (12). Furthermore, transfection of macrophages with purified EHEC RNA potently activated IL-1β production in an NLRP3-dependent manner (Fig. 2F), and transfection with RNase A protein [but not a control protein R-phycoerythrin (R-PE)] into the cytosol before EHEC infection resulted in a marked reduction in IL-1β production, but not TNF-α, consistent with a role for cytosolic EHEC RNA in NLRP3 activation by EHEC (Fig. 2G). Thus, EHEC RNA has a significant role in EHEC-induced inflammasome activation. Interestingly, although colocalization of EHEC RNA and NLRP3 was frequent (25% of cells), we also observed NLRP3 specks in ∼20% of EHEC-infected cells that were not colocalized with EHEC RNA (Fig. S2D), indicating the presence of an additional bacterial stimulus that likely elicits NLRP3 complex formation in cells.
Bacterial RNA:DNA Hybrids Access the Cytosol and Colocalize with the NLRP3 Inflammasome.
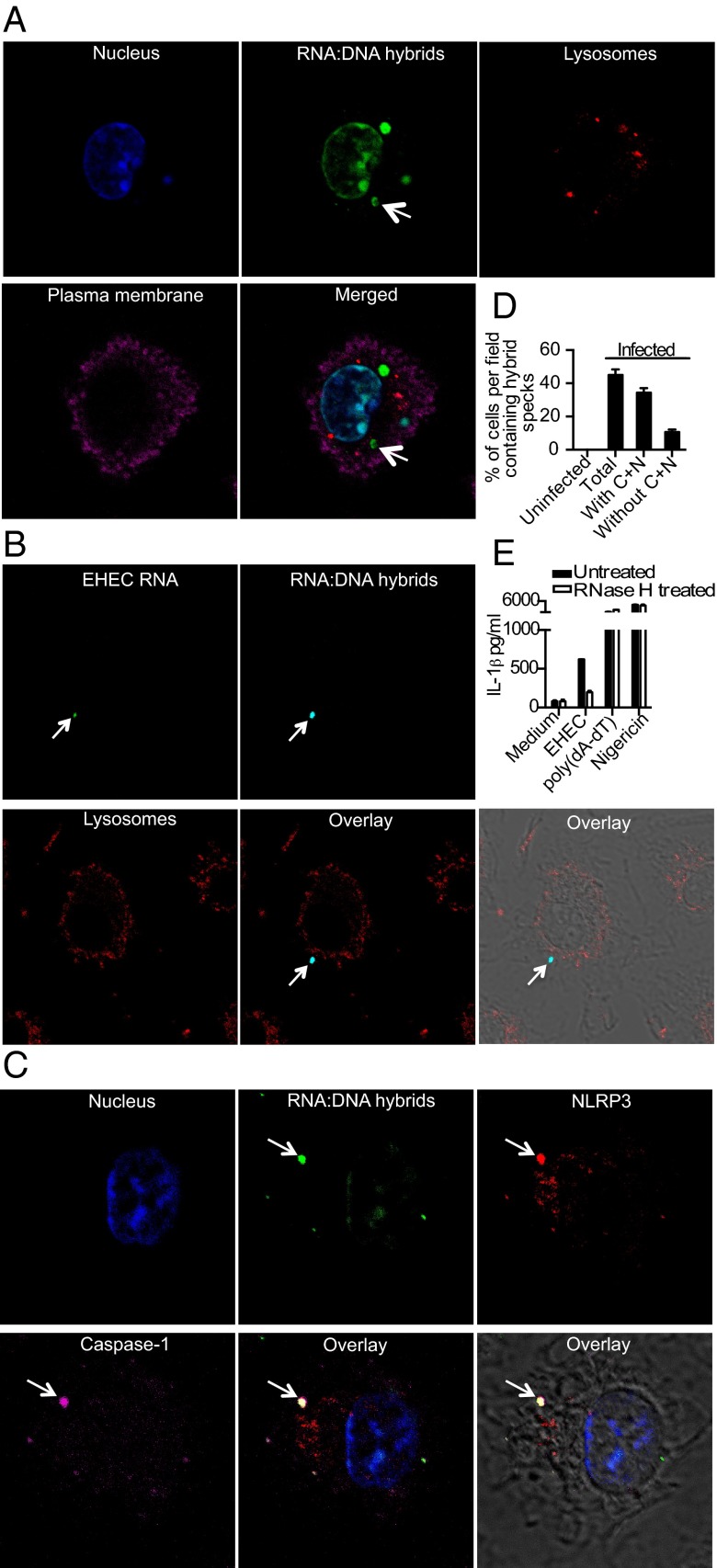
In addition to RNA and DNA species, prokaryotic cells possess RNA:DNA hybrids, which are generated during DNA replication and transcription (13, 14). RNA:DNA hybrid molecules are a relatively stable and abundant species of nucleic acids in prokaryotes. The presence of both bacterial RNA and DNA in the cytosol raised the possibility that EHEC RNA:DNA hybrids might be released into the cytoplasm from killed phagosomal bacteria to trigger an innate immune response. Using a well-characterized monoclonal antibody that specifically detects RNA:DNA hybrids (15), we detected RNA:DNA hybrids in the nucleus of almost all infected and uninfected cells, consistent with their generation and accumulation during normal eukaryotic DNA replication and transcription (Fig. 3A and Fig. S3A). In addition to eukaryotic hybrids in the nucleus, RNA:DNA hybrids were observed within the phagosomes of infected but not of uninfected cells (Fig. S3B). Notably, starting 1 h after infection, we detected bacterial RNA:DNA hybrids in the cytosol of infected macrophages (Fig. 3A). Importantly, both the anti-RNA:DNA hybrid antibody and the clickIT system, which stains newly synthesized bacterial RNA and thus the RNA in RNA:DNA hybrids formed during transcription, detected the same hybrid specks in the cytosol, indicating that these hybrids are in fact of bacterial, not eukaryotic, origin (Fig. 3B).
Fig. 3.
Bacterial RNA:DNA hybrids access the cytosol and colocalize with the NLRP3 inflammasome. (A and C) Confocal microscopy of EHEC-infected BMDMs stained with anti-RNA:DNA hybrid (S9.6), anti-LAMP-1, anti-NLRP3, and anti-caspase-1 p10 antibodies to visualize RNA:DNA hybrids (green), lysosomes (red in A), NLRP3 (red in C), and caspase-1 (magenta in C). Host cell nucleus was visualized using DAPI (blue) and plasma membrane by cholera toxin B (magenta in A). (B) Confocal microscopy of C57BL/6 BMDMs infected with EU-labeled EHEC at MOI = 10 for 2 h. EHEC RNA is visualized using clickIT reagent (green), RNA:DNA hybrids using S9.6 antibody (light blue), and lysosomes using cholera toxin B (red). (D) Total hybrid specks and hybrid specks with or without caspase-1 and NLRP3 (C+N) in uninfected and EHEC-infected BMDMs detected by confocal microscopy from 30 fields containing 10 cells each. (E) IL-1β in the supernatants of BMDMs pretreated with RNase H or left untreated for 2 h before infection with EHEC at MOI = 25 for 8 h or treatment with poly(dA:dT) for 6 h or nigericin for 1 h. Data presented as mean SEM are from one experiment representative 2–3 experiments.
To determine if bacterial RNA:DNA hybrids, like bacterial RNA species, colocalized with inflammasomes in the cytosol, we costained cells for hybrids, NLRP3, ASC, and caspase-1 (Fig. 3C and Fig. S4A). At 2 h of infection, ∼40% of the cells contained RNA:DNA hybrid specks in the cytosol and, notably, 75% of these RNA:DNA hybrid specks colocalized with NLRP3 and caspase-1, implicating RNA:DNA hybrids as important inflammasome activators in EHEC infection (Fig. 3D and Fig. S4B). Approximately 10% of the cells contained NLRP3-caspase-1 specks that did not colocalize with RNA:DNA hybrids. Given our finding that EHEC RNA is found in the cytoplasm, these hybrid-negative specks could be NLRP3 inflammasomes triggered by EHEC RNA.
To test whether RNA:DNA hybrids are required for the activation of NLRP3 during EHEC infection, we delivered into the macrophage cytosol RNase H, which specifically degrades the RNA in the RNA:DNA hybrid molecules (14, 16). This treatment abrogated IL-1β production in response to EHEC. The inhibitory effect of RNase H was specific for EHEC infection, because IL-1β production induced by the AIM2 ligand poly(dA:dT) or the NLRP3 activator nigericin was unaffected (Fig. 3E). Furthermore, RNase H did not affect the production of an inflammasome-independent cytokine, TNF-α, by EHEC (Fig. S4C), demonstrating that its inhibitory effect was specific for EHEC-induced IL-1β.
Synthetic RNA:DNA Hybrids Activate the NLRP3 Inflammasome.
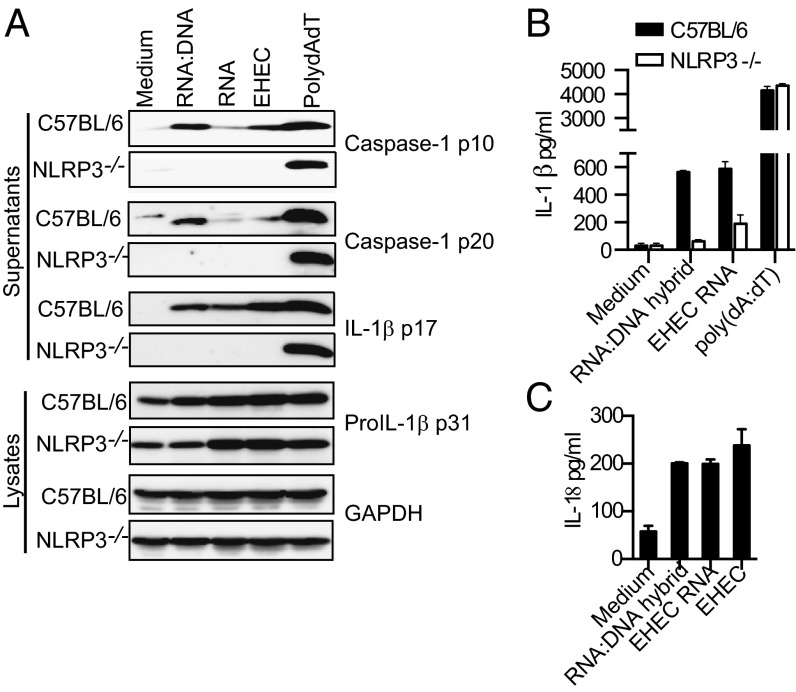
To further determine if RNA:DNA hybrids are capable of activating NLRP3 inflammasome in the absence of other bacterial components, we tested whether a 120-bp synthetic RNA:DNA hybrid homologous to a portion of the EHEC rimM gene could activate NLRP3 inflammasome in macrophages. The purity of the synthetic hybrid preparation was verified by agarose gel electrophoresis (Fig. S5A). Similar to a recent study by Rigby et al. (17), we observed that the synthetic RNA:DNA hybrids induced IFN (IFN)-β production, albeit at modest levels (Fig. S5B). Notably, RNA:DNA hybrids potently stimulated the activation of caspase-1 and production of IL-1β and IL-18 (Fig. 4 A–C). As predicted, AIM2 was dispensable for inflammasome activation by RNA:DNA hybrids (Fig. S5C). In contrast, all of these inflammasome-mediated responses triggered by synthetic bacterial RNA:DNA hybrids were NLRP3 dependent (Fig. 4 A and B).
Fig. 4.
Synthetic RNA:DNA hybrids activate NLRP3 inflammasome. (A) Immunoblots for caspase-1 p10, caspase-1 p20, IL-1β p17 in the supernatants, and proIL-1β p31 and GAPDH in the lysates of BMDMs transfected with 1 μg of RNA:DNA hybrids, EHEC RNA, EHEC DNA, or poly(dA:dT) or infected with EHEC at MOI = 25 for 8 h. (B and C) IL-1β or IL-18 in the supernatants of indicated BMDMs transfected with 1 μg of RNA:DNA hybrids, EHEC RNA, EHEC DNA, or poly(dA:dT) or infected with EHEC at MOI = 25 for 8 h.
RNase H Deficiency in Bacteria Enhances Inflammasome Activation.
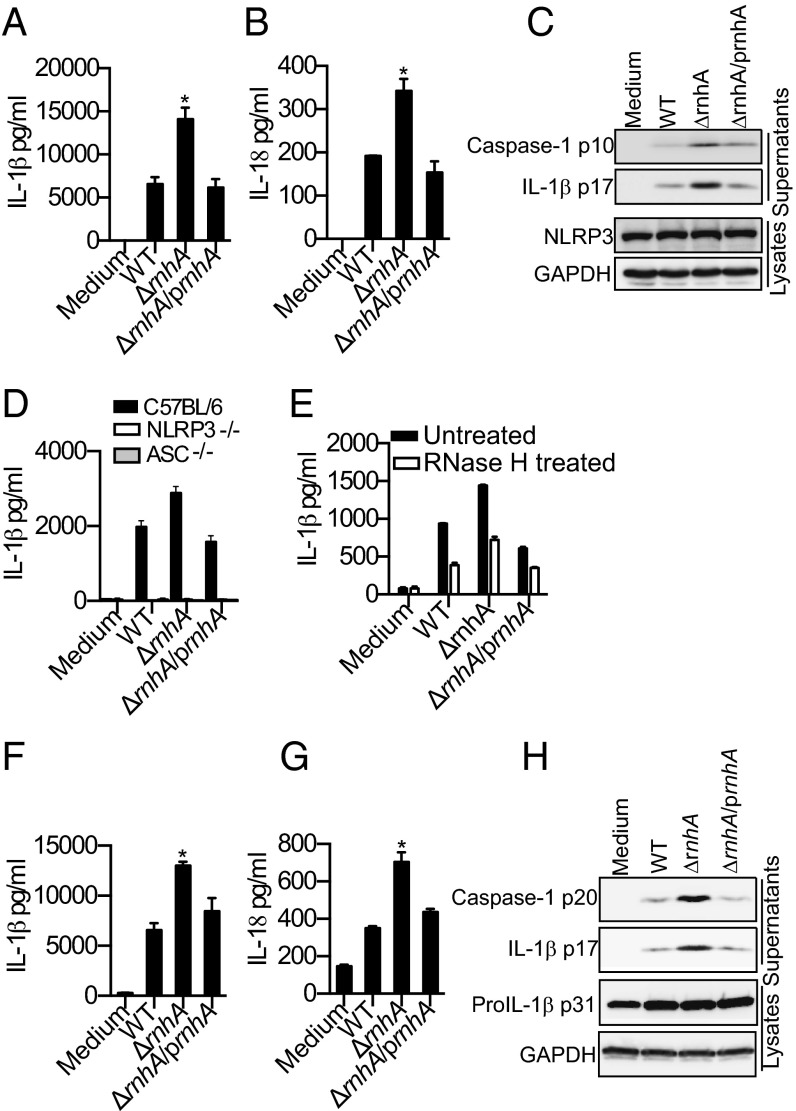
To directly test if bacterial RNA:DNA hybrids contribute to NLRP3 activation during infection, we used an E. coli strain deficient in RNase H enzyme (E. coli ΔrnhA), which specifically degrades RNA:DNA hybrids by degrading the RNA primer during DNA replication (14, 16). E. coli ΔrnhA strain accumulates higher levels of long and stable RNA:DNA hybrids (16, 18) but has growth kinetics and total RNA levels comparable to wild-type (WT) E. coli (Fig. S6 A and B). In addition, the rnhA mutant was killed within macrophages at a rate indistinguishable from WT (Fig. S6C). Supporting our hypothesis, the overproduction of RNA:DNA hybrids by E. coli resulted in enhanced NLRP3 activation as infection of macrophages with E. coli ΔrnhA resulted in a marked increase in the production of IL-1β and IL-18 as well as cleavage of caspase-1 and IL-1β relative to its parent strain (Fig. 5 A–C). This enhanced macrophage response was specific for NLRP3 activation, because E. coli ΔrnhA induce inflammasome-independent cytokines such as TNF and IL-6 at levels similar to WT (Fig. S6 D and E). The elevated IL-1β response elicited by ΔrnhA E. coli was completely dependent on ASC and NLRP3 (Fig. 5D). Importantly, complementation of E. coli ΔrnhA with plasmid-encoded RNase H (“prnhA”) attenuated the increase in caspase-1 and IL-1β processing and the secretion of IL-1β and IL-18 to WT levels (Fig. 5 A–C). Although the number of hybrid specks visualized after infection with E. coli ΔrnhA was not significantly higher than upon infection by WT E. coli (Fig. S6F), delivery of RNase H enzyme into ΔrnhA-infected reduced IL-1β production to levels equivalent to those of WT-infected macrophages, verifying that the enhanced inflammasome response induced by ΔrnhA E. coli was in fact due to higher levels of RNA:DNA hybrids (Fig. 5E). E. coli ΔrnhA appeared to induce greater than WT levels of oligomeric forms of ASC in an ASC oligomerization assay, raising the possibility that increased levels of RNA:DNA hybrids in the mutant may lead to increased inflammasome assembly (Fig. S6G). The recognition of RNA:DNA duplexes was not exclusive to macrophages; upon infection of dendritic cells, E. coli ΔrnhA induced greater inflammasome activation, characterized by IL-1β and IL-18 secretion and caspase-1 and IL-1β maturation (Fig. 5 F–H). As above, genetic complementation of E. coli ΔrnhA restored caspase-1 activation, IL-1β maturation, and IL-1β as well as IL-18 secretion to WT levels (Fig. 5 F–H). Overall, the bacterial genetic complementation and cytosolic RNase H delivery approaches provide direct evidence for RNA:DNA hybrids being bona fide bacterial components contributing to NLRP3 inflammasome activation during bacterial infection.
Fig. 5.
RNase H deficiency in bacteria enhances inflammasome activation. (A, B, F, and G) IL-1β and IL-18 in the supernatants of C57BL/6 BMDMs (A and B) or BMDCs (F and G) infected with indicated strains of E. coli at MOI = 25 for 8 h. (C and H) Immunoblots for cleaved caspase-1 and IL-1β in the supernatants or NLRP3, proIL-1β p31 and GAPDH in the lysates of C57BL/6 BMDMs (C) or BMDCs (H) infected with indicated E. coli strains for 8 h. (D) IL-1β in the supernatants of BMDMs indicated genotypes infected with indicated E. coli strains for 8 h. (E) IL-1β in the supernatants of BMDMs pretreated with RNase H for 2 h or left untreated before infecting with indicated E. coli strains for 8 h. Asterisk (*) indicates P < 0.05 for WT vs. ΔrnhA or WT vs. ΔrnhA/prnhA as determined by one-way ANOVA followed by Bonferroni test. Data presented as mean SEM are from one experiment representative 2–3 experiments.
Discussion
IL-1β is a potent inflammatory cytokine implicated in the outcome of EHEC disease (19). The production of bioactive IL-1β is regulated by cytosolic inflammasome complexes (6). In this study, we report that EHEC-induced IL-1β maturation and production occurred independently of known virulence factors; rather, induction of IL-1β secretion required uptake of EHEC and acidification of phagosomes, implicating EHEC breakdown products such as nucleic acids in this process.
Microbial nucleic acids have long been known to trigger host responses. Bacterial and viral RNA and DNA are potent activators of type I IFNs and IL-1 cytokines. Bacterial mRNA has been shown to induce NLRP3 inflammasome activation and IL-1 responses (7, 8). Using both microscopic and biochemical approaches, we have shown here that EHEC RNA was delivered into the host cell cytosol, colocalized with NLRP3 foci and triggered inflammasome activation in an NLRP3-dependent manner. Cytosolic delivery of exogenous RNase A attenuated the EHEC-driven IL-1β response.
Another class of nucleic acid species abundant in bacteria are RNA:DNA hybrids, which are formed during bacterial DNA replication and transcription (13, 14). During bacterial DNA synthesis, short RNA strands serve as primers for both leading and lagging strand (Okazaki fragments) synthesis, and RNA:DNA hybrids are formed every 100–200 nucleotides during lagging strand synthesis (20, 21). RNA:DNA hybrids are also formed during transcription, when a newly formed mRNA transcript is paired with one strand of duplex DNA (13). Notably, RNA:DNA hybrids have a relatively stable structure and withstand acidic conditions up to pH 5.5 (22, 23). Several lines of evidence in this study indicated that RNA:DNA hybrids of bacterial origin play a key role in NLRP3 inflammasome activation by EHEC. First, EHEC RNA:DNA hybrids were released into the cytosol of infected cells and colocalized with NLRP3 inflammasome. Second, depletion of EHEC RNA:DNA hybrids in the infected cells by the cytosolic delivery of RNase H enzyme specifically inhibited NLRP3 activation by EHEC. Third, an E. coli strain harboring higher levels RNA:DNA hybrids due to the lack of RNase H induced NLRP3 activation more efficiently than WT bacteria. The findings of this study thus collectively identify bacterial RNA:DNA hybrids as a unique microbe-associated molecular pattern triggering NLRP3 inflammasome activation. Interestingly, RNA:DNA hybrids have been recently shown to induce type I IFNs in a TLR9-MyD88-dependent manner (17).
Collectively, our findings highlight an emerging theme in host−pathogen interactions that the cytosolic immune surveillance mechanism is central to sensing extracellular or vacuolar bacteria through detection of microbial products that access the cytosol from phagosomes. Although EHEC is excluded from the cytosol owing to its susceptibility to the microbicidal nature of phagolysosomes, cytosolic sensing of EHEC is yet an integral part of innate immune responses to EHEC. The inflammasome-dependent cytosolic surveillance of EHEC relies heavily on the detection of bacterial nucleic acids in the cytosol rather than on virulence factors. Considering that nonpathogenic laboratory strains of E. coli, as well as other pathogenic strains of E. coli such as ETEC, stimulated NLRP3 inflammasome activation, the recognition of nucleic acids such as RNA or RNA:DNA hybrids may be a general mechanism by which the cytosolic innate immune system recognizes E. coli irrespective of virulence factor armament and pathogenic potential (8).
NLR inflammasomes such as NLRP1b and NLRC4/IPAF are activated by specific interaction with ligands such as anthrax toxin and bacterial T3SS/flagellin, respectively (6). In contrast, NLRP3 activation is triggered by a plethora of stimuli of diverse nature such as bacterial mRNA, pore forming toxins, crystalline particles, and cellular perturbations such as ionic imbalance. The prevailing consensus is that these disparate stimuli converge on one or more common elements or events upstream of NLRP3 (6). Although bacterial RNA has been shown to activate the NLRP3 inflammasome, the mechanism by which this occurs is not yet known. PKR represented an attractive candidate responsible for RNA-mediated inflammasome activation, but in contrast to published work (9), we found that PKR-deficient cells were not compromised for EHEC-induced NLRP3 inflammasome signaling. Similar observations were reported by Nuñez and colleagues (24). Interestingly, a recent study (25) demonstrated that DHX33, a DExD/H-box RNA helicase family member, is a sensor for viral dsRNA and it activates inflammasome by directly binding to NLRP3. It is possible therefore that either DHX33 or a similar cytosolic sensor(s) for RNA and RNA:DNA hybrids activates NLRP3 in EHEC infection, or that NLRP3 may sense these bacterial nucleic acids directly.
Our identification of bacterial RNA:DNA hybrids as activators of innate immunity has broad implications for microbial recognition and host immunity. RNA:DNA hybrids are also formed as replication intermediates in the life cycle of retroviruses (26), and it is possible that such hybrids generated during active retroviral replication also contribute to NLRP3 inflammasome-mediated immune responses. As with other nucleic acid sensing mechanisms, the ability of the innate immune system to detect RNA:DNA hybrids may also contribute to autoimmune disease. Eukaryotic cells generate RNA:DNA hybrids during transcription, genomic replication, mitochondrial DNA replication, and Ig class switching in B cells (13). Cytosolic exclusion of RNA:DNA hybrids, like that of self-DNA, by nuclear and mitochondrial compartmentalization could be a regulatory mechanism responsible for preventing self-RNA:DNA hybrids from activating cell-intrinsic innate immune responses. Degradation of self-RNA:DNA by RNase H in the nucleus may provide an additional mechanism to avoid recognition of this product. Indeed, Aicardi Goutieres syndrome (AGS), a hereditary neurodegenerative disorder characterized by autoimmune activation and type I IFN production, has been associated with mutations in the human RNASEH2B, which encodes an important subunit of the RNase H enzyme (27). Although the role of NLRP3 activation in AGS remains undefined, the identification of RNA:DNA hybrids as triggers of innate immunity provides new insights into cytosolic immune surveillance mechanisms with potentially important implications for host defense and autoimmunity.
Materials and Methods
A detailed materials and methods is given in SI Materials and Methods.
Briefly, BMDMs and BMDCs were generated from WT and knockout mice and were stimulated with bacteria (Table S1) or other activators as described before (28). Inflammasome responses were analyzed by ELISA and immunoblotting for IL-1β and caspase-1 or confocal microscopy as described before (28, 29).
Supplementary Material
Acknowledgments
The authors thank Andrew Wright (Tufts University) for the E. coli RNase H mutant, Steve Leppla and Clinton Leysath (National Institute of Allergy and Infectious Diseases) for anti-RNA:DNA hybrid antibody, and members of the J.M.L. and K.A.F. labs for helpful discussions. This work is supported by National Institutes of Health Grants AI46454 (to J.M.L.) and AI083713 (to K.A.F.) and a New England Regional Center for Excellence (NERCE) Post-Doctoral Fellowship Award U54 AI057159 (to S.K.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400075111/-/DCSupplemental.
References
- 1.Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010;8(1):26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 2.King AJ. Acute inflammation in the pathogenesis of hemolytic-uremic syndrome. Kidney Int. 2002;61(4):1553–1564. doi: 10.1046/j.1523-1755.2002.00281.x. [DOI] [PubMed] [Google Scholar]
- 3.Keepers TR, Psotka MA, Gross LK, Obrig TG. A murine model of HUS: Shiga toxin with lipopolysaccharide mimics the renal damage and physiologic response of human disease. J Am Soc Nephrol. 2006;17(12):3404–3414. doi: 10.1681/ASN.2006050419. [DOI] [PubMed] [Google Scholar]
- 4.Taneike I, Zhang HM, Wakisaka-Saito N, Yamamoto T. Enterohemolysin operon of Shiga toxin-producing Escherichia coli: A virulence function of inflammatory cytokine production from human monocytes. FEBS Lett. 2002;524(1-3):219–224. doi: 10.1016/s0014-5793(02)03027-2. [DOI] [PubMed] [Google Scholar]
- 5.Yu HB, Finlay BB. The caspase-1 inflammasome: A pilot of innate immune responses. Cell Host Microbe. 2008;4(3):198–208. doi: 10.1016/j.chom.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat Immunol. 2012;13(4):333–342. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanneganti TD, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440(7081):233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 8.Sander LE, et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474(7351):385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu B, et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488(7413):670–674. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perna NT, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409(6819):529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 11.Ramsby M, Makowski G. Proteomics Protocols Handbook. Springer, New York; 2005. [Google Scholar]
- 12.Manji GA, et al. PYPAF1, a PYRIN-containing Apaf1-like protein that assembles with ASC and regulates activation of NF-kappa B. J Biol Chem. 2002;277(13):11570–11575. doi: 10.1074/jbc.M112208200. [DOI] [PubMed] [Google Scholar]
- 13.Aguilera A, García-Muse T. R loops: From transcription byproducts to threats to genome stability. Mol Cell. 2012;46(2):115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Chapados BR, et al. Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication. J Mol Biol. 2001;307(2):541–556. doi: 10.1006/jmbi.2001.4494. [DOI] [PubMed] [Google Scholar]
- 15.Phillips DD, et al. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J Mol Recognit. 2013;26(8):376–381. doi: 10.1002/jmr.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kogoma T. RNase H-defective mutants of Escherichia coli. J Bacteriol. 1986;166(2):361–363. doi: 10.1128/jb.166.2.361-363.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rigby RE, et al. RNA:DNA hybrids are a novel molecular pattern sensed by TLR9. EMBO J. 2014;33(6):542–558. doi: 10.1002/embj.201386117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Meyenburg K, Boye E, Skarstad K, Koppes L, Kogoma T. Mode of initiation of constitutive stable DNA replication in RNase H-defective mutants of Escherichia coli K-12. J Bacteriol. 1987;169(6):2650–2658. doi: 10.1128/jb.169.6.2650-2658.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palermo M, et al. Pretreatment of mice with lipopolysaccharide (LPS) or IL-1beta exerts dose-dependent opposite effects on Shiga toxin-2 lethality. Clin Exp Immunol. 2000;119(1):77–83. doi: 10.1046/j.1365-2249.2000.01103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker TA, Kornberg A. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell. 1988;55(1):113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 21.Masukata H, Tomizawa J. A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell. 1990;62(2):331–338. doi: 10.1016/0092-8674(90)90370-t. [DOI] [PubMed] [Google Scholar]
- 22.Shaw NN, Xi H, Arya DP. Molecular recognition of a DNA:RNA hybrid: Sub-nanomolar binding by a neomycin-methidium conjugate. Bioorg Med Chem Lett. 2008;18(14):4142–4145. doi: 10.1016/j.bmcl.2008.05.090. [DOI] [PubMed] [Google Scholar]
- 23.Chien YH, Davidson N. RNA:DNA hybrids are more stable than DNA:DNA duplexes in concentrated perchlorate and trichloroacetate solutions. Nucleic Acids Res. 1978;5(5):1627–1637. doi: 10.1093/nar/5.5.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Y, Franchi L, Núñez G. The protein kinase PKR is critical for LPS-induced iNOS production but dispensable for inflammasome activation in macrophages. Eur J Immunol. 2013;43(5):1147–1152. doi: 10.1002/eji.201243187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitoma H, et al. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39(1):123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broecker F, Andrae K, Moelling K. Premature activation of the HIV RNase H drives the virus into suicide: A novel microbicide? AIDS Res Hum Retroviruses. 2012;28(11):1397–1403. doi: 10.1089/aid.2012.0067. [DOI] [PubMed] [Google Scholar]
- 27.Crow YJ, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38(8):910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 28.Rathinam VA, et al. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell. 2012;150(3):606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9(8):847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.