Significance
We report the crystal structures of precleavage and postcleavage forms of the lariat-capping (LC) ribozyme. The structures show how domains from an ancestral group I ribozyme have evolved due to loss of selection pressure for self-splicing. Instead, a branching activity has been selected, resulting in capping the downstream mRNA by a 3-nt lariat stabilized by the ribozyme core. The LC ribozyme constitutes an original ribozyme family with an unexpected 3D structure that departs significantly from that of group I introns. The structures also elucidate the regulatory domain’s role in transmitting a signal for cleavage to the ribozyme. The characterization of this natural evolutionary RNA speciation event is, to our knowledge, the first described at such an intricate level.
Keywords: RNA structure, RNA catalysis, GIR1, crystallography, SAXS
Abstract
The lariat-capping (LC) ribozyme is a natural ribozyme isolated from eukaryotic microorganisms. Despite apparent structural similarity to group I introns, the LC ribozyme catalyzes cleavage by a 2′,5′ branching reaction, leaving the 3′ product with a 3-nt lariat cap that functionally substitutes for a conventional mRNA cap in the downstream pre-mRNA encoding a homing endonuclease. We describe the crystal structures of the precleavage and postcleavage LC ribozymes, which suggest that structural features inherited from group I ribozymes have undergone speciation due to profound changes in molecular selection pressure, ultimately giving rise to an original branching ribozyme family. The structures elucidate the role of key elements that regulate the activity of the LC ribozyme by conformational switching and suggest a mechanism by which the signal for branching is transmitted to the catalytic core. The structures also show how conserved interactions twist residues, forming the lariat to join chemical groups involved in branching.
Various mechanisms critically regulate splicing, thus controlling the fate of host gene products. Most splicing that relies solely on autocatalytic RNA introns appears to be unregulated, but control can be achieved at the RNA level because the structural versatility of RNA allows for the fusing of functional modules that can work in concert (1). Examples of riboswitches adjoined to spliceosomal (2) or group I (3) introns illustrate this concept and suggest that uncharacterized splicing regulation mechanisms may exist.
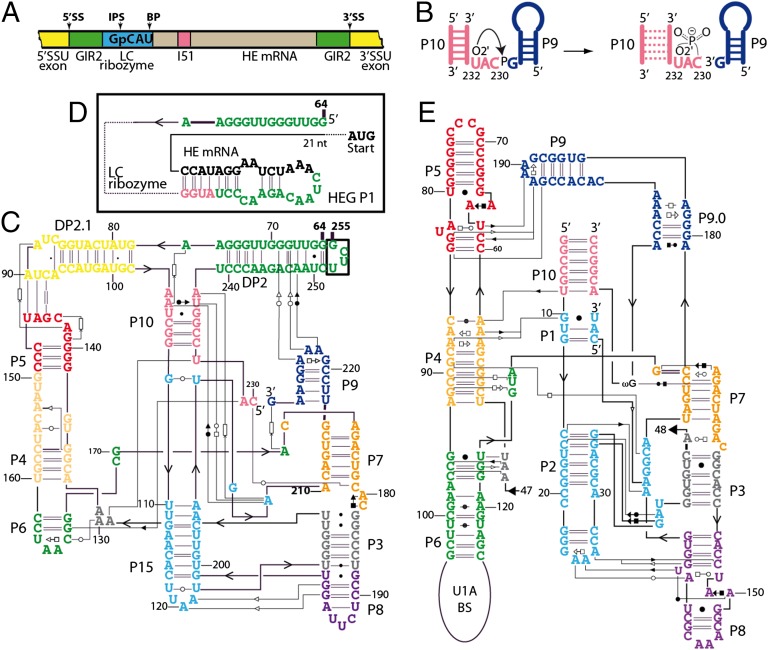
An elaborate example of RNA-regulated splicing is the group I twin-ribozyme introns found in the small subunit (SSU) ribosomal precursor in several protists (Fig. 1A). These twin-ribozyme introns are composed of a conventional group I splicing ribozyme (GIR2), into which is inserted a cassette composed of a branching ribozyme upstream from a homing endonuclease (HE) gene (4). The branching activity results in cleavage and concomitant formation of a 3-nt lariat capping the 5′ end of the HE pre-mRNA (Fig. 1B) (5), hence the name “lariat capping” (LC) ribozyme. The lariat cap appears to act as a substitute for a conventional mRNA m7G cap in a situation in which an mRNA is expressed from within a RNA polymerase I (polI) gene (6). The cross-talk between GIR2 and the LC ribozyme is supported by the existence of three distinct processing pathways of the rRNA precursor, depending on environmental conditions. Under favorable conditions, splicing by GIR2 takes place first, followed by branching by the LC ribozyme and further processing, and eventually translation of the HE mRNA (7, 8). The transcriptional order of the ribozymes implies that the activity of the branching ribozyme is specifically repressed until splicing has taken place. Cellular stress conditions induce formation of full-length intron circles by a circularization pathway (9) that leaves the ribosomal exons unligated. Finally, starvation conditions induce branching by the LC ribozyme without splicing taking place (8). Because splicing only enables ligation of ribosomal exons, the interplay between GIR2 and LC ribozymes influences the fate of both ribosomes and HE mRNA production.
Fig. 1.
Organization of the D. iridis SSU pre-rRNA reactivity and secondary structures of the active and inactive forms of the LC ribozyme compared with the Azoarcus group I intron (20). (A) The LC ribozyme is followed by a HE mRNA, which contains a 51-nt spliceosomal intron (I51). Nonobligatory introns organized identically have also been identified in Naegleria (44) and Allovahlkampfia species. This composite RNA is embedded in a canonical group I intron (GIR2) interrupting the SSU rRNA precursor. The internal processing site (IPS) of LC ribozyme is only 3 nt away from the branch point (BP). Positions of 5′ and 3′ splice sites (SS) are indicated. (B) Schematic representation of the branching reaction leading to the formation of the 3-nt lariat. (C–E) Secondary structure diagrams of the active form of the DirLC ribozyme (C), the alternative structure of the 3′ domain of the LC ribozyme involved in cleavage inhibition (D), and group IC3 ribozyme from Azoarcus (E). (D) Black nucleotides correspond to the HE mRNA. A PCR artifact introduced a C residue in place of U203, which may stabilize the ribozyme as shown previously for the Naegleria LC ribozyme (45). Tertiary interactions are displayed using the Leontis–Westhof nomenclature (46). Stacking interactions are indicated with an arrow pointing to a rectangle.
Here, we report the crystal structures of two different forms of the LC ribozyme from Didymium iridis (DirLC). The structure of the wild-type unreacted form of the LC ribozyme that allowed characterization of the branching reaction (5) was solved to a resolution of 3.85 Å. A higher level of detail was achieved in the 2.5-Å crystal structure of an inactive form of the LC ribozyme based on a circular permutation (CP) over the scissile bond and closure of the natural ends using a stable 5′-UUCG-3′ loop (Fig. 1C). Both structures adopted the same overall architecture (Fig. S1), as supported by the presence of the same structural features within the core, as well as the same tertiary interactions. Originally, the LC ribozyme was described as a group I-like ribozyme (GIR1) based on striking similarities at both sequence and secondary structure levels (10–12) (Fig. 1 C–E). However, the present fold departs so drastically from group I intron structures that it justifies the creation of an original ribozyme family for LC ribozymes (Fig. 2). The structures reveal how the DP2–DP2.1 domain connecting the LC ribozyme to GIR2 is organized and interacts with the catalytic core by mediating two sets of tertiary interactions. This domain has been shown to exert a regulatory role by switching on the LC ribozyme when a 3′ hairpin named HEG P1 (homing endonuclease gene paired segment 1), formed during transcription to prevent branching before splicing by GIR2 (Fig. 1D and Fig. S2) (13), undergoes a conformational change leading to the formation of DP2. The structures also highlight how a residue from the core docks within an RNA pocket that was folded after the formation of DP2, to transmit a signal for branching to the catalytic center. Moreover, we uncover the way in which residues forming the lariat are twisted by means of conserved interactions, to join chemical groups involved in branching. Finally, we demonstrate that most of the LC ribozyme structural elements have diverged from group I introns, and, although they have apparently equivalent secondary structures, they have acquired alternate functions and structures in the course of evolution.
Fig. 2.
Different views of the structure of the CP and wild-type forms of the DirLC ribozyme. (A–C) Three different views rotated by 90° clockwise on the vertical axis. (B) DirLC is represented according to the orientation used for group I introns with P9 in the upper right corner and P15 (equivalent to the P1 and P2 stems in group I ribozymes) in the lower left corner. (D) Rotation of DirLC from B rotated by 90° on a horizontal axis shows the signet ring shape resulting from the orientation of P4–P6 and DP2–DP2.1 domains. Discontinuities in the backbone are due to the absence of some residues in electron density maps. (E and F) Weighted electron-density maps of a representative region corresponding to the P15 loop contoured at 1.0 σ for the 2.5-Å (E) and 3.85-Å (F) resolution crystal structures. Statistics on data collection and refinement are summarized in Table S1.
Results
The Wild-Type and CP Forms of the LC Ribozyme Adopt Identical Folds.
The CP and wild-type forms of the DirLC crystallize in space group P212121 and present one molecule per asymmetric unit (Table S1). Because the wild-type ribozyme crystals only diffracted at low resolution, a CP construct with optimal conformational homogeneity was used. To achieve this goal, residues on either side of the cleavage site were defined as the 5′ and 3′ ends. To circumvent the poor transcription initiation capacity of the new 5′ end sequence (5′-CAU-3′) by T7 RNA polymerase, flanking ribozymes were added to the construct (14), and segments connecting DirLC to the splicing ribozyme were joined by a UUCG loop (Fig. S2). The 5′ hammerhead and the 3′ hepatitis delta virus ribozymes accurately generate 5′-hydroxyl and 2′,3′ cyclic phosphodiester ends (15). The structure of the CP DirLC ribozyme was solved by multiwavelength anomalous diffraction (MAD) experiments and was further used to solve the structure of the wild-type DirLC by the molecular replacement method. The observed pairing of DP2 strands in both the wild-type and CP forms of DirLC crystal structures demonstrated that the UUCG loop tethers the ends of DP2 without forcing its formation. The presence of the UUCG loop does not perturb the structure because another construct permutated on the loop of P8 retains catalytic activity (Fig. S3). The construct is thus locked into the active fold (13), although the opening of the scissile bond prevents the branching reaction from occurring.
The LC Ribozyme Structure Presents Unique Structural Features.
Like group I introns, DirLC is organized around stacks of helices that form three distinct domains (Fig. 1 C and E). DirLC harbors the group I typical P3–P7 pseudoknot that, together with an additional pseudoknot (P15–P3) and the associated three-way junction (3wj; P15–P3–P8), leads to a highly constrained, double pseudoknotted (P15–P3–P7) core (16). Distal from P15, the DP2.1 and DP2 elements, tethered to P10, reach P5 and P9, respectively (Fig. 1C). The curvature imposed by these tertiary interactions provokes stretching of the P4–P6 domain and results in an overall characteristic and striking signet ring shape in which the DP2.1 and the P4–P6 domains align perpendicular to the core (Fig. 2).
Two specific tertiary interactions are responsible for this characteristic shape. The GAAA tetraloop at the tip of P9 (L9) interacts with the shallow groove of the DP2 stem where two Watson–Crick purine–purine (R–R) pairs are observed (Figs. 2B and 3A). The tertiary interaction propagates through a ribose zipper between G221 and G68. The functional importance of this interaction is supported by an experiment in which a base pair was inserted or deleted within P9, which modifies the way the A residues from L9 interact in the shallow groove from DP2 (Fig. S4). Analysis of an analogous interaction between L9 and the regulatory domain in the Naegleria LC ribozyme (17) corroborates these observations. The second tertiary interaction involves the DL2.1 and the L5 loops, which form three cis WC base pairs between residues 5′-A90UC-3′ from DL2.1 and 5′-G144AU-3′ from L5 (Figs. 2D and 3B). This kissing complex is conserved at the sequence level among the known examples of LC ribozymes (Fig. S5). Moreover, both the CP and wild-type DirLC ribozyme forms show very similar shapes in solution as deduced from small angle X-ray scattering experiments (SAXS) (Fig. S6).
Fig. 3.
Tertiary interactions promoted by a zigzag of the backbone in J5/4 stabilize the 3wj involving P10. (A) The DP2/L9 tertiary interaction involves the Watson–Crick edge of residues A222, A223, and A224 from the L9 tetraloop (deep blue), which interact in the shallow groove of A248, G68, and G69 in DP2 (green), respectively. (B) The DL2.1 loop (yellow) and the L5 loop (red) form Watson–Crick base pairs between residues 5′-A90UC-3′ from DL2.1 and 5′-G144AU-3′ from L5. (C) Overall view of the P4/P6 zigzag motif. A penta-hydrated Mg2+ ion (Mg1) is bound to the O2P atom from G170 with the coordinated water molecules H-bonding with O6 positions of bases G134 and G135. The second Mg2+ ion (Mg2) is hexa-hydrated and bridges the bases of C159 and G169 from P4 and J6/7, respectively. The iridium hexammine ion in the deep groove of P5 stabilizes the loop by binding to the unpaired G141 and the preceding G═C base pair. Detailed interactions taking place in the zigzag are represented in Fig. S7. The inflection of the backbone corresponds to the A-minor interactions mediated by A152 and A153 to the G═C pairs within P4.
The Fold of the Internal Loop Between P4 and P5 Allows for the L5–L2.1 Loop–Loop Interaction.
The J5/4 junction between P4 and P5 adopts a zigzag motif, which is distant from the core and bends the P4–P6 domains so that L5 can form a kissing interaction with the loop of DP2.1 (Figs. 2 C and D and 3C). The J5/4 zigzag is made by two loop-like conformations that cap and unstack the flanking helices. The opposite J4/5 strand adopts a typical A-form helical conformation. U136 locks the zigzag by forming a Watson–Crick/Watson–Crick trans base pair with A155 that is stacked on the unpaired G137 (Fig. S7). Mutational analysis shows that A153 from the zigzag motif is critical for catalysis despite its distant location relative to the active site (12). A153 resides at the hinge of the motif where it participates in a tertiary contact through an A-minor interaction (18) to the apical G═C pair of P4, with A152 providing the second A-minor contact with the second base pair of P4 (Fig. S7C).
The DP2–DP2.1 Bridge Participates in the Folding of the Ribozyme Core.
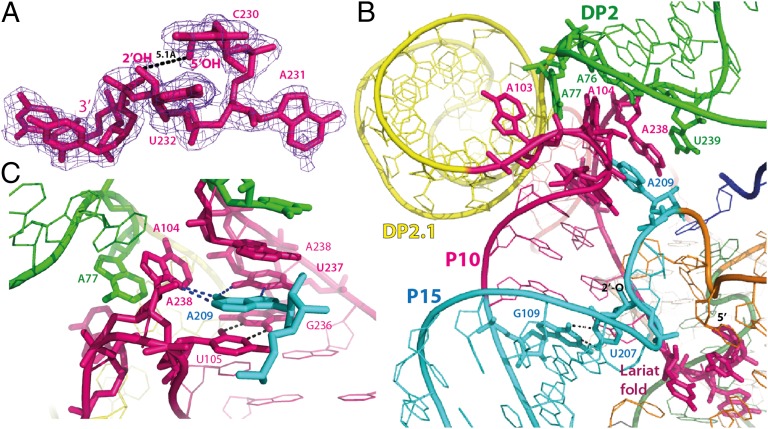
The tertiary interactions between the regulatory domain and the core lock the 3wj involving P10. P10 folds only in the active state of the LC ribozyme. Indeed, the folding of the HEG P1 hairpin characteristic of the inactive state relies on residues from P10 as well as residues downstream from DP2 (Fig. 1 C and D and Fig. S3). The absence of the 3′ residues necessary for HEG P1 folding in the crystallized constructs explains why only DP2 can be formed. At the P10 immediate 5′ end, the residues from the lariat fold are stabilized by formation of the 3wj. Specific interactions (Fig. S8) position the nucleophile U232(O2′) 5.1 Å away from the C230(O5′) (Fig. 4A), despite the absence in the CP construct of the phosphate group involved in branching in the wild-type LC ribozyme. However, the lack of electron density for C230 and A231 in the 3.85-Å crystal structure prevents a comparison from being made with what is occurring in the wild-type LC ribozyme.
Fig. 4.
Structure of the lariat fold and of the P10 3wj. (A) The lariat fold inscribed in a weighted 2Fo − Fc map contoured at 1.0 σ shows a very constrained backbone with bases popping out to interact with specific residues (Fig. S8). The 5′-OH group of C230 is at 5.1 Å from the O2′ group of U232 despite the absence of the phosphate group missing because of the design of the CP construct. (B) Overall view of the ribozyme centered on the P10 3wj shows how five adenosines (76, 77, 103, 104, and 238) and a U residue (U239) mediate stacking continuity to stabilize the A209 binding pocket (cyan). The 2′ and 5′ hydroxyl groups from U207 and from C230 are labeled. (C) A close-up shows how A209 mediates stacking with neighboring residues (U105, A238) as well as how its Hoogsteen and Watson–Crick edges form H-bonds with residues in the same plane (A104, U237).
Stacking interactions between five unpaired adenosines (residues 76, 77, 103, 104, and 238) mostly stabilize the 3wj at the P10 interface (Fig. 4B). One side of A76 and A103 stacks on the last base pair of DP2 and DP2.1, respectively. A77 stacks with A104, which stabilizes the sharp turn between DP2.1 and P10, and A238 stacks upon the unpaired U239 at the inlet of the 3′ strand of DP2. This architecture forms a cavity where a residue from the single strand joining P15 to P7 (J15/7), A209, fits snugly (Fig. 4C). A209 is intercalated between the G⋅U pair of P10 and A238. The Hoogsteen edge of A209 contacts the Watson–Crick edge of U237. Moreover, the A209 syn orientation permits additional H-bonds with A104(O2′, N3). Thus, the 3wj acts as a recognition receptor for A209. A209 is transferred to its recognition pocket by means of a S-turn (19) mediated by the trans conformation of the wobble pair G109oU207 closing P15. This unusual geometry places the O2′ group from U207 in a position opposite to the scissile bond (Fig. 4B). Downstream from A209, A210 loops back toward P7 and contacts the lariat fold as it passes. Moreover, an iridium hexammine ion in the vicinity of the lariat fold could indicate the presence of a metal ion site involved in catalysis (Fig. S8). Thus, the binding of A209 in its recognition pocket seems to signal to the core that the tertiary interactions mediated by the regulatory domain have taken place.
Structural Comparison of the LC and Group I Ribozymes.
In group I ribozymes, the three domains from the core are invariably clamped by interactions of the loops closing P2 and P9 with stems P8 and P5, respectively (20). Strikingly, P3 and P7 have the same number of base pairs in both the LC and group I ribozymes, and the manner in which P3, P7, P8, and P9 are connected to other domains is identical. In DirLC, the P2/P8 interaction is replaced by the 3wj encompassing P15–P3–P8, resulting from the double pseudoknot P3–P7–P15. This 3wj shows how two Watson–Crick U⋅U pairs provide stacking continuity. In this context, P15 is equivalent to stems P2 and P1 from group I introns. Both P1 and P15 have as a first base pair a wobble G⋅U, albeit cis or trans, respectively (Fig. 5A). At the second step of splicing, the P1 substrate docks along the junction between P4 and P5 (J4/5) to allow the O3′ group from the catalytic U to attack the bond between the intron and the 3′ exon. Consequently, in group I ribozymes, the P4–P6 domain docks along the core and forms a tertiary interaction with P9, due to the conserved presence of a structural module that bends P9 toward P5 (Fig. 5B). In DirLC, P4, P5, and P6 present the same topology as in group I ribozymes, although the domain they form adopts a completely different structure. The reorganization of P4–P6 together with the L2.1/L5 loop–loop interaction prevent any contact between P15 and J4/5, which is averted by the absence of the characteristic triple interactions which take place in group I introns between P4 and J6/7.
Fig. 5.
Global comparison of group I intron from Azoarcus (Lower) and DirLC (Upper) ribozymes. To compare 3D structures, color schemes were made identical, and the Azoarcus ribozyme was oriented similarly to DirLC by superimposing their P3 and P7 stems in program lsqman (47) (rmsd = 3.14 Å for 153 backbone atoms). The orientations of the domains left out from the rmsd calculation can be compared pairwise. (A) P3 and P7 are displayed as cartoon filled rings to show the orientation resulting from their superimposition. P15 is equivalent to the Azoarcus P1/P2 stack. The G⋅U base pair of each ribozyme is shown as sticks (cyan). (B) A 90° rotation on a vertical axis shows that the P4–P6 domain leans on the Azo core, contrarily to the situation in DirLC, where it sits perpendicularly, ensuing base pairing between L5 and DL2.1, conferring to DirLC its signet-ring shape. (C) The G⋅U trans Watson–Crick base pair at the top of P15 allows A209 to interact with A104 and U237 in the 3wj involving DP2, DP2.1, and P10. C230, A231, and U232 together form a properly folded lariat. In the postcatalytic state, G229, homologous to ωG in Azo, does not occupy the G-binding pocket of P7 as in the group I intron. It is noteworthy that A210 can adopt two conformations, one of which is pointing at the lariat, indicating that it may play a role in the catalysis. In Azo, ωG is in the G-binding pocket of P7 and close to the G⋅U base pair of P1. It forms a base triple with G130 and C177 of Azo. G128 from J6/7 forms a base triple with A129 and C178 from P7.
In the second step of splicing, the scissile phosphate is brought in close proximity of the nucleophile by the docking of ωG to the P7 G-binding site (Fig. 5C). Mutational analysis of the LC ribozyme has shown G229, which is homologous to ωG, to be a critical nucleotide (16). G229 was expected to bind at the G-binding site before branching. However, in the ribozyme crystal structures, G229 is not bound to P7 and is instead located ∼10 Å from the nucleophile in the CP crystals. G216 does not form the first base triple with either C171 or A172, a standard characteristic of P7 in group I splicing (Fig. 5C). P7 is also relaxed in group I ribozymes after cleavage (21), which could indicate that the crystal structure of the CP form reflects the postcatalytic state of the LC ribozyme.
Discussion
We have solved the crystal structures of two versions of the DirLC comprising the core and the appended regulatory domain DP2–DP2.1. The catalytically active form, solved to a resolution of 3.85 Å, harbors the same fold as the inactive CP version, solved to 2.5 Å. The similarity between the two forms is also observed in solution by SAXS studies (Fig. S6). In the two structures, the formation of the DP2 stem results from the absence of the nucleotides forming the 3′ strand of the HEG P1 hairpin (Fig. 1D) and supports the neutral effect of the UUCG loop appended to DP2 in the CP construct (Fig. 1C). Thus, the CP construct corresponds to the postcleavage state because the 3′ terminal G229 resides at 10 Å from the 5′ start C230 (Fig. 5C). It is apparent from the present structures that the ionic conditions in the crystals and the stabilization of the lariat fold by a set of precise interactions fully conserved in LC ribozymes (Fig. S5) compensate for the absence of the phosphate group closing the lariat fold. Moreover, the absence of the scissile phosphate group could influence metal ion binding in the active site. Folding of the HEG P1 hairpin in the inactive state of the LC ribozyme precludes P10 formation. Our structures reveal how the two tertiary interactions, mediated by the activated regulatory domain DP2–DP2.1, are critical for assembly of the active site of the ribozyme through folding of the 3wj including P10. The latter serves as a receptor for the internal residue A209 to nucleate folding of the catalytic core. We now better understand why the core undertakes catalysis only upon DP2 formation. It is reasonable to hypothesize that the flexibility conferred to the regulatory domain by HEG P1 may release the structural constraints applied to the core of the LC ribozyme to inhibit cleavage.
Sequence and secondary structure similarities between the LC and the Azoarcus group I ribozymes are striking (11). From a structural point of view, the similarity to the P3–P7 pseudoknot domain typical of all group I introns and the divergence from the P4–P6 domain suggest that LC ribozymes were originally derived from group I introns but then rapidly diverged, which would explain their fairly recent appearance [on a ∼106 to 107 years time scale (22)]. The high level of constraints imposed by the double pseudoknot was already used to identify the organization of the P3–P7 domain with respect to P10–P15 by molecular modeling (12). However, because of a lack of understanding concerning the tertiary interactions mediated by DP2–DP2.1, the regulatory domain was not constructed, and P4–P6 was built alongside the core as in group I introns, a feature observed neither in the crystal structures nor in the SAXS shapes presented herein. Nevertheless, the prominence of junctions J15/7 and J9/10, stressed as critical in the present study, was highlighted, albeit improperly modeled.
The appearance of this original ribozyme family could have been driven by (i) the insertion of one group I ribozyme within another (23), (ii) sequence drift due to the absence of selection pressure toward splicing (12), and (iii) the possibility that intrachain 2′ OH groups can be activated at any place of the RNA chain (24), making them statistically more reactive than 3′ OH groups. Indeed, because 2′,5′ bonds emerge readily in in-vitro-selected ribozymes (25, 26), splicing may have developed more efficiently in group II introns or in the spliceosome than from group I introns due to a more flexible lariat-based mechanism in catalytic RNAs (27). Our structures reveal the surprising speciation of elements known from two-step 3′,5′ splicing group I intron ribozymes into a structure that supports a one-step branching reaction. Group I-derived ribozymes can perform hydrolytic cleavage and splicing both in cis and in trans (28), act as a ligase (29), and work as an allosteric enzyme regulated by a second messenger (3). This ribozyme can assemble itself from pieces (30) and can even form cooperative networks that support self-replication (31). Thus, the present structure of the LC ribozyme adds to the versatility of group I ribozyme scaffolds and completes the collection of crystal structures of large ribozyme classes that comprise group I (32–34) and group II (35) splicing ribozymes and RNase P (36).
Experimental Procedures
Preparation of the Circularly Permutated Form of the LCrz Ribozyme.
RNA transcripts were obtained by in vitro transcription using T7-RNA polymerase (37). Cleavage by the 5′ hammerhead ribozyme in the course of the transcription reaction was observed while further incubation at 60 °C was necessary to complete HδV cleavage. Thus, the RNA of interest harbors a 5′ hydroxyl and a 2′,3′ cyclic phosphodiester ends.
Crystallization, X-Ray Data Collection, Structure Determination, and Refinement.
Crystal growth was completed between a few weeks and few months at 20 °C in 400-nL droplets composed of 200 nL of a 100 µM solution of purified LC RNA transcripts mixed with the same volume of crystallization solutions from various high-throughput (HT) kits from Hampton or Jena BioSciences set up by using a mosquito robot (TPP Labtech). The CP LC ribozyme crystallized in IndexHT-D11-F11-F12-H6, NatrixHT-E11, and PEGRxHT-F1-F3, and the wild-type ribozyme crystallized in IndexHT-F12 and PEGRxHT-A10-C1-F3.
Diffraction data were collected at 100 K at the macromolecular crystallography beamlines X06DA at the Swiss Light Source and PROXIMA1 at Synchrotron SOLEIL. XDS was used for processing (38). Experimental phase information of the CP LC ribozyme was obtained (Table S1) by a four-wavelength MAD experiment around the Iridium edge using the SHELX package (39). The wild-type LC ribozyme was solved by the molecular replacement method using Molrep (40) and the CP LC ribozyme crystal structure as a search model. The models were refined with Phenix (41) or Buster (42) and iteratively built by using Coot (43). Values of final Rwork/Rfree of 18.80%/23.57% and 20.99%/25.64% were obtained for the native and iridium derivative CP LC ribozymes, respectively. The wild-type form was refined to Rwork/Rfree of 24.55%/29.77%.
Supplementary Material
Acknowledgments
We thank scientists at SOLEIL, Pierre Legrand, Andrew Thompson, and Jean Cavarelli for support in the synchrotron Block Allocation Group. X-ray data were collected at the Swiss Light Source–Paul Scherrer Institut and SOLEIL. We thank Marat Yusupov for the gift of osmium hexamine; Stéphane Bellemin-Laponaz for the gift of iridium hexamine; and Kelle Freel for English-language editing. This work was supported by the Centre National de la Recherche Scientifique. M.M. was supported by a University of Strasbourg PhD grant. This work was supported in part by L'Agence Nationale de la Recherche Project ANR-10-BLAN-1502-02 “GRP2CONF” (to E.W.), le Laboratoire d'Excellence Project ANR-11-LABX-0057-MITOCROSS, and the Danish Council for Independent Research in Natural Sciences (to H.N.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4P9R, 4P95, and 4P8Z).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322248111/-/DCSupplemental.
References
- 1.Masquida B, Beckert B, Jossinet F. Exploring RNA structure by integrative molecular modelling. New Biotechnol. 2010;27(3):170–183. doi: 10.1016/j.nbt.2010.02.022. [DOI] [PubMed] [Google Scholar]
- 2.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447(7143):497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 3.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329(5993):845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen H, Beckert B, Masquida B, Johansen SD. The GIR1 branching ribozyme. In: Lilley DMJ, Eckstein F, editors. Ribozymes and RNA catalysis. London: Royal Society of Chemistry; 2008. pp. 229–252. [Google Scholar]
- 5.Nielsen H, Westhof E, Johansen S. An mRNA is capped by a 2′, 5′ lariat catalyzed by a group I-like ribozyme. Science. 2005;309(5740):1584–1587. doi: 10.1126/science.1113645. [DOI] [PubMed] [Google Scholar]
- 6.Johansen SD, Haugen P, Nielsen H. Expression of protein-coding genes embedded in ribosomal DNA. Biol Chem. 2007;388(7):679–686. doi: 10.1515/BC.2007.089. [DOI] [PubMed] [Google Scholar]
- 7.Vader A, Nielsen H, Johansen S. In vivo expression of the nucleolar group I intron-encoded I-dirI homing endonuclease involves the removal of a spliceosomal intron. EMBO J. 1999;18(4):1003–1013. doi: 10.1093/emboj/18.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vader A, Johansen S, Nielsen H. The group I-like ribozyme DiGIR1 mediates alternative processing of pre-rRNA transcripts in Didymium iridis. Eur J Biochem. 2002;269(23):5804–5812. doi: 10.1046/j.1432-1033.2002.03283.x. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen H, et al. The ability to form full-length intron RNA circles is a general property of nuclear group I introns. RNA. 2003;9(12):1464–1475. doi: 10.1261/rna.5290903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansen S, Vogt VM. An intron in the nuclear ribosomal DNA of Didymium iridis codes for a group I ribozyme and a novel ribozyme that cooperate in self-splicing. Cell. 1994;76(4):725–734. doi: 10.1016/0092-8674(94)90511-8. [DOI] [PubMed] [Google Scholar]
- 11.Johansen S, Einvik C, Nielsen H. DiGIR1 and NaGIR1: Naturally occurring group I-like ribozymes with unique core organization and evolved biological role. Biochimie. 2002;84(9):905–912. doi: 10.1016/s0300-9084(02)01443-8. [DOI] [PubMed] [Google Scholar]
- 12.Beckert B, et al. Molecular modelling of the GIR1 branching ribozyme gives new insight into evolution of structurally related ribozymes. EMBO J. 2008;27(4):667–678. doi: 10.1038/emboj.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen H, Einvik C, Lentz TE, Hedegaard MM, Johansen SD. A conformational switch in the DiGIR1 ribozyme involved in release and folding of the downstream I-DirI mRNA. RNA. 2009;15(5):958–967. doi: 10.1261/rna.669209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer M, Masquida B. cis-Acting 5′ hammerhead ribozyme optimization for in vitro transcription of highly structured RNAs. Methods Mol Biol. 2014;1086:21–40. doi: 10.1007/978-1-62703-667-2_2. [DOI] [PubMed] [Google Scholar]
- 15.Price SR, Ito N, Oubridge C, Avis JM, Nagai K. Crystallization of RNA-protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J Mol Biol. 1995;249(2):398–408. doi: 10.1006/jmbi.1995.0305. [DOI] [PubMed] [Google Scholar]
- 16.Einvik C, Nielsen H, Westhof E, Michel F, Johansen S. Group I-like ribozymes with a novel core organization perform obligate sequential hydrolytic cleavages at two processing sites. RNA. 1998;4(5):530–541. doi: 10.1017/s1355838298971758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Y, Nielsen H, Birgisdottir AB, Johansen S. A natural fast-cleaving branching ribozyme from the amoeboflagellate Naegleria pringsheimi. RNA Biol. 2011;8(6):997–1004. doi: 10.4161/rna.8.6.16027. [DOI] [PubMed] [Google Scholar]
- 18.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci USA. 2001;98(9):4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Correll CC, et al. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc Natl Acad Sci USA. 1998;95(23):13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430(6995):45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 21.Lipchock SV, Strobel SA. A relaxed active site after exon ligation by the group I intron. Proc Natl Acad Sci USA. 2008;105(15):5699–5704. doi: 10.1073/pnas.0712016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proc Natl Acad Sci USA. 1999;96(24):13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hafez M, et al. The mtDNA rns gene landscape in the Ophiostomatales and other fungal taxa: Twintrons, introns, and intron-encoded proteins. Fungal Genet Biol. 2013;53:71–83. doi: 10.1016/j.fgb.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Zaher HS, Shaw JJ, Strobel SA, Green R. The 2′-OH group of the peptidyl-tRNA stabilizes an active conformation of the ribosomal PTC. EMBO J. 2011;30(12):2445–2453. doi: 10.1038/emboj.2011.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee CS, Mui TP, Silverman SK. Improved deoxyribozymes for synthesis of covalently branched DNA and RNA. Nucleic Acids Res. 2011;39(1):269–279. doi: 10.1093/nar/gkq753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlomagno T, et al. Structural principles of RNA catalysis in a 2′-5′ lariat-forming ribozyme. J Am Chem Soc. 2013;135(11):4403–4411. doi: 10.1021/ja311868t. [DOI] [PubMed] [Google Scholar]
- 27.Anokhina M, et al. RNA structure analysis of human spliceosomes reveals a compact 3D arrangement of snRNAs at the catalytic core. EMBO J. 2013;32(21):2804–2818. doi: 10.1038/emboj.2013.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue T, Sullivan FX, Cech TR. Intermolecular exon ligation of the rRNA precursor of Tetrahymena: Oligonucleotides can function as 5′ exons. Cell. 1985;43(2 Pt 1):431–437. doi: 10.1016/0092-8674(85)90173-4. [DOI] [PubMed] [Google Scholar]
- 29.Vicens Q, Cech TR. A natural ribozyme with 3′,5′ RNA ligase activity. Nat Chem Biol. 2009;5(2):97–99. doi: 10.1038/nchembio.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Draper WE, Hayden EJ, Lehman N. Mechanisms of covalent self-assembly of the Azoarcus ribozyme from four fragment oligonucleotides. Nucleic Acids Res. 2008;36(2):520–531. doi: 10.1093/nar/gkm1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaidya N, et al. Spontaneous network formation among cooperative RNA replicators. Nature. 2012;491(7422):72–77. doi: 10.1038/nature11549. [DOI] [PubMed] [Google Scholar]
- 32.Adams PL, et al. Crystal structure of a group I intron splicing intermediate. RNA. 2004;10(12):1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golden BL, Kim H, Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat Struct Mol Biol. 2005;12(1):82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- 34.Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: Base triple sandwich and metal ion at the active site. Mol Cell. 2004;16(3):351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320(5872):77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reiter NJ, et al. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468(7325):784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichetovkin IE, Abramochkin G, Shrader TE. Substrate recognition by the leucyl/phenylalanyl-tRNA-protein transferase. Conservation within the enzyme family and localization to the trypsin-resistant domain. J Biol Chem. 1997;272(52):33009–33014. doi: 10.1074/jbc.272.52.33009. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 40.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bricogne G, et al. BUSTER. Cambridge, U.K.: Global Phasing Ltd; 2011. Version 2.11.5. [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Einvik C, Decatur WA, Embley TM, Vogt VM, Johansen S. Naegleria nucleolar introns contain two group I ribozymes with different functions in RNA splicing and processing. RNA. 1997;3(7):710–720. [PMC free article] [PubMed] [Google Scholar]
- 45.Jabri E, Cech TR. In vitro selection of the Naegleria GIR1 ribozyme identifies three base changes that dramatically improve activity. RNA. 1998;4(12):1481–1492. doi: 10.1017/s1355838298981237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7(4):499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleywegt GJ, Jones TA. A super position. CCP4/ESF-EACBM Newslett Protein Crystallogr. 1994;31:9–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.