Significance
The unfolded protein response (UPR) mitigates endoplasmic reticulum (ER) stress. In this regard, ER stress-inducing agents were found to be highly active in a clinical trial for children with relapsed acute lymphoblastic leukemia (ALL), a disease derived from transformed pre-B cells. To understand the efficacy of ER stress-inducing agents in pre-B ALL, we studied the relevance of the UPR pathway in genetic and patient-derived (xenograft) models of human pre-B ALL. Our studies revealed an unrecognized vulnerability of both normal pre-B cells and pre-B cell-derived ALL cells to genetic or pharmacological blockade of the UPR pathway. Our results establish a mechanistic rationale for the treatment of children with pre-B ALL with agents that block the UPR pathway and induce ER stress.
Abstract
The unfolded protein response (UPR) pathway, a stress-induced signaling cascade emanating from the endoplasmic reticulum (ER), regulates the expression and activity of molecules including BiP (HSPA5), IRE1 (ERN1), Blimp-1 (PRDM1), and X-box binding protein 1 (XBP1). These molecules are required for terminal differentiation of B cells into plasma cells and expressed at high levels in plasma cell-derived multiple myeloma. Although these molecules have no known role at early stages of B-cell development, here we show that their expression transiently peaks at the pre–B-cell receptor checkpoint. Inducible, Cre-mediated deletion of Hspa5, Prdm1, and Xbp1 consistently induces cellular stress and cell death in normal pre-B cells and in pre–B-cell acute lymphoblastic leukemia (ALL) driven by BCR-ABL1- and NRASG12D oncogenes. Mechanistically, expression and activity of the UPR downstream effector XBP1 is regulated positively by STAT5 and negatively by the B-cell–specific transcriptional repressors BACH2 and BCL6. In two clinical trials for children and adults with ALL, high XBP1 mRNA levels at the time of diagnosis predicted poor outcome. A small molecule inhibitor of ERN1-mediated XBP1 activation induced selective cell death of patient-derived pre-B ALL cells in vitro and significantly prolonged survival of transplant recipient mice in vivo. Collectively, these studies reveal that pre-B ALL cells are uniquely vulnerable to ER stress and identify the UPR pathway and its downstream effector XBP1 as novel therapeutic targets to overcome drug resistance in pre-B ALL.
Terminal B-cell differentiation is regulated through two sets of antagonizing transcription factors: paired box gene 5 (PAX5), BTB and CNC homology 1, basic leucine zipper transcription factor 2 (BACH2), and BCL6 maintain B-cell identity of postgerminal center B cells (1), whereas the transcription factor PR domain containing 1, with ZNF domain (PRDM1) (also known as B-lymphocyte-induced maturation protein 1; BLIMP1) and X-box binding protein 1 (XBP1) drive plasma cell differentiation (2–4). The plasma cell transcription factor XBP1 and its upstream regulator PRDM1 have been extensively studied in plasma cell differentiation and the plasma cell malignancy multiple myeloma (5, 6), but not in early B-cell development or leukemias and lymphomas representing early stages of B-cell differentiation. Surprisingly, endoplasmic reticulum (ER) stress-inducing agents were recently found to be highly active in a clinical trial for children with relapsed acute lymphoblastic leukemia (ALL) (7), a disease derived from transformed pre-B cells.
XBP1 is highly expressed in multiple myeloma and plasma cells, where it mitigates ER stress through engagement of the unfolded protein response (UPR). The UPR network consists of three major branches including Inositol-requiring enzyme 1a (IRE1α, ERN1), PKR-like ER kinase, and activating transcription factor 6 (ATF6) (8). ERN1 is activated by ER stress through autophosphorylation and oligomerization and induces cleavage and splicing of XBP1 by its endoribonuclease (RNase) domain, resulting in the removal of a 26-nucleotide intron. This frame shift modification leads to expression of a longer, highly active splice variant (XBP1-s) (9), responsible for the regulation of a variety of downstream targets to relieve ER stress. Activation of the UPR by ER stress has been linked to the transition of mature surface Ig-dependent B cells to Ig-secreting plasma cells that no longer express Ig on the surface. An important role in this transition is played by Ig heavy chain-binding protein (BiP)—also known as heat shock 70-kDa protein 5 (HSPA5) and Grp78—which chaperones folding of Ig heavy, but not light, chain proteins (10). A previous study also demonstrated that IRE1 (ERN1) is required during V(D)J recombination at the transition from pro- to pre-B cells (11).
Here we provide genetic evidence for the emerging concept that the UPR molecules ERN1 and HSPA5, as well as their downstream effectors PRDM1 and XBP1, are not only critical for the transition from surface Ig-dependent B cells to Ig-secreting plasma cells, but also regulate the pre–B-cell stage, when Ig heavy-chain variable region genes are rearranged and Ig heavy chains are expressed for the first time. Whereas recent studies have identified XBP1 as a therapeutic target in multiple myeloma, we demonstrate that pre-B ALL cells are distinctly sensitive to ER stress and pharmacological inhibition of XBP1. Pre-B ALL arises from a transformed pre-B cell and represents the most frequent type of cancer in children (12). Frequent oncogenic lesions in pre-B ALL include BCR-ABL1 (present in ∼30% of adult ALL) (13) and hyperactivating mutations of RAS genes (in ∼50% of childhood pre-B ALL) (14). Small-molecule inhibitors targeting the RNase domain of ERN1 prevent activation of XBP1 and have disease-modifying activity in multiple myeloma (15, 16). Here we show that small-molecule inhibition of ERN1/XBP1 represents a promising novel approach to overcome drug resistance in pre-B ALL.
Results
Hspa5 (Ig Heavy-Chain Binding Protein) Is Required for the Survival and Malignant Transformation of Pre-B Cells.
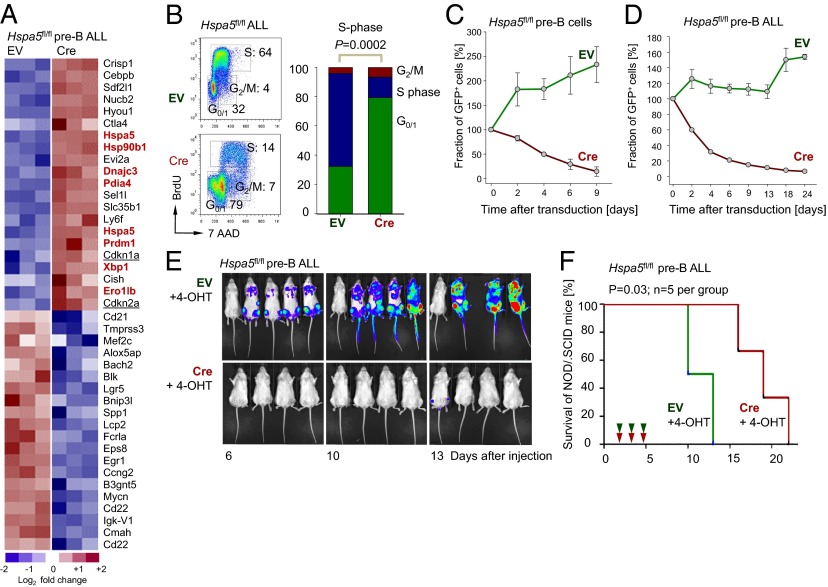
To elucidate the function of UPR-related molecules in both normal pre-B and transformed pre-B ALL cells, we first focused on the ER chaperone heat shock 70-kDa protein 5 (HSPA5) (17), which mitigates ER stress upstream of the ER sensor ERN1 (IRE-1). In a genetic experiment, we studied the consequences of acute ablation of Hspa5 function in normal pre-B cells and BCR-ABL1–transformed pre-B ALL cells that were generated from bone marrow B-cell progenitor cells of Hspa5fl/fl mice (18) (Fig. 1). The 4-hydroxytamoxifen (4-OHT)-mediated activation of Cre for 2 d resulted in excision of Hspa5 exons 5–7 and compensatory up-regulation of Hspa5 mRNA (Fig. 1A), whereas Hspa5 protein was destabilized and no longer detectable by Western blot (SI Appendix, Fig. S1A). Interestingly, acute ablation of Hspa5 function results in strong up-regulation of the ER chaperones Hsp90b1, Dnajc3, and others as well as Prdm1 and Xbp1 (highlighted in Fig. 1A). Inducible deletion of Hspa5 has profound effects and rapidly induced cell cycle arrest (Fig. 1B) in parallel with strong up-regulation of the cell-cycle checkpoint regulators Arf (Cdkn2a) and p21 (Cdkn1a; underlined in Fig. 1A). Microarray gene expression analysis was validated by single-gene quantitative RT (qRT)-PCR (SI Appendix, Fig. S2). Within 4 d of Hspa5 deletion, ∼50% of IL-7–dependent pre-B cells (Fig. 1C) and >70% of BCR-ABL1–transformed pre-B ALL cells (Fig. 1D) underwent cell death. Deletion of Hspa5 in BCR-ABL1–transformed pre-B ALL cells in vivo resulted in a drastic reduction of leukemia burden, as measured by luciferase bioimaging (Fig. 1E) and prolonged overall survival of transplant recipient mice (Fig. 1F). In these experiments, 1 million luciferase-labeled Hspa5fl/fl BCR-ABL1 pre-B ALL cells carrying either tamoxifen-inducible GFP-tagged Cre (Cre; n = 9) or a GFP-tagged empty vector (EV) control (n = 9) were injected (i.v.) into each sublethally irradiated (2 Gy) NOD/SCID mouse. Although deletion of Hspa5 significantly extended overall survival of recipient mice (P = 0.028; Fig. 1F), recipient mice in both groups eventually developed terminal disease and had to be killed. In the analysis of bone marrows and spleens from transplant recipient mice we distinguished infiltrating CD45.1− CD45.2+ leukemia cells from CD45.1+ CD45.2− host cells. Interestingly, virtually all CD45.2+ ALL cells expressed the GFP-tagged EV control, whereas expression of GFP-tagged Cre was silenced in a fraction of leukemia cells (SI Appendix, Fig. S1B). Consistent with these findings, ALL clones in all nine NOD/SCID mice receiving Cre-transduced Hspa5fl/fl leukemia cells retained Hspa5 floxed alleles in contrast to complete deletion in vitro (SI Appendix, Fig. S1C). These findings imply that Hspa5fl/fl ALL in this transplant model is strongly selected for clones that escaped Hspa5 deletion. Fatal disease developing in NOD/SCID mice receiving Cre-transduced Hspa5fl/fl leukemia cells might originate from clones that had escaped complete Cre-mediated deletion.
Fig. 1.
Hspa5 (Ig heavy-chain binding protein) is required for the survival and malignant transformation of pre-B cells. (A) Gene expression profiling was assessed in Hspa5fl/fl pre-B ALL cells transduced with EV control and 4-OHT–inducible retroviral Cre (Cre) after 2 d of 4-OHT treatment. Highlighted are ER-related molecules such as Hsp90b1, Dnajc3, Pdia4, and Ero1lb as well as Prdm1 and Xbp1 in red and cell-cycle checkpoint regulators such as Arf (Cdkn2a) and p21 (Cdkn1a) (GEO accession no. GSE53684). (B) Hspa5fl/fl ALL cells with EV and Cre were stained with BrdU and 7AAD and the percentages of cells in the G0/1, S, and G2/M cell cycles are indicated after 1 d of 4-OHT treatment and shown in bar graph (n = 3). (C) Hspa5fl/fl pre-B cells were transduced with MSCV-GFP (EV) and Cre-GFP (Cre) and the relative fraction of GFP+ cells was measured up to 9 d (n = 3). (D) Similarly, the relative fraction of GFP+ cells in Hspa5fl/fl pre-B ALL cells with EV and Cre was measured up to 24 d (n = 3). (E) One million luciferase-labeled Hspa5fl/fl pre-B ALL cells carrying either tamoxifen-inducible GFP-tagged Cre (CreGFP) or a GFP-tagged empty vector control (EVGFP) were injected (i.v.) into sublethally irradiated (2 Gy) NOD/SCID mice and leukemic expansion was tracked by luciferase bioimaging. (F) The overall survival of transplant recipient mice is shown in a Kaplan–Meier analysis (n = 5 per group, P = 0.03).
Expression of Plasma Cell-Specific UPR Molecules ERN1, HSPA5, PRDM1, and XBP1 in Normal Pre-B Cells and Pre-B ALL.
To study the function of the ER chaperone HSPA5 in the context of other UPR-related genes [ERN1 (IRE1), HSPA5 (BIP), PRDM1 BLIMP, and XBP1] we tracked expression of these genes throughout early and late stages of B-cell development (SI Appendix, Fig. S3). As expected, plasma cells express these molecules at the highest levels (SI Appendix, Fig. S3A). Consistent with our findings for HSPA5, promoter regions of ERN1, PRDM1, and XBP1 were hypomethylated in human pre-B ALL (SI Appendix, Fig. S3 A, E, and I) compared with normal pre-B cells or mature B-cell lymphomas. Similarly, mRNA levels of ERN1, PRDM1, and XBP1 were high in pre-B ALL and also peaked during normal early B-cell development at the pre–B-cell receptor (pre-BCR) checkpoint in human bone marrow (SI Appendix, Fig. S3A). The XBP1 microarray probeset (200670_at) does not distinguish between the unspliced XBP1-u and the more active, spliced form XBP1-s (9); therefore, specific up-regulation of Xbp1-s at the pre-BCR checkpoint was confirmed by qRT-PCR (SI Appendix, Fig. S3B), using sorted Lin− Sca-1+ c-Kit+ multilineage progenitors, bone marrow B-cell precursor populations, as well as mature B cells from spleens of C57BL/6 mice (Hardy fractions A–F; SI Appendix, Fig. S4A). Previous studies of ERN1, HSPA5, PRDM1, and XBP1 largely focused on their critical role in plasma cell differentiation (i.e., the transition from surface Ig-dependent B cells to Ig-secreting plasma cells) (19, 20). Specific up-regulation at early pre–B-cell stages suggests that these molecules are also functional at the pre-BCR checkpoint, when Ig heavy-chain variable genes are assembled and Ig heavy chains are expressed for the first time. For instance, HSPA5 was first described as “Ig heavy chain-binding protein” (BiP), which binds to Ig heavy-chain proteins in pre-B cells (10). In mice lacking the transmembrane exon of the Ig μ-heavy chain (Ighm−/−) (21), B-cell development is arrested at the pre-BCR checkpoint. Interestingly, early pre-B cells from Ighm−/− mice express high levels of Xbp1-s; however, reconstitution of Ig μ-heavy-chain expression reinstated differentiation of pre-B cells past the pre-BCR checkpoint and induced reduction of Xbp1-s expression (SI Appendix, Fig. S3C).
The Plasma Cell Transcription Factor XBP1 Is Demethylated and Up-Regulated in Pre-B ALL Cells.
In patient-derived pre-B ALL samples including various ALL subsets, the promoter regions of ERN1, HSPA5, PRDM1, and XBP1 genes were consistently demethylated compared with mature B-cell non-Hodgkin lymphomas (SI Appendix, Fig. S3 D–G). Low levels of CpG methylation in promoter regions correlated with high mRNA levels of ERN1, HSPA5, PRDM1, and XBP1, and also for BCR-ABL1–driven Ph+ ALL (SI Appendix, Fig. S3 H–K). High levels of promoter CpG methylation correlated with low expression levels in mature B-cell lymphoma. Overall, relative mRNA levels of ERN1, HSPA5, PRDM1, and XBP1 were higher in ALL cells compared with normal pre-B cells but lower compared with plasma cells (SI Appendix, Fig. S3 H–K). Similarly, Western blot analysis showed that both PRDM1 and XBP1-s are expressed in CD19+ magnetic activated cell sorting (MACS)-enriched pre-B cells (SI Appendix, Fig. S4B); however, expression levels in patient-derived pre-B ALL samples are significantly higher for XBP1-s whereas only a slight increase was observed in the case of PRDM1 as verified by densitometry analysis (SI Appendix, Fig. S5A).
Prdm1 (Blimp1) Is Required for Proliferation and Survival of Pre-B ALL Cells.
To study the role of Prdm1 in pre-B ALL cells using a genetic experiment, we transformed bone marrow pre-B cells from Prdm1fl/fl mice (22) with the human BCR-ABL1 tyrosine kinase that drives Ph+ ALL. Inducible deletion of Prdm1 was achieved by retroviral activation of 4-OHT–inducible Cre in Prdm1fl/fl BCR-ABL1 pre-B ALL cells and deletion was verified after 1, 2, and 5 d (SI Appendix, Fig. S5 B and C), significantly reducing the expression and activity of its target Xbp1, indicating that Prdm1 is functionally active in BCR-ABL1 pre-B ALL cells (SI Appendix, Fig. S5D). Interestingly, inducible activation of the oncogenic TEL-SYK tyrosine kinase was shown to result in Prdm1-dependent terminal differentiation of B cells into plasma cells (23). Nevertheless, activation of Prdm1 downstream of the BCR-ABL1 tyrosine kinase in human Ph+ ALL cells was not sufficient to induce terminal differentiation and did not affect expression levels of CD19 and CD138 as in plasma cells and multiple myeloma (2) (SI Appendix, Fig. S5E). Interestingly, Cre-mediated ablation of Prdm1 induced apoptosis (day 2: P = 0.004; day 5: P = 0.0002; SI Appendix, Fig. S5F) and cell-cycle arrest (SI Appendix, Fig. S5G) in BCR-ABL1 pre-B ALL cells. We conclude that Prdm1 is important for survival and proliferation in a mouse model of human Ph+ ALL.
Oncogenic Tyrosine Kinase Activity Drives Expression of XBP1-s in Ph+ ALL Cells.
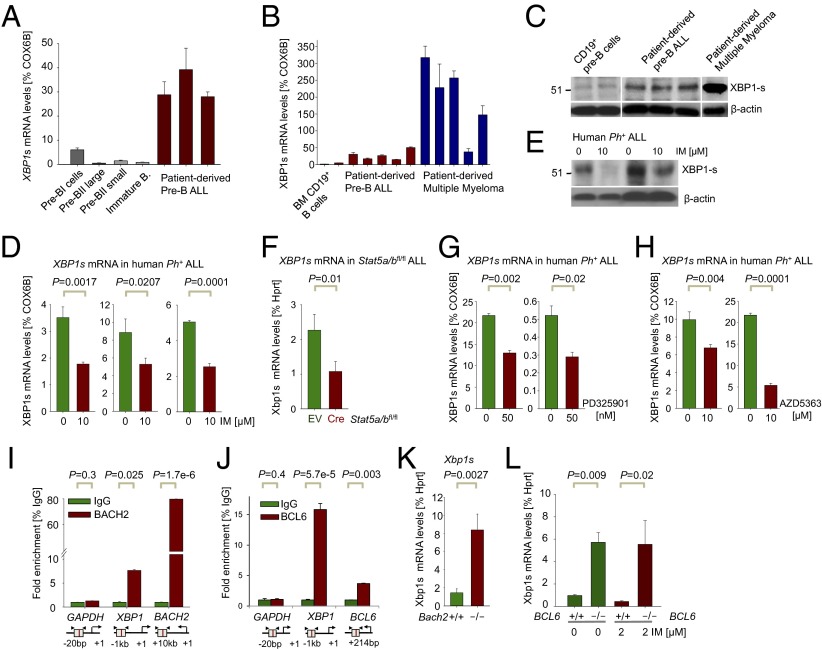
As highlighted in our gene expression analysis (Fig. 1A), increased ER stress owing to acute ablation of Hspa5 results in strong up-regulation of Xbp1 mRNA in BCR-ABL1 pre-B ALL cells, as also suggested by our microarray analyses (SI Appendix, Fig. S3K), while not distinguishing XBP1-u and XBP1-s (9). Therefore, using qRT-PCR primers that specifically amplify XBP1-s, we confirmed that XBP1-s mRNA levels were 5- to 41-fold higher in patient-derived pre-B ALL cells compared with normal bone marrow B-cell precursor populations (Fig. 2A; P = 0.001), albeit up to sevenfold lower than in some cases of multiple myeloma (Fig. 2B). At the protein level, similar differences were observed (Fig. 2C). Because the oncogenic BCR-ABL1 tyrosine kinase accounts for ∼30% of cases of adult ALL (13), we tested whether oncogenic BCR-ABL1 tyrosine kinase activity is responsible for increased levels of Xbp1-s in Ph+ ALL compared with normal pre-B cells and other ALL subsets lacking an oncogenic tyrosine kinase (SI Appendix, Fig. S3K). Consistent with this hypothesis, tyrosine kinase inhibitor (TKI) (imatinib, IM) treatment of human Ph+ ALL cells reduced mRNA (Fig. 2D) and protein levels (Fig. 2E; 54 kDa) of XBP1-s. Besides activation of Ras-MAPK and PI3K/AKT pathways, STAT5 represents a critical mediator of BCR-ABL1 downstream signaling (24). Inducible deletion of Stat5a and Stat5b (25) in BCR-ABL1–transformed Stat5a/bfl/fl ALL cells reduced Xbp1-s mRNA levels to a degree similar to inhibition of BCR-ABL1 itself (Fig. 2F). Besides STAT5, oncogenic downstream signaling from BCR-ABL1 is also involves the PI3K/AKT and RAS-MAPK pathways. For this reason, we treated patient-derived pre-B ALL cells with either an AKT (AZD5363) or MEK inhibitor (PD325901). Both inhibition of MEK (SI Appendix, Fig. S6A) and AKT (SI Appendix, Fig. S6B) significantly reduced XBP1s mRNA levels to a comparable degree as seen after genetic deletion of STAT5 (Fig. 2 G and H). We conclude that BCR-ABL1 oncogenic tyrosine kinase activity up-regulates expression and activity of XBP1 via STAT5, ERK, and AKT.
Fig. 2.
Regulation of XBP1 by STAT5, ERK, and AKT (positive) and BACH2 and BCL6 (negative) in Ph+ ALL. (A) XBP1-s mRNA levels were measured by qRT-PCR in sorted B-cell fractions from healthy human donors compared with patient-derived pre-B ALL cells (ALL-Q5, ALL-O2, ALL-X2, BLQ11, and LAX9; n = 3) and (B) in two CD19+ MACS-enriched pre–B-cell samples and five patient-derived multiple myeloma samples (MM1–5; n = 3). (C) Western blot analysis of spliced XBP1-s for MACS-enriched normal human CD19+ pre-B cells compared with patient-derived pre-B ALL cells (as in B) and one patient-derived multiple myeloma (MM6) sample using β-actin as loading control. (D) XBP1-s mRNA levels were measured in Ph+ ALL cell lines (NALM1, TOM1, and BV173) treated with or without the TKI IM for 16 h (10 µM IM) by qRT-PCR (n = 3). (E) Protein levels of XBP1-s were measured by Western blot analysis in patient-derived Ph+ ALL cases (LAX9 and PDX59) treated with IM for 16 h (10 µM IM) using β-actin as loading control. (F) Xbp1-s mRNA levels were measured in Stat5a/bfl/fl ALL cells with EV control and 4-OHT–inducible Cre (Cre) after 24 h by qRT-PCR. (G and H) XBP1-s mRNA levels were measured in patient-derived pre-B ALL cases (PDX2 and LAX7R) treated with or without PD325901 or AZD5363 for 5 h (50 nM PD325901; 10 µM AZD5363) by qRT-PCR (n = 3). (I) Quantitative ChIP validation of BACH2 binding to the XBP1 promoter in a human Ph+ ALL patient sample (ICN1), GAPDH as a negative and BACH2 as a positive control.(J) Quantitative ChIP validation of BCL6 binding to the XBP1 promoter in human pre-B ALL cells. (K) XBP1 expression was assessed in Bach2−/− ALL cells compared with WT controls (Bach2+/+) by qRT-PCR using specific primers for spliced Xbp1-s (n = 3). (L) Xbp1s mRNA levels were measured by qRT-PCR in Bcl6−/− ALL cells compared with WT controls (Bcl6+/+) treated either with or without the TKI IM for 16 h (2 µM IM).
XBP1 Is a Target and Transcriptionally Repressed by Both BACH2 and BCL6 in Ph+ ALL.
Recent work from our laboratory had identified BCL6 (26) and BACH2 (27) as transcriptional repressors that oppose activation signals downstream of BCR-ABL1 and STAT5 in Ph+ ALL. Contrary to down-regulation of XBP1-s expression, both BCL6 (26) and BACH2 (SI Appendix, Fig. S6C) are strongly up-regulated by TKI (IM) treatment of Ph+ ALL cells. ChIP sequencing (ChIP-seq) and ChIP-on-chip analysis revealed strong binding of both BACH2 and BCL6 to the XBP1 promoter (SI Appendix, Fig. S6D), which was further enhanced by TKI treatment (IM) (SI Appendix, Fig. S6E). Specific binding of BACH2 and BCL6 to XBP1 promoter regions in Ph+ ALL cells was confirmed by single-locus quantitative ChIP (Fig. 2 I and J). Genetic loss of function models for Bach2−/− and BCL6−/− demonstrated that Xbp1-s mRNA levels in Bach2−/− and BCL6−/− BCR-ABL1 ALL cells were eightfold (Bach2−/−) and sixfold (BCL6−/−) higher compared with WT controls (Fig. 2 K and L). These findings are consistent with our observation of down-regulation of Xbp1-s levels during normal pre–B-cell differentiation after the pre-BCR checkpoint (SI Appendix, Fig. S3 B and C). De novo expression of Ig μ-heavy chains results in rapid down-regulation of Xbp1-s in parallel with up-regulation of its transcriptional repressors Bach2 (27) and BCL6 (28) at this stage.
Inducible Ablation of Xbp1 Causes UPR-Related Gene Expression Change in Transformed Pre-B Cells.
To test the function of the plasma-cell transcription factor XBP1 in normal pre-B cells and transformed pre-B ALL (BCR-ABL1 and NRASG12D) cells, we developed a genetic system for inducible deletion of Xbp1 in BCR-ABL1– (29) and NRASG12D- transformed pre-B ALL cells. To this end, we isolated bone marrow B-cell precursors from Xbp1fl/fl mice (30) and cultured the cells in the presence of IL-7. To study the role of Xbp1 in B-cell precursor ALL, we transduced IL-7–dependent Xbp1fl/fl pro-B and pre-BI cells with retroviral BCR-ABL1 to model human Ph+ ALL (13) and NRASG12D to model RAS-driven childhood ALL (14).
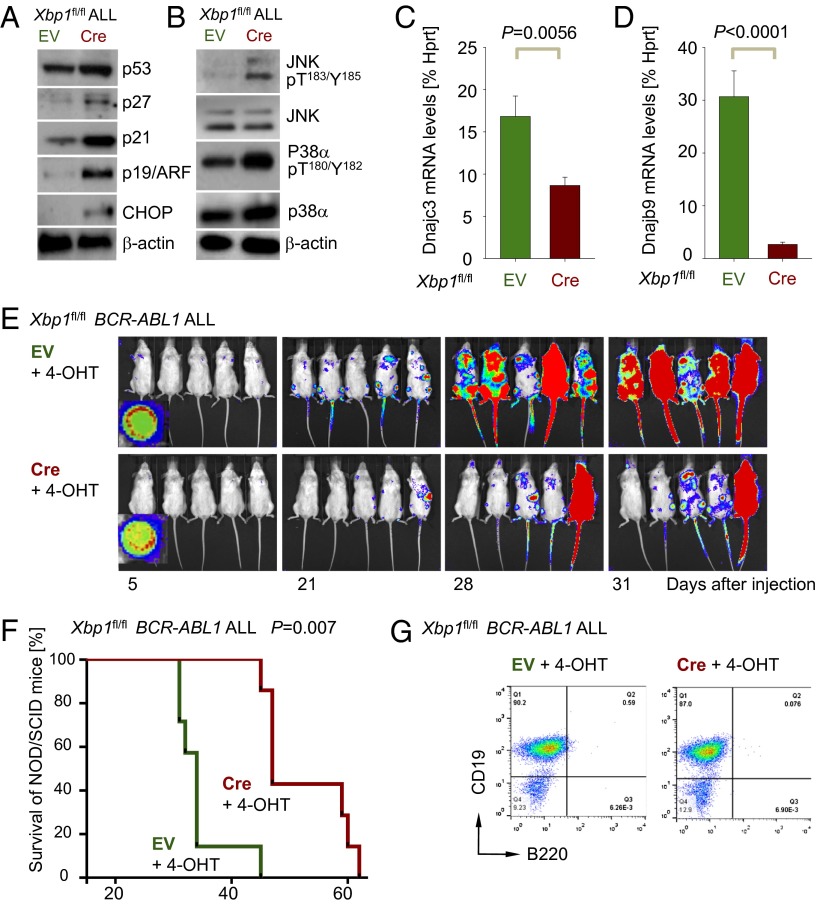
We first examined gene expression changes in response to Cre-mediated deletion of the Xbp1 transcription factor (Fig. 3A and SI Appendix, Fig. S7) in BCR-ABL1–transformed pre-B ALL cells (Fig. 3B). Genetic deletion of Xbp1 function resulted in decreased expression of molecules related to the secretory and Golgi apparatus (Sirpa, B3gnt5, Cst7, Gldc, Hs3st2, and Slc16a3, underlined in Fig. 3B) as well as Ig light chains and the early B-cell antigens Enpep and Cd22 (highlighted in blue, Fig. 3B). Conversely, ablation of Xbp1 induced compensatory up-regulation of Hsp90b1, Derl3, and Atf6 (highlighted in red, Fig. 3B) to potentially mitigate increased ER stress following Xbp1 deletion. This interpretation would be consistent with up-regulation of the stress/activation markers Il2ra (Cd25), Cd80, and Ly6f (Cd59). The ER-stress sensor ERN1 initiates splicing and activation of Xbp1 and is negatively regulated by the ER chaperone Hspa5 (BiP) (31). Given the importance of ERN1-mediated splicing for Xbp1 function, we hypothesized that inducible deletion of Hspa5 will result in a net gain of Xbp1 function. On this basis, we predicted that observed gene expression changes upon Xbp1 deletion (Fig. 3B, Left), would be reversed by Hspa5 deletion (Fig. 3B, Right). In fact, only two molecules (Pbx3 and Evi2a) were regulated in opposite direction by deletion of Xbp1 and Hspa5. Of the 41 most significant Xbp1-dependent gene expression changes (P < 0.001), 27 were recapitulated in the same direction by inducible ablation of Hspa5, indicating that most of Xbp1- and Hspa5-dependent gene expression changes are not direct targets of Xbp1 but are affected indirectly through increased levels of ER stress.
Fig. 3.
Inducible ablation of Xbp1 causes cell-cycle arrest and apoptosis in transformed pre-B cells. (A) Genomic deletion of Xbp1 was verified using specific primers to amplify floxed alleles in Xbp1fl/fl ALL cells transduced with EV control or 4-OHT–inducible Cre (Cre) after in vitro deletion or in vivo (three mice after treatment with 4-OHT) WT and H2O controls. (B) Gene expression profiling of Cre-mediated deletion of Xbp1- or Hspa5-deletion in BCR-ABL1–transformed Xbp1fl/+ or Hspa5fl/fl pre-B ALL cells. Highlighted are molecules related to the secretory and Golgi apparatus (Sirpa, B3gnt5, Cst7, Gldc, Hs3st2, and Slc16a3,underlined), Ig light chains and the early B-cell antigens Enpep and Cd22 (highlighted in blue) and ER chaperons (Hsp90b1, Derl3 and Atf6, highlighed in red; GEO accession nos. GSE53683 and GE53684). (C) Apoptosis was measured by Annexin-V and 7AAD staining in Xbp1fl/fl ALL cells with EV or Cre after 1 and 2 d 4-OHT treatment (n = 3). (D) Xbp1fl/fl ALL cells with EV or Cre were stained with BrdU and 7AAD and the percentages of cells in the G0/1, S, and G2/M cell cycles are indicated after 24 h of 4-OHT treatment and shown in bar graph (n = 3). (E) Xbp1fl/fl ALL cells with EV or Cre were stained with ER tracker green after 24 h of 4-OHT treatment (n = 3). (F) Xbp1fl/fl ALL cells with EV or Cre were stained in a senescence-associated β-galactosidase assay after 24 h of 4-OHT treatment and percentages of positively senescent cells are indicated and shown in bar graph (n = 3).
Inducible Ablation of Xbp1 Causes Cell Cycle Arrest and Apoptosis in Transformed Pre-B Cells.
Inducible deletion of Xbp1 resulted in increased apoptosis (Fig. 3C) and cell-cycle arrest (Fig. 3D) and induced ER stress in the vast majority of BCR-ABL1 ALL cells as measured with a recently established ER tracker dye (32) (Fig. 3E). Furthermore, Cre-mediated deletion of Xbp1 resulted in an approximately eightfold increased propensity of leukemia cells to cellular senescence (P = 0.0001; Fig. 3F) as measured by staining for senescence-associated β-galactosidase (26).
Loss of Xbp1 Function Activates Proapoptotic Pathways and Prolongs Survival of BCR-ABL1 Pre-B ALL Transplant Recipient Mice.
Consistent with induction of cellular senescence upon loss of Xbp1 function, we observed that Cre-mediated deletion of Xbp1 resulted in up-regulation of the checkpoint molecules Arf (Cdkn2a), p53, p21 (Cdkn1a), and p27 (Cdkn1b; Fig. 4A). Activation of C/EBP homologous protein (Chop) is the result of prolonged ER stress, ultimately leading to apoptosis (33). Cre-mediated deletion of Xbp1 led to increased expression of Chop (Fig. 4A) and also caused increased phosphorylation of the proapoptotic MAP kinase Jnk1/2 (T183/Y185), whereas increased phosphorylation of p38α (T180/Y182) may be due to an increase of global p38α protein levels (Fig. 4B). Cre-mediated deletion of Xbp1 demonstrated that Xbp1 is also required to relieve ER stress through activation of DnajC3 (p58IPK) and DnjaB9 (Erdj4; Fig. 4 C and D). DnajC3 is known to reduce the overall substrate influx (34) and DnjaB9 is important in the stabilization of Hspa5 and thereby prevents the accumulation of unfolded proteins (35). We next tested whether Cre-mediated deletion of Xbp1 affects the course of pre-B ALL disease in an in vivo transplant model. To this end, 1 million BCR-ABL1–transformed Xbp1fl/fl pre-B ALL cells carrying 4-OHT–inducible Cre (Cre-ERT2) or an EV control were injected into sublethally irradiated NOD/SCID mice. Pre-B ALL cells were labeled with firefly luciferase and leukemic expansion was monitored by luciferase bioimaging (Fig. 4E). Xbp1fl/fl leukemia cells transduced with EV rapidly expanded and caused lethal disease in all recipient mice within 45 d. By contrast, inducible Cre-mediated deletion of Xbp1 delayed the onset of disease and substantially prolonged overall survival of recipient mice (Fig. 4 E and F; P = 0.007). Analysis of infiltrating BCR-ABL1 pre-B ALL cells in mice that succumbed to disease revealed that deletion of Xbp1 did not cause a significant phenotypic change of the ALL cells (Fig. 4G).
Fig. 4.
Loss of Xbp1 function activates proapoptotic pathways and prolongs survival of BCR-ABL1 pre-B ALL transplant recipient mice. (A) Protein levels of Arf (Cdkn2a), p53, p21 (Cdkn1a), p27 (Cdkn1b), and Chop were studied by Western blot analysis in Xbp1fl/fl ALL cells with (Cre) or without (EV) deletion of Xbp1 using β-actin as loading control (n = 3). (B) Likewise, phosphorylation of MAP kinases p38α (T180/Y182) and JNK1/2 (T183/Y185) were assessed (n = 3). (C and D) mRNA levels of DnajC3 (p58IPK) and DnajB9 (Erdj4) were measured in Xbp1fl/fl ALL cells with (Cre) or without (EV) deletion of Xbp1 by qRT-PCR (n = 3). (E) One million luciferase-labeled Xbp1fl/fl ALL cells with EV or Cre were injected (i.v.) into sublethally irradiated (2.5 Gy) NOD/SCID mice and leukemic expansion was tracked by luciferase bioimaging. (F) Overall survival of transplant recipient mice is shown in a Kaplan–Meier analysis (n = 7 per group). (G) A representative FACS analysis for surface markers CD19 and B220 (n = 4) is shown for Xbp1fl/fl ALL cells isolated from killed mice.
Xbp1 Function Is Critical for the Survival and Proliferation of Normal and NRASG12D Transformed Pre-B Cells.
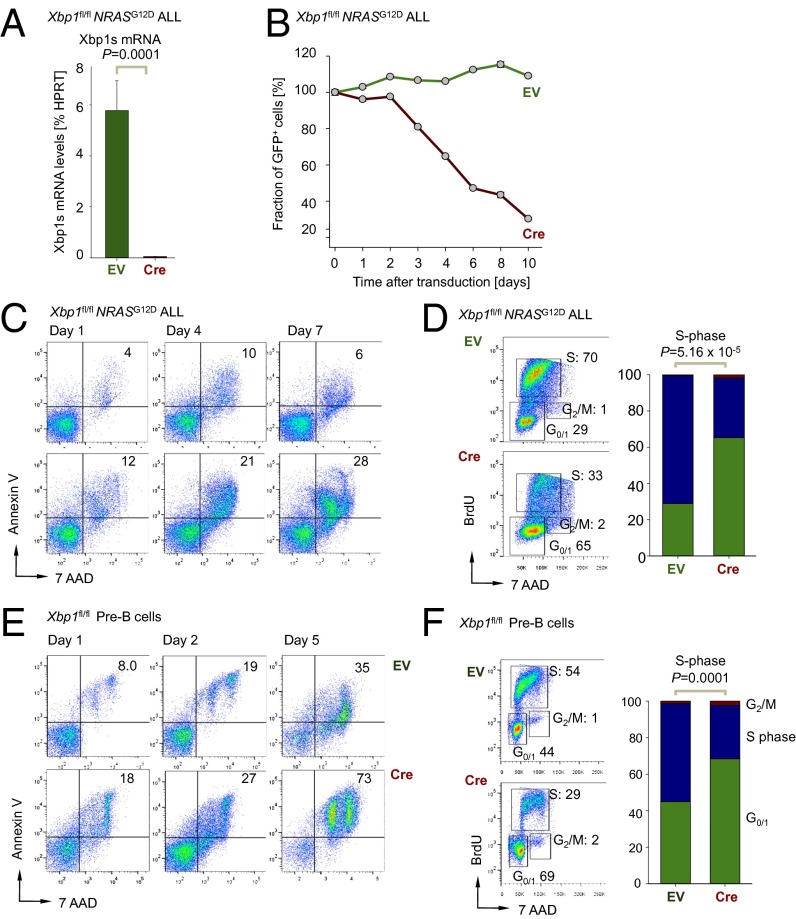
Oncogenic lesions that result in hyperactivation of the RAS pathway occur in ∼50% of childhood ALL cases (14) and are particularly frequent in MLL-rearranged and low hypodiploid ALL (36). To investigate whether our observations in BCR-ABL1–transformed pre-B ALL and Ph+ ALL (∼30% of ALL cases in adults) also pertain to RAS-driven ALL, we used as described above a NRASG12D -driven ALL model and induced deletion of Xbp1 using 4-OHT–inducible Cre (Fig. 5A). As observed for BCR-ABL1–transformed ALL, NRASG12D-driven ALL cells also underwent cell cycle arrest and apoptosis (Fig. 5 B–D and SI Appendix, Fig. S7E), although the kinetics of cell death induction following ablation of Xbp1 were slower compared with BCR-ABL1 ALL cells (Fig. 5B). We conclude that Xbp1 function is critical for survival and proliferation in two major subsets of high-risk ALL, namely Ph+ ALL and ALL driven by hyperactive mutant RAS. Compared with mature B-cell subsets, normal pre-B cells express XBP1-s at increased levels, albeit 5- to 41-fold lower than in pre-B ALL cells (Fig. 2A and SI Appendix, Fig. S3G). We, therefore, tested how Cre-mediated inducible deletion of Xbp1 affects normal IL-7–dependent pro- and pre-BI cells that were transduced with a 4-OHT–inducible Cre and EV controls. Although effects of Xbp1 deletion were less profound than in pre-B ALL cells, Cre-mediated deletion of Xbp1 caused increased apoptosis and G0/G1 cell-cycle arrest in normal pro- and pre-BI cells (Fig. 5 E and F and SI Appendix, Fig. S7F). These findings indicate that high expression levels of Xbp1 at the pro-B and pre-BI stages of B-cell development (SI Appendix, Fig. S3B) reflect an important function of Xbp1 in regulating pre–B-cell survival. Therapeutic targeting of Xbp1 in human ALL will most likely also affect normal B lymphopoiesis.
Fig. 5.
Xbp1 function is critical for the survival and proliferation of normal and NRASG12D transformed pre-B cells. (A) Xbp1 mRNA levels were measured in Xbp1fl/fl NRASG12D ALL cells transduced with EV control or 4-OHT–inducible retroviral Cre (Cre) after 48 h of 4-OHT treatment by qRT-PCR (n = 3). (B) Xbp1fl/fl NRASG12D ALL were transduced with GFP-tagged EV control or 4-OHT–inducible GFP-tagged Cre (Cre) and the relative fraction of GFP+ cells was measured (n = 3). (C) Apoptosis was assessed by Annexin-V and 7AAD staining in Xbp1fl/fl NRASG12D ALL cells with EV and Cre after 1, 4, and 7 d of treatment with 4-OHT (n = 3). (D) Xbp1fl/fl NRASG12D ALL cells with EV and Cre were stained with BrdU and 7AAD and the percentages of cells in the G0/1, S, and G2/M cell cycles are indicated after 5 d of 4-OHT treatment and shown in a bar graph (n = 3). (E) Apoptosis was assessed by Annexin-V and 7AAD FACS staining in IL-7–dependent Xbp1fl/fl pre-B cells with (Cre) or without (EV) deletion of Xbp1 after 1, 2, and 5 d of treatment with 4-OHT (n = 3). (F) IL-7–dependent Xbp1fl/fl pre-B cells with EV and Cre were stained with BrdU and 7AAD and the percentages of cells in the G0/1, S, and G2/M cell cycles are indicated after 5 d of 4-OHT treatment and shown in bar graph (n = 3).
Effects of Pharmacological ERN1/XBP1 Inhibition in Patient-Derived Pre-B ALL Cells in Vitro and in Vivo.
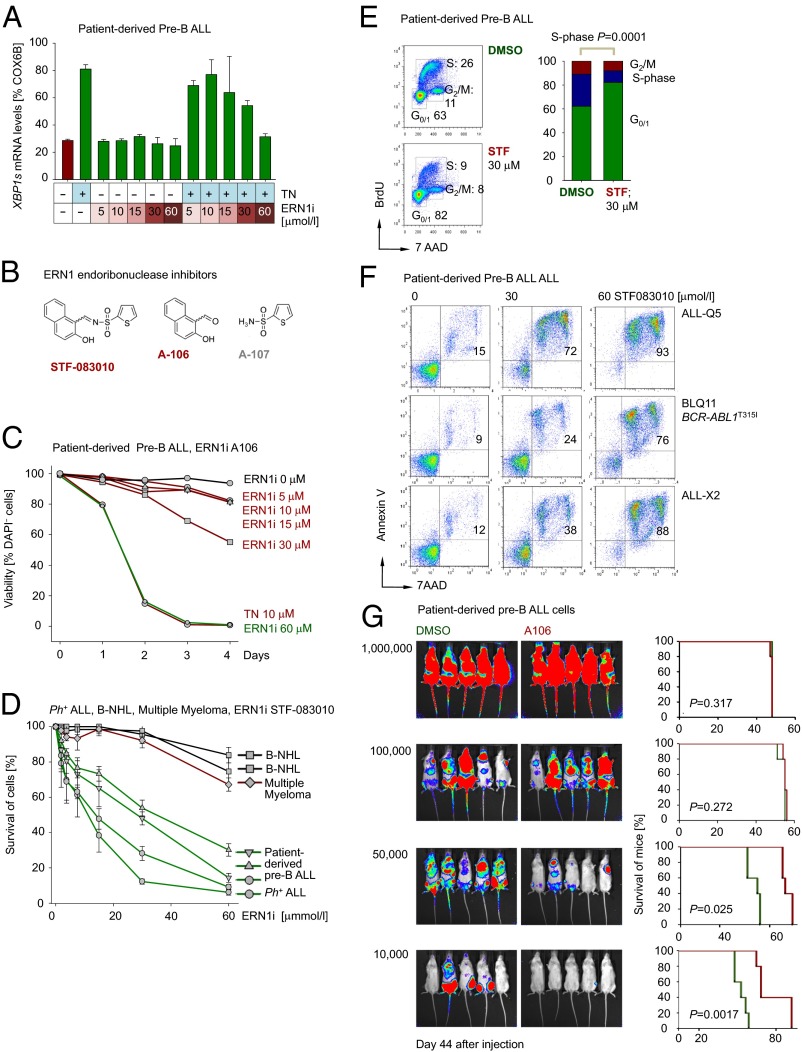
These findings collectively suggest that ERN1 and XBP1 represent potential therapeutic targets in patients with pre-B ALL. For this reason, we tested the ability of a recently developed small-molecule inhibitor of the ERN1 endoribonuclease activity (STF-083010) (15) required for XBP1 activation. First, we tested the specificity of STF-083010 after treatment with the ER-stress inducer thapsigargin (TN) in patient-derived pre-B ALL cells (Fig. 6A). TN treatment alone induced up-regulation of XBP1-s, whereas STF-083010 (ERN1i) reduced XBP1-s in a dose-dependent manner (Fig. 6A and SI Appendix, Fig. S8A). A recent study showed that spontaneous hydrolysis of STF-083010 results in two fragments, termed A106 and A107 (37) (Fig. 6B), and the A106 moiety retained full inhibitory activity on ERN1 endoribonuclease activity (37). Indeed, dose–response curves for A106 and STF-083010 were virtually undistinguishable (SI Appendix, Fig. S8 B and C). Removal of the A107 moiety results in a significantly smaller molecule and hence increases inhibitory specificity of the remaining A106 compound (37).
Fig. 6.
Efficacy of pharmacological inhibition of XBP1 activation on patient-derived pre-B ALL cells in vitro and in vivo. (A) Patient-derived pre-B ALL cells (ALL-Q5) were treated with either the ER-stress inducer TN (10 µg) to activate XBP1 splicing (XBP1-s) alone, increasing concentrations of the ERN1 inhibitor STF-083010 (0–60 µM; ERN1i), or in combination for 24 h and mRNA levels of XBP1-s were measured by qRT-PCR (n = 3). (B) Chemical structures of the STF-083010 or its two hydrolysis products, A106 and A107, are shown. (C) Patient-derived pre-B ALL (ALL-Q5) were treated with A106 (0–60 µM) and TN (10 µg) and viability was assessed by DAPI staining up to 4 d and relative cell viability is shown (n = 3). (D) Patient-derived pre-B ALL (ALL-O2, ALL-Q5, and ALL-X2), Ph+ ALL (ICN1), B-cell non-Hodgkin lymphoma (B-NHL; JEKO1 and TOLEDO) or multiple myeloma (JJN3) cells were treated with STF-083010 (0–60 µM) and the relative viability was assessed by CCK-8 assay. (E) Patient-derived pre-B ALL cells (ALL-Q5) were stained with BrdU and 7AAD and the percentages of cells in G0/1, S, and G2/M cell cycle phases are indicated after 24 h of treatment with STF-083010 (30 µM) and shown in bar graph (n = 3). (F) Apoptosis was assessed by Annexin-V and 7AAD staining in patient-derived pre-B ALL cells (ALL-Q5, BLQ11, and ALL-X2) after 3 d of treatment with STF-083010 at two concentrations (30 and 60 µM; n = 3). (G) Patient-derived pre-B ALL cells (ALL-Q5) were treated for 24 h with A106 (30 µM) or DMSO control, labeled with firefly luciferase, and injected (i.v.) into sublethally irradiated (2.5 Gy) NOD/SCID mice at different dose levels (106, 105, 50,000, and 10,000 cells per mouse) and leukemic expansion was tracked by luciferase bioimaging. The overall survival of transplant recipient mice is shown in a Kaplan–Meier analysis (n = 5 per group).
Both A106 and STF-083010 also affected proliferation and survival (A106, Fig. 6C and SI Appendix, Fig. S8D; STF-083010, Fig. 6D and SI Appendix, Fig. S9A) of patient-derived pre-B ALL cells in a dose-dependent manner. Interestingly, mature B-cell lymphoma and multiple myeloma cells were significantly less sensitive than patient-derived pre-B ALL and Ph+ ALL cells (Fig. 6D and SI Appendix, Fig. S9A). Cell cycle analyses revealed that treatment of patient-derived pre-B ALL cells with STF-083010 caused G0/G1 arrest (Fig. 6E). Interestingly, one case of Ph+ ALL carrying mutant BCR-ABL1T315I, which confers resistance to most currently available TKIs, was equally as sensitive as other pre-B ALL cases to STF-083010 treatment at concentrations previously achieved in vivo (15) (Fig. 6F). In a manner comparable to Cre-mediated deletion of Xbp1 (Fig. 3E), treatment with A106 caused significant increased uptake of ER tracker dye in a time- and dose-dependent manner (SI Appendix, Fig. S9B).
To validate potential usefulness of pharmacological inhibition of ERN1/XBP1 in vivo, we performed a limiting dilution experiment to determine the impact of A106 on leukemia-initiating cells in a xenotransplantation setting. We injected patient-derived pre-B ALL cells at different dose levels (106, 105, 50,000, and 10,000 cells) into sublethally irradiated NOD/SCID mice (n = 5 per group). Single-agent inhibition of ERN1/XBP1 had no significant effect on established leukemia when pre-B ALL cells were injected at high cell numbers (106 and 105; Fig. 6G). At lower cell numbers (50,000 and 10,000), however, single-agent treatment with A106 was sufficient to significantly prolong survival of xenotransplant recipients (P = 0.0017; Fig. 6G).
These findings suggest that pharmacological inhibition of ERN1/XBP1 may be useful to overcome drug resistance and to treat pre-B ALL to prevent relapse arising from a small number of drug-resistant leukemia-initiating cells.
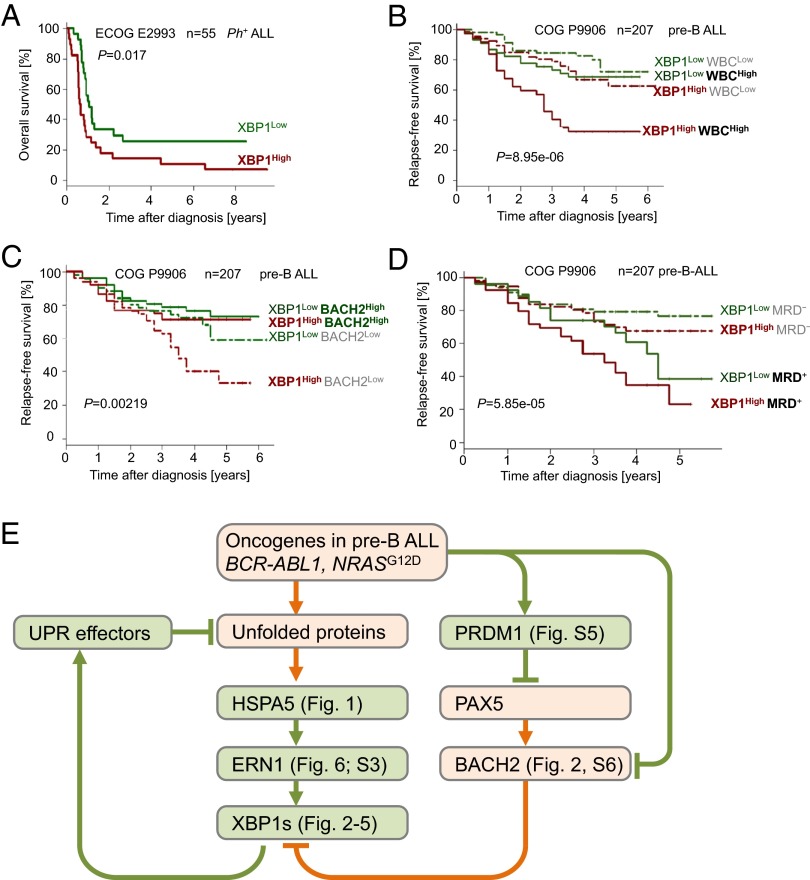
High XBP1 Expression at the Time of Diagnosis Predicts Poor Outcome in Patients with Pre-B ALL.
Based on genetic studies in mouse models for Ph+ and RAS-driven ALL, we demonstrated that expression and activity of the plasma cell transcription factor XBP1 is critical for survival and proliferation of ALL cells in these two subsets. For this reason, we analyzed mRNA levels of XBP1 in ALL samples at the time of diagnosis from two clinical trials, ECOG E2993 for Ph+ ALL in adults (n = 55) and COG P9906 for high-risk childhood ALL (n = 207). One caveat of this analysis is the fact that currently available XBP1 microarray probesets used in these analyses bind to the 3′UTR of the XBP1 mRNA and do not distinguish between XBP1-u and XBP1-s (SI Appendix, Fig. S10 A and B). qRT-PCR and Western blot (Fig. 2 A–C and SI Appendix, Fig. S5A) show, however, that patient-derived pre-B ALL cells, like multiple myeloma cells, express higher levels of XBP-s compared with XBP-u.
For a clinical trial (ECOG E2993) including 55 patients with Ph+ ALL, we segregated patients into two groups based on whether XBP1 mRNA levels in Ph+ ALL cells were higher (XBP1High) or lower (XBP1Low) than the median expression value in this trial. Comparing these two groups, we found that patients with Ph+ ALL in the XBP1High group had significantly worse overall survival (P = 0.017; Fig. 7A). Likewise, multivariate analyses for patients in a pediatric trial with high-risk childhood ALL (COG 9906) revealed that high mRNA levels of XBP1 at the time of diagnosis was a strong and independent predictor of poor clinical outcome (SI Appendix, Fig. S10C) relative to WBC counts (P = 8.95e-06; Fig. 7B), BACH2 mRNA levels (P = 0.0022; Fig. 7C and SI Appendix, Fig. S10D), and minimal residual disease status 29 d after initiation of chemotherapy (P = 5.85e-05; Fig. 7D). Similarly, higher than median mRNA levels of the ER-stress sensor and activator of XBP1-s, ERN1, were predictive of failure to achieve complete remission in patients with Ph+ ALL (P = 0.01; n = 53, ECOG 2993; SI Appendix, Fig. S10 E and F).
Fig. 7.
XBP1 expression and activity is a predictor of poor clinical outcome and a therapeutic target in pre-B ALL. (A) In an analysis, Ph+ ALL patients (ECOG E2993, n = 55, log rank test P = 0.017) were segregated into two groups based on high or low mRNA levels in respect to the median mRNA value of the XBP1 probeset and the overall survival was assessed. (B) In a multivariate analysis, pre-B ALL patients (COG clinical trial P9906, n = 207) were segregated into four groups based on high or low mRNA levels of XBP1 (higher or lower than median mRNA levels of XBP1) and WBC counts, (C) mRNA levels of BACH2 (27), or (D) minimal residual disease (MRD) status. (E) A schematic overview summarizing the key findings of this study.
Discussion
Here we show that molecules that typically mitigate ER stress in plasma cells (ERN1, HSPA5, PRDM1, and XBP1) are expressed at surprisingly high levels specifically at the pre-BCR checkpoint during early B-cell development. Genetic studies of inducible deletion of Hspa5, Prdm1, and Xbp1 demonstrated that these molecules are not only critical for survival and proliferation of early pre-B cells but in particular in pre-B ALL cells that are typically arrested at developmental stages before or at the pre-BCR checkpoint. Ph+ ALL cells carrying the BCR-ABL tyrosine kinase exhibit particularly high levels of XBP1, which is induced by oncogenic tyrosine kinase activity via downstream targets such as Stat5, AKT, and ERK signaling (schematic in Fig. 7E). In mature B cells and B-cell lymphoma, XBP1 levels are low, owing to transcriptional repression by BACH2 and BCL6. Whereas Ph+ ALL is particularly frequent in adults (∼30%), RAS-driven leukemias account for ∼50% of childhood ALL (14). Modeling both diseases with BCR-ABL1 and NRASG12D oncogenes, genetic ablation experiments established that Xbp1 is critical to enable oncogenic signaling in both major subtypes of pre-B ALL.
Plasma Cell-Specific Gene Expression As a Result of Genetic Lesions Targeting B-Cell Lineage Transcription Factors.
Both adult Ph+ ALL and childhood pre-B ALL cells express high levels of PRDM1, a key transcription factor that orchestrates terminal differentiation into plasma cells by extinguishing B-cell–specific gene expression (2), for instance through repression of PAX5, IKZF1, EBF1, and BACH2 (http://lymphochip.nih.gov/blimp/). Conversely, PAX5 (38) and, as we show here, BACH2, are transcriptional repressors of XBP1 (schematic in Fig. 7E). In this context, it seems relevant that adult Ph+ ALL (39) and childhood pre-B ALL (40) are characterized by frequent genetic lesions in B-cell lineage transcriptions factors (IKZF1, PAX5, EBF1, and BACH2) (27) that oppose plasma cell differentiation, including expression of XBP1. Although Ph+ ALL and childhood pre-B ALL cells clearly do not exhibit a plasma cell-like phenotype, genetic lesions targeting IKZF1, PAX5, EBF1, and BACH2 may facilitate expression of PRDM1 and XBP1 and, hence, enable pre-B ALL cells to mitigate ERstress resulting from strong oncogenic signaling. Here we studied the dependency of pre-B ALL cells transformed by BCR-ABL1 and NRASG12D oncogenes on XBP1 expression and activity. A previous study showed that overexpression of TCL1 results in increased ER stress (37). In addition, results by us and others demonstrate that oncogenic tyrosine kinase activity results in ectopic expression of Prdm1 (23) and XBP1 (this study; Fig. 7E).
B-cell–Specific Transcription Factors Limit the Ability of Pre-B ALL Cells to Relieve Oncogene-Induced ER Stress.
Further studies will be needed to show whether genetic lesions targeting IKZF1, PAX5, EBF1, and BACH2 are required to enable surprisingly high levels of PRDM1 and XBP1 expression observed in pre-B ALL cells compared with normal pre-B cells. However, the unexpected finding of high expression levels of ERN1, HSPA5, PRDM1, and XBP1 in the context of known deletions of IKZF1, PAX5, EBF1, and BACH2 in pre-B ALL support the concept that pre-B ALL cells are distinctly vulnerable to ER stress. Our proof-of-principle studies using pharmacological inhibitors of ERN1/XBP1 in patient-derived pre-B ALL cells support this scenario and indicate that combinations between conventional chemotherapy and novel agents targeting ER stress and UPR may improve clinical outcomes for patients with drug-resistant pre-B ALL.
Materials and Methods
Primary Human Samples and Human Cell Lines.
We obtained primary cases (SI Appendix, Table S1) in compliance with the institutional review board of the University of California, San Francisco. The cells were cultured as described in SI Appendix, Supplemental Material and Methods.
Extraction of Bone Marrow Cells from Mice.
Bone marrow cells were extracted from young age-matched mice with different genetic backgrounds (SI Appendix, Table S2) and further processed as described in SI Appendix, Supplemental Material and Methods. All mouse experiments were subject to approval by the Children's Hospital Los Angeles Institutional Animal Care and Use Committee.
Mouse Model of Human Ph+ ALL.
We collected bone marrow cells from the mice and retrovirally transformed them using BCR-ABL1 or NRASG12V in the presence of 10 ng⋅mL−1 IL-7 (Peprotech) in RetroNectin- (Takara) coated Petri dishes as described below. The cells were maintained as previously described (26). A detailed description is given in SI Appendix, Supplemental Material and Methods.
Retroviral Transduction.
We performed transfections of retroviral constructs and their corresponding empty vector controls (SI Appendix, Table S5) as previously described (26, 27). A detailed description is given in SI Appendix, Supplemental Material and Methods.
qRT-PCR and Genomic PCR.
Total RNA from cells was extracted using RNeasy isolation kit from Qiagen. cDNA was generated using a poly(dT) oligonucleotide and the SuperScript III Reverse Transcriptase (Invitrogen). Quantitative real-time PCR was performed with the SYBRGreenER mix (Invitrogen) and the ABI7900HT real-time PCR system (Applied Biosystems) according to standard PCR conditions. Primers for qRT-PCR or genomic PCR are listed in SI Appendix, Table S3.
Western Blotting.
After harvest, cells were washed twice with PBS and lysed in CelLytic MT buffer (Sigma-Aldrich) supplemented with Mini Complete protease inhibitor (Roche), 1% phosphatase inhibitor mixture (Calbiochem), and 1 mM PMSF. After 10 min of incubation on ice and centrifugation at 11,000 × g for 10 min at 4 °C, the protein concentration was determined by Coomassie blue assay (Thermo Scientific). Protein samples were loaded on 4–20% Bis-Tris gradient gels and transferred on nitrocellulose membranes or PVDF (BioRad). The primary antibodies used are listed in SI Appendix, Table S4. For protein detection, the WesternBreeze immunodetection system (Invitrogen) was used and light emission was either detected by film exposure or by the BioSpectum imaging system (UPV).
Cell Sorting.
For magnetic bead sorting, peripheral blood or bone marrow mononuclear cells were purchased from AllCells LLC and CD19-enrichment was performed by magnetic bead cell sorting according to the manufacturer’s instructions (Miltenyi Biotech). Cell sorting by flow cytometry was performed using a BD FACSAriaII (BD Biosciences). Gating strategies and analysis of the population purity are shown in SI Appendix, Fig. S1A.
Flow Cytometry.
About 1 million cells per sample were resuspended in PBS blocked using Fc blocker for 10 min on ice, followed by staining with the appropriate dilution of the antibodies for 15 min on ice. Cells were washed and resuspended in PBS with propidium iodide (0.2 µg/mL) as a dead cell marker. The antibodies used for flow cytometry are listed in SI Appendix, Table S4. For Annexin V stainings, Annexin V binding buffer (BD Biosciences) was used instead of PBS and 7AAD (BD Biosciences) instead of propidium iodide.
Cell-Cycle Analysis.
For cell-cycle analysis in BCR-ABL1 ALL cells, the BrdU flow cytometry kit for cell-cycle analysis (BD Biosciences) was used according to the manufacturer’s instructions. BrdU incorporation (FITC-labeled anti-BrdU antibodies) was measured along with DNA content (7-amino-actinomycin-D) in fixed and permeabilized cells. The analysis was gated on viable cells that were identified based on scatter morphology.
ER Tracker Green.
ER tracker green was used according to the manufacturer’s instructions (Life Technology). For cytospin experiments, cells were incubated for 15 min with ER tracker green and washed and cytospin preparations performed.
MTT/CCK8 Proliferation Assays.
Cell proliferation was determined by CCK8 proliferation kit (Dojindo) according to the manufacturer’s instructions.
Inhibitor Studies.
STF-083010 and A106 were previously described (15, 37) and obtained from Axon Medchem and VWR. AZD5363 and PD325901 were obtained from Axon Medchem and dissolved in DMSO and stored at −20 °C for further experiments. The cells were treated with the inhibitors for specific time points as mentioned in discussion of the experiments.
Senescence-Associated β-Galactosidase Assay.
Senescence-associated β-galactosidase activity was performed on cytospin preparations as described (26). For a detailed description see SI Appendix, Supplemental Material and Methods.
Single-Locus Quantitative ChIP, ChIP-on-Chip, and ChIP-Seq Analysis.
Single-locus quantitative ChIP, ChIP-on-chip, and ChIP-seq analysis were performed as previously described (26, 41). A detailed protocol and primer sequences are available in SI Appendix, Supplemental Material and Methods.
Gene Expression Microarray and RNA Sequencing Analysis.
Total RNA from cells used for microarray was isolated by RNeasy purification. Biotinylated cRNA was generated and fragmented according to the Affymetrix protocol and hybridized to mouse 430A 2.0 Array (Affymetrix). A detailed description is given in SI Appendix, Supplemental Material and Methods.
Patient Outcome and Gene Expression Microarray Data.
Gene expression microarray and patient outcome data were obtained from the GEO database [accession nos. GSE5314 (42) and GSE34941 (41)] of the Eastern Cooperative Oncology Group Clinical Trial E2993 for adult pre-B ALL, from GSE11877 and GSE28460 (43) of the Children’s Oncology Group Clinical Trial P9906 for pediatric pre-B ALL (the National Cancer Institute TARGET Data Matrix, http://targetnci.nih.gov/dataMatrix/TARGET_DataMatrix.html), and from the St. Jude Research Hospital pediatric ALL clinical trial (raw data can be downloaded from www.stjuderesearch.org/site/data/ALL3/).
Array-Based Methylation Analysis Using HELP.
The HELP (HpaII tiny fragment enrichment by ligation mediated PCR) assay was performed as previously published (41). A detailed description is given in SI Appendix, Supplemental Material and Methods.
Survival Analysis.
Kaplan–Meier survival analysis was used to estimate overall survival. Basically, patients were segregated into two groups according whether they had above or below the median expression level of the XBP1 probeset. Log rank test was used to compare survival differences between patient groups. R package “survival” version 2.35-8 was used for the survival analysis.
Supplementary Material
Acknowledgments
We thank Laurie H. Glimcher (Weill Cornell College of Medicine) for Xbp1fl/fl mice, Lothar Hennighausen (National Institute of Diabetes and Digestive and Kidney Diseases) for Stat5a/bfl/fl mice, and Amy S. Lee (University of Southern California) for Hspa5fl/fl mice. This work is supported by National Institutes of Health/National Cancer Institute Grants R01CA137060, R01CA139032, R01CA157644, R01CA169458, and R01CA172558 (to M.M.), the William Lawrence and Blanche Hughes Foundation, Stand Up To Cancer American Association for Cancer Research Innovative Research Grant IRG00909 (to M.M.), California Institute for Regenerative Medicine Grant TR2-01816 (to M.M.) and Leukaemia and Lymphoma Research (M.M.). M.M. is a Scholar of the Leukemia and Lymphoma Society and a Senior Investigator of the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data are available from the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE5314, GSE34941, GSE28460, GSE11877, GSE20987, GSE30883, GSE19599, GSM488979, GSE13411, GSE12453, GSE53684, and GSE53683). See also Supplemental S3h-k: www.stjuderesearch.org/data/ALL3.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400958111/-/DCSupplemental.
References
- 1.Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol Cell. 2009;35(5):551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaffer AL, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17(1):51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 3.Shaffer AL, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21(1):81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Hetz C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 5.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 6.Leung-Hagesteijn C, et al. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013;24(3):289–304. doi: 10.1016/j.ccr.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messinger YH, et al. Therapeutic Advances in Childhood Leukemia & Lymphoma (TACL) Consortium Bortezomib with chemotherapy is highly active in advanced B-precursor acute lymphoblastic leukemia: Therapeutic Advances in Childhood Leukemia & Lymphoma (TACL) Study. Blood. 2012;120(2):285–290. doi: 10.1182/blood-2012-04-418640. [DOI] [PubMed] [Google Scholar]
- 8.Scheuner D, et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7(6):1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 9.Lee K, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16(4):452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306(5941):387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang K, et al. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115(2):268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pui C-H, et al. Total Therapy Study XIIIB at St Jude Children’s Research Hospital Improved outcome for children with acute lymphoblastic leukemia: Results of Total Therapy Study XIIIB at St Jude Children’s Research Hospital. Blood. 2004;104(9):2690–2696. doi: 10.1182/blood-2004-04-1616. [DOI] [PubMed] [Google Scholar]
- 13.Westbrook CA, et al. Clinical significance of the BCR-ABL fusion gene in adult acute lymphoblastic leukemia: A Cancer and Leukemia Group B Study (8762) Blood. 1992;80(12):2983–2990. [PubMed] [Google Scholar]
- 14.Zhang J, et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: A report from the Children’s Oncology Group. Blood. 2011;118(11):3080–3087. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papandreou I, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117(4):1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mimura N, et al. Blockade of XBP1 splicing by inhibition of IRE1α is a promising therapeutic option in multiple myeloma. Blood. 2012;119(24):5772–5781. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brewer JW, Cleveland JL, Hendershot LM. A pathway distinct from the mammalian unfolded protein response regulates expression of endoplasmic reticulum chaperones in non-stressed cells. EMBO J. 1997;16(23):7207–7216. doi: 10.1093/emboj/16.23.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006;26(15):5688–5697. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412(6844):300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 20.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77(2):297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 21.Kitamura D, Roes J, Kühn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350(6317):423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 22.Shapiro-Shelef M, et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19(4):607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 23.Hug E, Hobeika E, Reth M, Jumaa H. 2013. Inducible expression of hyperactive Syk in B cells activates Blimp-1-dependent terminal differentiation. Oncogene, 10.1038/onc.2013.326. [DOI] [PubMed]
- 24.Hoelbl A, et al. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med. 2010;2(3):98–110. doi: 10.1002/emmm.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Y, et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24(18):8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duy C, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473(7347):384–388. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swaminathan S, et al. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med. 2013;19(8):1014–1022. doi: 10.1038/nm.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nahar R, et al. Pre-B cell receptor-mediated activation of BCL6 induces pre-B cell quiescence through transcriptional repression of MYC. Blood. 2011;118(15):4174–4178. doi: 10.1182/blood-2011-01-331181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189(9):1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Todd DJ, et al. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J Exp Med. 2009;206(10):2151–2159. doi: 10.1084/jem.20090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2(6):326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 32.Dengler MA, et al. Oncogenic stress induced by acute hyper-activation of Bcr-Abl leads to cell death upon induction of excessive aerobic glycolysis. PLoS ONE. 2011;6(9):e25139. doi: 10.1371/journal.pone.0025139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11(4):381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 34.Yan W, et al. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci USA. 2002;99(25):15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurisu J, et al. MDG1/ERdj4, an ER-resident DnaJ family member, suppresses cell death induced by ER stress. Genes Cells. 2003;8(2):189–202. doi: 10.1046/j.1365-2443.2003.00625.x. [DOI] [PubMed] [Google Scholar]
- 36.Holmfeldt L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45(3):242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kriss CL, et al. Overexpression of TCL1 activates the endoplasmic reticulum stress response: A novel mechanism of leukemic progression in mice. Blood. 2012;120(5):1027–1038. doi: 10.1182/blood-2011-11-394346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nera K-P, et al. Loss of Pax5 promotes plasma cell differentiation. Immunity. 2006;24(3):283–293. doi: 10.1016/j.immuni.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453(7191):110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 40.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 41.Geng H, et al. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discov. 2012;2(11):1004–1023. doi: 10.1158/2159-8290.CD-12-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juric D, et al. Differential gene expression patterns and interaction networks in BCR-ABL-positive and -negative adult acute lymphoblastic leukemias. J Clin Oncol. 2007;25(11):1341–1349. doi: 10.1200/JCO.2006.09.3534. [DOI] [PubMed] [Google Scholar]
- 43.Hogan LE, et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood. 2011;118(19):5218–5226. doi: 10.1182/blood-2011-04-345595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.