Significance
Protein–protein interactions are attractive targets for drug design due to their fundamental role in biological function. However, small molecules that selectively target the intended interactions have been difficult to access using traditional drug discovery approaches. We show that compounds that reproduce key functionality at the interface between transcription factor hypoxia-inducible factor 1α (HIF1α) and coactivator p300 (or CREB binding protein, CBP) can inhibit expression of a multitude of genes under hypoxic environments. The designed inhibitors target the chosen protein–protein interaction in a predictable manner and reduce tumor growth in mouse xenograft models.
Keywords: helix mimics, hypoxia signaling, synthetic inhibitors of transcription
Abstract
Development of small-molecule inhibitors of protein–protein interactions is a fundamental challenge at the interface of chemistry and cancer biology. Successful methods for design of protein–protein interaction inhibitors include computational and experimental high-throughput and fragment-based screening strategies to locate small-molecule fragments that bind protein surfaces. An alternative rational design approach seeks to mimic the orientation and disposition of critical binding residues at protein interfaces. We describe the design, synthesis, biochemical, and in vivo evaluation of a small-molecule scaffold that captures the topography of α-helices. We designed mimics of a key α-helical domain at the interface of hypoxia-inducible factor 1α and p300 to develop inhibitors of hypoxia-inducible signaling. The hypoxia-inducible factor/p300 interaction regulates the transcription of key genes, whose expression contributes to angiogenesis, metastasis, and altered energy metabolism in cancer. The designed compounds target the desired protein with high affinity and in a predetermined manner, with the optimal ligand providing effective reduction of tumor burden in experimental animal models.
Protein–protein interactions (PPIs) represent attractive yet largely unexplored groups of targets for drug design (1). Despite their fundamental importance in biological signaling, relatively few small-molecule inhibitors have been discovered, underscoring the difficulty in inhibiting large interfaces (2). Recent analyses suggest that PPIs may be categorized as those that are amenable to inhibition by small molecules and those that will require large molecules (3–5). Key to these analyses is the recognition that although PPIs encompass larger surface areas than enzyme–substrate complexes, a handful of key residues dominate the binding energy landscape (6, 7). The design of PPI inhibitors, then, requires mimicry of the relative positioning and disposition of these important residues, termed “hotspot residues,” on synthetic scaffolds (3, 4, 8–11). Based on these hypotheses, we describe a rationally designed compound that down-regulates hypoxia-inducible signaling in cell culture by targeting a PPI predicted to be suitable for small inhibitors. The designed inhibitor reduces tumor burden in mouse tumor xenografts where hypoxia-inducible proteins are overexpressed.
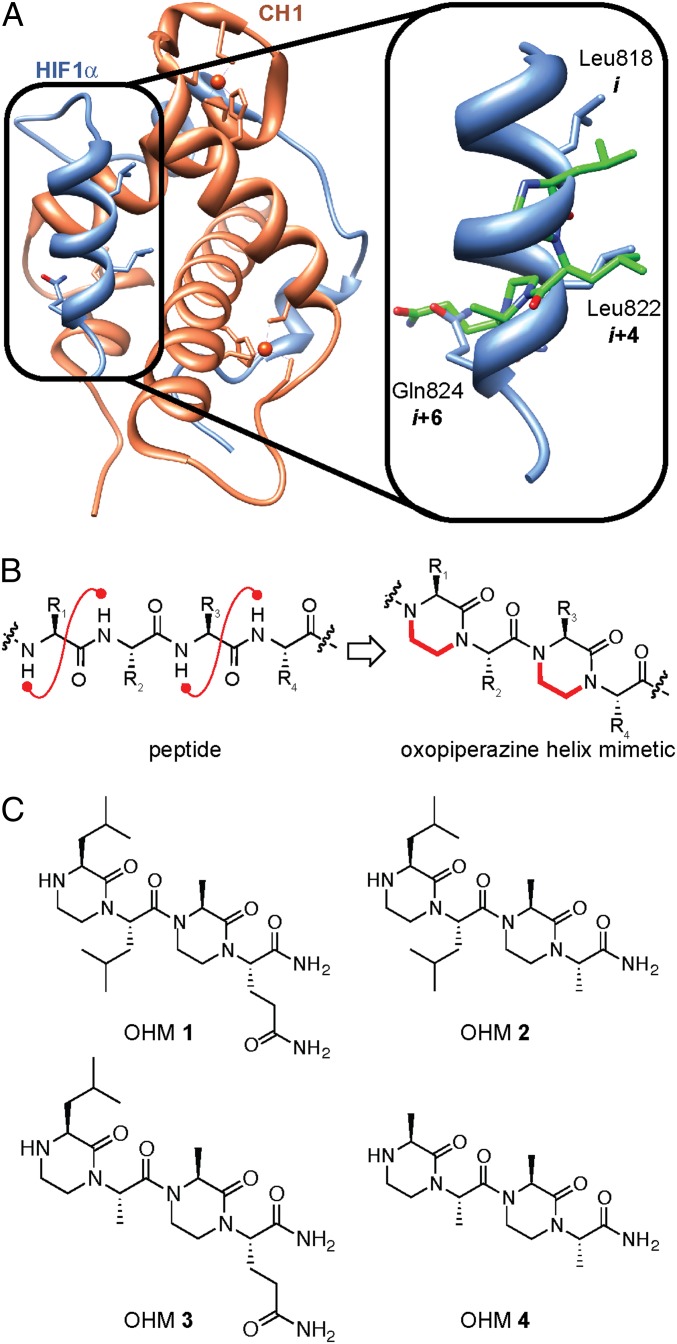
Metazoan cells possess an intricate signaling network that responds to changes in the oxygen tension in their immediate surroundings (12). Under hypoxia, the state of reduced oxygen levels, cells express a family of hypoxia-inducible transcription factors (HIFs), which are heterodimeric basic helix–loop–helix proteins composed of a regulatory α (HIF1α) and a constitutively expressed β (HIF1β; also termed ARNT) subunit. The C-terminal transactivation domain (CTAD, aa 786–826) of HIF1α interacts with the cysteine–histidine rich 1 (CH1) domain of the coactivator protein p300 (or its ortholog CREB binding protein, CBP; Fig. 1A) (13, 14). The HIF/p300 complex mediates transactivation of hypoxia-inducible genes, which are important contributors to angiogenesis, invasion, and altered energy metabolism in cancer (15).
Fig. 1.
Design of HIF1α mimetics as modulators of hypoxia-inducible gene expression. (A) Model depicting complex of HIF1α and the CH1 domain of p300/CBP. Structural data was retrieved from the Protein Data Bank (PDB), ID code 1L8C. The key residues Leu818, Leu822, and Gln824 of HIF1α CTAD (shown in blue) are located in the binding pocket of the p300/CBP CH1 domain (depicted in orange). Magnified is an overlay of the HIF1α helix spanning residues 816–824 (blue) and OHM 1 (green). (B) OHMs were designed to mimic the key helical region. OHMs feature ethylene bridges between adjacent amino acid residues; the bridges lock the side chain groups in orientations that mimic α-helices. (C) OHM derivatives—positive and negative controls—designed to inhibit the target complex.
Inhibitors of hypoxia-inducible gene expression would serve as unique tools for dissecting hypoxia signaling in tumors, as well as leads for cancer therapeutics (16–25). The transcription factor–coactivator interaction presents an intriguing target for controlling hypoxia signaling because it is a critical node directing downstream expression of various genes that work in concert to modulate cancer progression. From a ligand design perspective, transcriptional PPIs are often challenging because of their transient existence and relatively low binding affinities (26–28). Our work supports the hypothesis that topographical mimics of energetically important residues on protein secondary structures offer a rational approach for discovery of PPI inhibitors (3, 4, 8–11, 29, 30).
Results
Design and Synthesis of Topographical HIF1α Mimics.
The C-terminal activation domain of HIF1α uses two short α-helices to bind to the CH1 domain of p300/CBP (Fig. 1A). Small molecules that mimic the structural arrangement of the key residues on these helices should afford competitive inhibitors of HIF1α/p300 complex formation (19, 22). We used a recently described strategy from our groups to mimic the interacting face of an α-helix on a small-molecule oxopiperazine scaffold (31). Oxopiperazine helix mimetics (OHMs) are assembled from naturally occurring amino acids with the nitrogen atoms of neighboring backbone amides constrained with ethylene bridges to afford a nonpeptidic chiral scaffold displaying protein-like functionality (Fig. 1B). Oxopiperazines are attractive scaffolds for discovery of PPI inhibitors because of their rich history in drug design (32). Molecular modeling suggests that the low-energy conformation of the oxopiperazine scaffold arrays side chain functionality to mimic the arrangement of the i, i+4, and the i+6 or i+7 residues on canonical α-helices (31). OHMs add to a growing class of nonpeptidic helix mimetics that have been shown to have biological activity (8, 9, 11, 29). The appeal of OHMs derives from their chiral backbone. A majority of nonpeptidic helix mimetics are based on achiral aromatic scaffolds (8, 31). Chiral scaffolds are expected to interact with molecular binding pockets with higher specificity.
High-resolution structures and computational analyses (33) of the HIF1α/p300 complex reveal that four helical residues from the HIF1α helix816–824 (Leu818, Leu822, Asp823, and Gln824) make close contacts with the CH1 domain of p300/CBP (SI Appendix, Table S1). Three of these residues—Leu818, Leu822, and Gln824—can be mimicked by oxopiperazine dimers consisting of the appropriate building blocks (Fig. 1C). We designed and synthesized four analogs of HIF1α CTAD to inhibit its binding with p300/CBP. OHM 1 contains projections representing all three residues from HIF1α: R1 as Leu818, R2 as Leu822, and R4 as Gln824. The R3 position of the oxopiperazine scaffold is not predicted to make contacts with the target protein; an alanine residue was inserted at this position. OHMs 2 and 3 are single mutants of 1 with R4 and R2 positions, respectively, substituted with alanine residues. Based on computational analysis and the relative contributions of Leu822 and Gln824, OHM 2 would be expected to bind p300–CH1 with a much higher affinity than OHM 3. We also prepared OHM 4 containing alanine residues at all four positions as a negative control.
OHMs 1–4 were synthesized using standard Fmoc amino acids and coupling reagents on Rink amide resin (SI Appendix, Fig. S1). The oxopiperazine ring is obtained via the application of the Fukuyama–Mitsunobu strategy, which involves activating the amine functionality by reaction with o-nitrobenzenesulfonyl chloride followed by N-alkylation with 2-bromoethanol. Typical syntheses are performed at a 0.25 mmol scale on standard solid phase matrices and afford overall yields of 10–20% after HPLC purification.
Binding Affinities of OHMs for the p300–CH1 Domain.
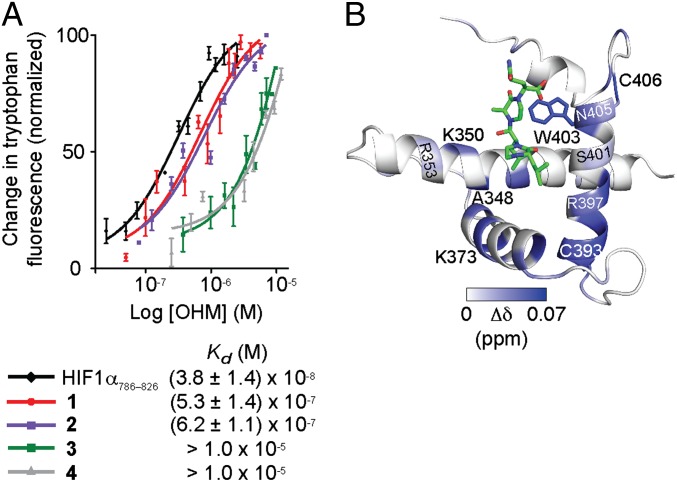
The binding affinities of OHMs for the p300–CH1 domain were evaluated using intrinsic tryptophan fluorescence spectroscopy, as described previously (19, 34). Because Trp403 is located in the binding cleft of p300/CBP where a native HIF1α816–824 helix binds, it offers a probe for investigating mimetics of this helix. Tryptophan fluorescence spectroscopy provides a dissociation constant, Kd, of (3.8 ± 1.4) × 10−8 M for HIF1α CTAD786–826 to p300 CH1, which is consistent with the values obtained from a fluorescence polarization assay using fluorescein-labeled HIF1α CTAD (SI Appendix, Fig. S2) and those reported in the literature with isothermal titration microcalorimetry (14, 19). OHM 1 targets CH1 with an affinity of (5.3 ± 1.4) × 10−7 M (Fig. 2A). OHM 2, which contains the two critical leucine residues but lacks Gln824, binds with a slightly reduced affinity Kd = (6.2 ± 1.1) × 10−7 M. The binding affinity of 2 confirms the computational prediction that Gln824 is a weak contributor to binding (SI Appendix, Table S1). The negative controls OHMs 3 and 4 displayed very weak affinities for p300–CH1, with Kd values of >>1.0 × 10−5 M in each case. The results signify that the designed scaffolds are able to target the protein of interest in a predetermined manner.
Fig. 2.
Binding affinities of designed compounds for p300–CH1. (A) The affinity of OHMs 1–4 and HIF1α C-TAD786–826 for the CH1 domain was determined by tryptophan fluorescence spectroscopy; the binding affinity of HIF–CTAD for p300–CH1 measured using the tryptophan fluorescence assay is in agreement with that obtained in a fluorescence polarization assay with the fluorescein-labeled derivative of the peptide (SI Appendix, Fig. S2). (B) Molecular model that depicts the results of a 1H-15N HSQC NMR titration experiment. The p300–CH1 residues undergoing chemical shift perturbations upon addition of OHM 1 are color-mapped, matching the magnitude of the chemical shift changes. The structure of the HIF1α/CH1 complex (PDB ID code 1L8C) was used to construct the model.
We further characterized the interaction of OHM 1 with the p300–CH1 domain using 1H-15N heteronuclear single quantum correlation (HSQC) nuclear magnetic resonance (NMR) titration experiments with the uniformly 15N-labeled CH1 (19). Concentration-dependent shifts of several residues were observed upon addition of OHM 1 to 170 μM CH1 in CH1:OHM 1 ratios of 1:0.6, 1:1.2, 1:3.5, and 1:7 (SI Appendix, Fig. S3). Addition of OHM 1 leads to consistent shifts in resonances of residues corresponding to the HIF1α816–824 binding surface, as expected from the design and the tryptophan fluorescence spectroscopy assay. The magnitude of the resonance shifts is consistent with the observation that CH1 has a stable conformation that does not reorganize substantially, at least upon binding of small ligands (19, 35). Fig. 2B depicts a model of OHM 1 docked to CH1 residues that undergo chemical shift perturbation. The NMR results, along with the fluorescence binding experiments, provide strong evidence that rationally designed topographical mimics of protein α-helices can bind the predicted binding surfaces of their intended targets.
Designed Mimetics Down-Regulate Hypoxia-Inducible Gene Expression.
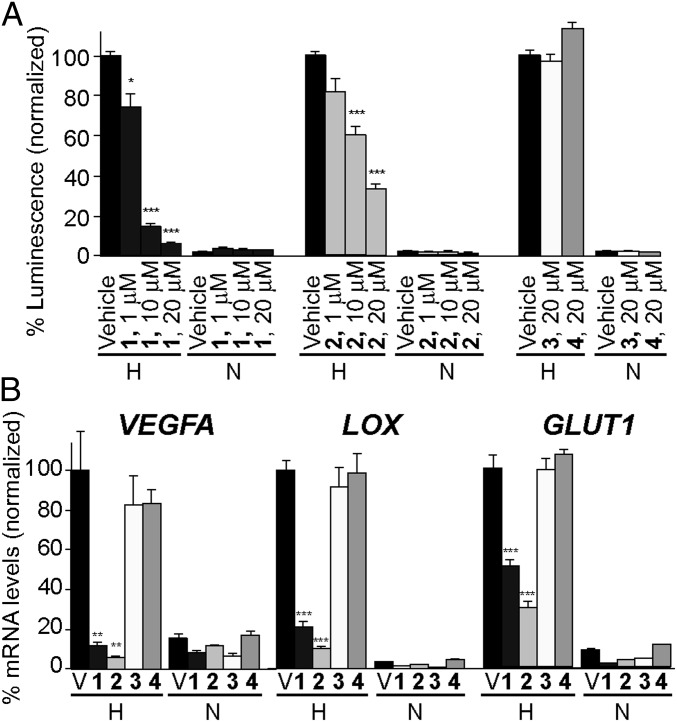
We next assessed the potential of OHMs to modulate the target interaction in cell culture and suppress hypoxia-inducible gene expression. We began by measuring the effect of compounds on the viability of human alveolar adenocarcinoma cells (A549) using the MTT assay. All four compounds showed dose-dependent decrease in cell viability with EC50 values of 30–40 μM, suggesting low cytotoxicity (SI Appendix, Fig. S4). We determined the potential of these compounds to inhibit HIF transcriptional activity using a luciferase-based reporter assay (19, 36). A triple-negative breast cancer cell line MDA-MB-231 was stably transfected with a construct designed to express a firefly luciferase protein under hypoxic conditions, which were generated by O2 deprivation with a GasPak EZ pouch (BD). Under these conditions, treatments with OHM 1 and OHM 2 resulted in a dose-dependent reduction in the promoter activity (Fig. 3A). OHM 1 at 20 μM reduces the level of HIF1α transcriptional activity under hypoxia to that observed under normoxia. OHM 2 is slightly less effective in this assay, whereas the designed negative controls OHM 3 and OHM 4 do not cause a decrease in HIF1α activity at 20 μM concentrations. The activity of OHM 1 in MDA-MB-231 is encouraging, as this cell line often exhibits confluence-dependent resistance to anticancer drugs (37). Importantly, the mimetics did not lead to a decrease in HIF1α protein levels, as measured by Western blots (SI Appendix, Fig. S5).
Fig. 3.
Transcriptional regulation of hypoxia-inducible genes by helix mimetics. OHMs 1 and 2 down-regulate hypoxia-induced promoter activity in luciferase assays (A) and transcription of VEGFA, LOX, and GLUT1 genes in cell culture as measured by real-time qRT-PCR (B). OHMs 3 and 4 show reduced inhibitory activities at the same concentrations. Error bars are ±SEM of four independent experiments. ***P < 0.001, **P < 0.01, *P < 0.05, t test. H, hypoxia; N, normoxia; V, vehicle.
The ability of OHMs to inhibit transcription of three selected HIF target genes—VEGFA, LOX, and GLUT1—was assessed using real-time quantitative RT-PCR (qRT-PCR) assays in A549 cells. OHMs 1 and 2 at 10 μM concentrations down-regulate the mRNA expression levels of the critical angiogenesis regulator vascular endothelial growth factor (VEGFA) (38) by 80% and 90%, respectively (Fig. 3B). In comparison, control compounds had no effect on VEGFA mRNA levels at these concentrations. Similar levels of decrease were observed for lysyl oxidase (LOX) and glucose transporter 1 (GLUT1) expression. OHM 2 is slightly more active than OHM 1 in the qRT-PCR experiments compared with the luciferase assays, reflecting the inherent variance in quantification of gene expression of endogenous genes and transfected constructs by two different methods.
Gene Expression Profiling of OHMs in A549 Cells.
The interaction of HIF1α with p300/CBP controls multiple downstream genes beyond the three genes interrogated with RT-PCR. To comprehensively assess the effect of the mimetics on global gene expression, we used Affymetrix Human Gene ST 1.0 arrays containing oligonucleotide sequences representing over 28,000 transcripts. The OHMs are small scaffolds (MW < 500) with an inherently restricted set of interaction points, and as such, they could have nonspecific off-target effects. In addition, p300 and CBP are multidomain coactivator proteins known to interact with many different transcription factors (39). We sought to assess the genome-wide effects of the two successful inhibitors of the hypoxia-inducible transcription OHM 1 and OHM 2 and compare these to the negative control OHM 4 (Fig. 4).
Fig. 4.
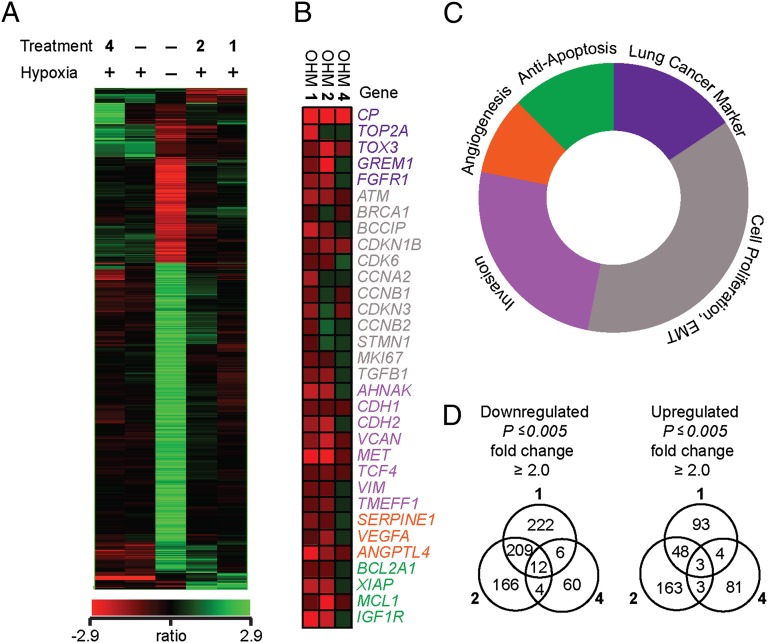
Results from gene expression profiling obtained with Affymetrix Human Gene ST 1.0 arrays. (A) Hierarchical agglomerative clustering of transcripts induced or repressed twofold or more (one-way ANOVA, P ≤ 0.005) by hypoxia (GasPak EZ pouch) under the three specified conditions: -, no treatment; 1, OHM 1 (10 μM); 2, OHM 2 (10 μM); 4, OHM 4 (10 μM). Clustering was based on a Pearson-centered correlation of intensity ratios for each treatment compared with hypoxia-induced cells (controls) using average linkage as a distance. (B) Select tumor-promoting genes affected upon OHM treatment. (C) Schematic representation of genes affected by OHM 1 treatment in B color-coded in relation to hallmarks of cancer. (D) Venn diagrams representing transcripts up- and down-regulated (|fold change| ≥ 2.0, P ≤ 0.005) by OHMs 1, 2, and 4. Numbers inside the intersections represent hypoxia-induced transcripts affected by corresponding treatments.
Analysis of oncogenic signaling pathways in cells treated with OHM 1 revealed down-regulation in expression of multiple genes responsible for accelerated tumor progression. Examples of several HIF gene targets involved in glucose and lipid metabolism and iron transport are shown in Fig. 4B. The targeted genes and the affected pathways can be classified into functional groups commonly attributed to as “hallmarks of cancer” (Fig. 4C) (40, 41). OHM 1 down-regulates multiple genes implicated in angiogenesis, apoptosis, cell proliferation, and invasion, along with several cancer-specific markers in the A549 non-small-cell lung cancer cell line.
In A549 cells, several genes are up-regulated that constitute a signature of lung cancer. These include ceruloplasmin (CP); DNA topoisomerase 2-α (TOP2A); the high mobility group box family member 3 (TOX3) that modifies the chromatin structure; Gremlin 1 (GREM1), a 184-amino-acid glycoprotein that is important in the survival of cancer stroma and cancer cell proliferation; and FGFR1, which is overexpressed in squamous cell lung cancers. All these markers were down-regulated in A549 cells treated by OHM 1, suggesting that in addition to suppression of hypoxia-inducible transcription, the oxopiperazine also exerts a general antitumor growth effect. The microarray data also reveal down-regulation of angiogenesis genes, such as VEGFA and SERPINE1, and genes that encode proteins promoting invasion, such as E-cadherin (CHD1), vimentin, and MET proto-oncogene. Expression of antiapoptosis genes such as BCL2A1, XIAP, and MCL1R (Fig. 4B) was also modulated.
Treatment with OHM 1 resulted in the change of 32 transcripts by at least fourfold (P ≤ 0.005) and 597 transcripts by at least twofold (P ≤ 0.005), 11 of which are validated as HIF1α target genes (Fig. 4C) (42). Similar results were observed for OHM 2, in which 41 transcripts were affected by treatment at least fourfold (P ≤ 0.005) and 608 transcripts at least twofold. OHM 1 and OHM 2 were found to have overlap in the down-regulation of nine transcripts and up-regulation of 10 transcripts by at least fourfold (SI Appendix, Fig. S6). In contrast, no overlap was found between OHM 1 or OHM 2 with negative control OHM 4 for down-regulated genes, and only one transcript overlapped for up-regulation by fourfold. In total, treatment with OHM 4 changed expression of 27 transcripts by fourfold and 91 transcripts by twofold.
We compared our microarray results with oxopiperazines with other direct-acting inhibitors of the hypoxia-inducible transcription reported in the literature. Dimeric epidithiodiketopiperazines (ETPs) are recent examples of allosteric small-molecule inhibitors of the HIF complex that also target the p300 CH1 domain (20, 23). Treatment of hypoxic MCF7 cells with dimeric ETP 2 resulted in at least twofold expression changes in 329 genes (SI Appendix, Table S2) (20). A structurally similar ETP 3 alters the expression of 409 genes by at least twofold (23). A sequence-specific DNA-binding oligomer, polyamide 1, has been reported to change expression levels of 2,284 genes by at least twofold in hypoxic U251 cells (17). In contrast, echinomycin, a DNA-binding cyclic peptide, under the same conditions and cell line produces changes in expression of 10,918 genes by at least twofold. RNA interference directed at HIF1α leads to expression changes in 3,194 genes at least twofold (SI Appendix, Table S2). Taken together, these data suggest that the designed oxopiperazine α-helix mimetic, despite its low molecular weight and a limited number of contacts with the intended target proteins p300/CBP, shows remarkably high specificity on a genome-wide scale.
In Vivo Assessment of the Efficacy of OHM in a Mouse Tumor Xenograft Model.
We next sought to investigate the potential of oxopiperazine 1 to reduce the tumor growth rate in a mouse xenograft model. We began by determining the maximum tolerated dose of OHM 1 in BALB/c mice. Escalating doses of 2–100 mg/kg OHM 1 were injected intraperitoneally every other day. No signs of toxicity were found, as assessed by daily weight measurements and visual inspections of the appearance and the behavior of treated mice. The animals were then injected i.v. with 10 mg/kg and then 100 mg/kg of OHM 1, again without measurable weight loss or changes in appearance or behavior.
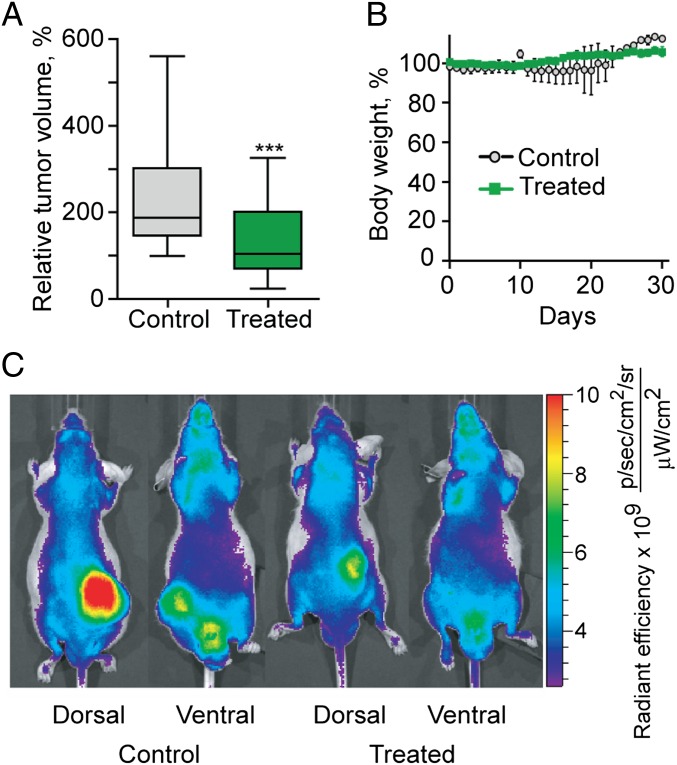
Xenograft models of MDA-MB-231 cells were used for the in vivo efficacy studies. Mice bearing xenografts were randomly assigned to treated and control groups when the tumor volumes reached 200 mm3. The treated groups received i.p. injections of 15 mg/kg OHM 1 in PBS, whereas control groups received injections of PBS (100 μL per animal). Tumor sizes were monitored daily for both groups of mice. In the treated group, the median tumor volume was smaller (103 mm3) compared with the control group (186 mm3), indicating 45% median tumor volume reduction at the conclusion of the experiment (Fig. 5A). Treated and control mice maintained their weights at 100 ± 15% before euthanasia (Fig. 5B). At the endpoint of the experiment, mice were injected with near-infrared (NIR) tumor-targeting contrast agent IR-783 and imaged using the Xenogen IVIS 200 system. The intensity of the NIR signal in the OHM 1-treated mice was consistently lower than vehicle-treated mice (Fig. 5C).
Fig. 5.
Effect of OHM 1 treatment on tumor growth rate in MDA-MB-231 xenografts. (A) Box- and Whisker plots of the percentages of tumor volumes measured throughout the duration of the experiment: boxes represent the upper and lower quartiles and the median, and the error bars show maximum and minimum tumor volumes. ***P < 0.001. (B) Weight measurements of control (–O–) and OHM 1-treated (–■–) mice engrafted with MDA-MB-231 tumors through the course of the study. Error bars are ±SEM of the weight measurements of the mice within each experimental group. (C) Localization of the NIR contrast agent IR-783 in the tumors of the control and treated mice. The fluorescence output was processed with Living Image software with one representative sample for each group presented above. Mice from the OHM 1-treated group show lower intensity of the signal originating from the tumor-accumulated contrast agent compared with the control group.
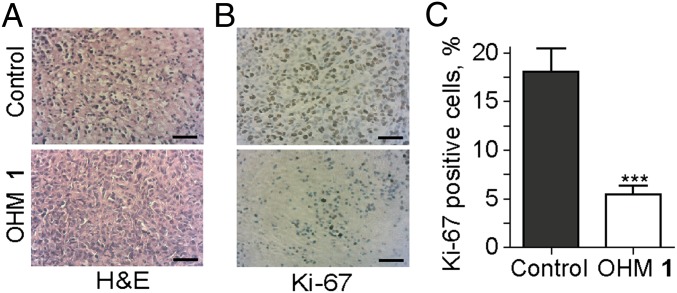
The tumors from control and treated mice were excised, dissected, and used for histopathology evaluations. We used hematoxylin and eosin (H&E) stain to evaluate cell morphology (Fig. 6). Cells in the treated tumors appear more differentiated with a greater cytoplasm-to-nucleus ratio. Cell proliferation marker Ki-67 was also used to determine the pattern of proliferating cells. Tumors in the untreated mice exhibit threefold higher levels of cell proliferation (Fig. 6B) as assessed by quantification of the Ki-67 stained images (Fig. 6C) (43).
Fig. 6.
(A) H&E-stained sections of MDA-MB-231 xenograft (purple, nuclei; pink, cytoplasm) treated with vehicle or OHM 1. (B) Anti–Ki-67–stained MDA-MB-231 xenografts (brown stain), treated with vehicle or OHM 1. (Scale bar, 50 μm.) (C) Quantification of Ki-67–stained images using ImageJ with the ImmunoRatio plugin (43). ***P < 0.001, t test.
Discussion
In this study, we describe rational design of peptidomimetics that target transcriptional PPIs in a predetermined manner in biochemical, cell culture, and in vivo assays. Oxopiperazine analogs, designed to mimic side chain orientations of protein helices, are shown to inhibit the interaction of HIF1α with the coactivator p300/CBP. The successful candidate, OHM 1, is a small molecule with a molecular weight of less than 500 Daltons. Small molecules are often considered better candidates as inhibitors of enzyme function rather than PPIs; however, analysis of PPIs that have been inhibited by small molecules suggests that they contain hotspot residues clustered within relatively small volumes—a feature that is reminiscent of enzyme active sites (1, 3, 4, 44).
The HIF–p300/CBP interface features a high concentration of hotspot residues, making it an attractive candidate for inhibition by small molecules. Although other inhibitors of this transcription factor–coactivator interaction have been described (21), the best known example, chetomin (45), inhibits the interaction by chelating essential zinc metals and denaturing the coactivator. We aimed to discover small-molecule orthosteric inhibitors that target the interaction with high specificity. The designed compounds bind the target protein in a predictable manner, as the single residue mutant OHM 3 shows an expected weaker affinity for p300 compared with OHM 1. The p300 binding site for OHM 1 was confirmed by HSQC titration experiments.
OHM 1 significantly reduces HIF promoter activity and down-regulates the expression of hypoxia-inducible genes responsible for promoting angiogenesis, invasion, and glycolysis. Despite its low molecular weight and a limited number of contacts with the target protein, OHM 1 shows high specificity on a genome-wide scale, especially compared with other reported inhibitors of HIF1α-mediated transcription. Comparative analysis of the genome-wide effects of OHMs 1, 2, and 4 provide important insights into the ability of related compounds to disrupt transcriptional activity of hypoxia-inducible genes. The microarray gene expression heat map observed with the control tetraalanine compound 4 is similar to that of the vehicle under normoxic conditions, underscoring the levels of specificity offered by the display of the correct side chains on the scaffold.
To assess the in vivo potential of OHM 1, murine tumor xenografts derived from the triple-negative breast cancer cell line MDA-MB-231 were treated with the compound. Injections of OHM 1 reduced the median tumor volume by roughly 50% compared with the untreated group. Importantly, the OHM 1 treatment did not cause measurable changes in animal body weight or other signs of toxicity in tumor-bearing animals, nor increased the metastasis rate. The efficacy and specificity of the OHM derivatives, in genome-wide analysis, supports our hypothesis that PPIs that offer clefts can be targeted by judiciously designed small molecules (4, 44).
Methods
Synthesis and characterization of OHMs 1–4, 1H-15N HSQC titration spectra, protocols for cloning and expression of the p300–CH1–GST protein, binding assays, cell viability assays, luciferase expression assays, Western blot analysis of HIF1α levels, gene expression profiling, in vivo toxicity dose-finding and efficacy studies, mouse tumor imaging and immunohistochemistry are described in SI Appendix. Computational alanine scanning mutagenesis energies were calculated with Rosetta (33) v. 3.3, and are available in SI Appendix. The gene expression profiling data have been deposited in the Gene Expression Omnibus database, www.ncbi.nlm.nih.gov/GEO (accession no. GSE48134).
Supplementary Material
Acknowledgments
The authors thank Nathaniel Traaseth for help with p300 NMR and Daniela Buccella at New York University (NYU) for the use of the spectrofluorometer. This work was financially supported by the National Science Foundation (CHE-1161644 to B.Z.O. and CHE-1151554 to P.S.A.). The microarray analysis was performed at the Genome Technology Center, NYU School of Medicine, partially supported by NYU Cancer Center Support Grant 5P30CA16087-33.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The gene expression profiling data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/GEO (accession no. GSE48134).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402393111/-/DCSupplemental.
References
- 1.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450(7172):1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 2.Higueruelo AP, et al. Atomic interactions and profile of small molecules disrupting protein-protein interfaces: The TIMBAL database. Chem Biol Drug Des. 2009;74(5):457–467. doi: 10.1111/j.1747-0285.2009.00889.x. [DOI] [PubMed] [Google Scholar]
- 3.Bullock BN, Jochim AL, Arora PS. Assessing helical protein interfaces for inhibitor design. J Am Chem Soc. 2011;133(36):14220–14223. doi: 10.1021/ja206074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jochim AL, Arora PS. Systematic analysis of helical protein interfaces reveals targets for synthetic inhibitors. ACS Chem Biol. 2010;5(10):919–923. doi: 10.1021/cb1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson AD, Dugan A, Gestwicki JE, Mapp AK. Fine-tuning multiprotein complexes using small molecules. ACS Chem Biol. 2012;7(8):1311–1320. doi: 10.1021/cb300255p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280(1):1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 7.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267(5196):383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 8.Azzarito V, Long K, Murphy NS, Wilson AJ. Inhibition of α-helix-mediated protein-protein interactions using designed molecules. Nat Chem. 2013;5(3):161–173. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- 9.Cummings CG, Hamilton AD. Disrupting protein-protein interactions with non-peptidic, small molecule alpha-helix mimetics. Curr Opin Chem Biol. 2010;14(3):341–346. doi: 10.1016/j.cbpa.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Ko E, Liu J, Burgess K. Minimalist and universal peptidomimetics. Chem Soc Rev. 2011;40(8):4411–4421. doi: 10.1039/c0cs00218f. [DOI] [PubMed] [Google Scholar]
- 11.Jayatunga MKP, Thompson S, Hamilton AD. α-helix mimetics: Outwards and upwards. Bioorg Med Chem Lett. 2014;24(3):717–724. doi: 10.1016/j.bmcl.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Fraisl P, Aragonés J, Carmeliet P. Inhibition of oxygen sensors as a therapeutic strategy for ischaemic and inflammatory disease. Nat Rev Drug Discov. 2009;8(2):139–152. doi: 10.1038/nrd2761. [DOI] [PubMed] [Google Scholar]
- 13.Freedman SJ, et al. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci USA. 2002;99(8):5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha/CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci USA. 2002;99(8):5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirota K, Semenza GL. Regulation of angiogenesis by hypoxia-inducible factor 1. Crit Rev Oncol Hematol. 2006;59(1):15–26. doi: 10.1016/j.critrevonc.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Semenza GL. Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33(4):207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Modulating hypoxia-inducible transcription by disrupting the HIF-1-DNA interface. ACS Chem Biol. 2007;2(8):561–571. doi: 10.1021/cb700110z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olenyuk BZ, et al. Inhibition of vascular endothelial growth factor with a sequence-specific hypoxia response element antagonist. Proc Natl Acad Sci USA. 2004;101(48):16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kushal S, et al. Protein domain mimetics as in vivo modulators of hypoxia-inducible factor signaling. Proc Natl Acad Sci USA. 2013;110(39):15602–15607. doi: 10.1073/pnas.1312473110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubey R, et al. Suppression of tumor growth by designed dimeric epidithiodiketopiperazine targeting hypoxia-inducible transcription factor complex. J Am Chem Soc. 2013;135(11):4537–4549. doi: 10.1021/ja400805b. [DOI] [PubMed] [Google Scholar]
- 21.Xia Y, Choi H-K, Lee K. Recent advances in hypoxia-inducible factor (HIF)-1 inhibitors. Eur J Med Chem. 2012;49(0):24–40. doi: 10.1016/j.ejmech.2012.01.033. [DOI] [PubMed] [Google Scholar]
- 22.Henchey LK, et al. Inhibition of hypoxia inducible factor 1-transcription coactivator interaction by a hydrogen bond surrogate alpha-helix. J Am Chem Soc. 2010;132(3):941–943. doi: 10.1021/ja9082864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Block KM, et al. Direct inhibition of hypoxia-inducible transcription factor complex with designed dimeric epidithiodiketopiperazine. J Am Chem Soc. 2009;131(50):18078–18088. doi: 10.1021/ja807601b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheuermann TH, et al. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol. 2013;9(4):271–276. doi: 10.1038/nchembio.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miranda E, et al. A cyclic peptide inhibitor of HIF-1 heterodimerization that inhibits hypoxia signaling in cancer cells. J Am Chem Soc. 2013;135(28):10418–10425. doi: 10.1021/ja402993u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: Structural, functional, and network properties. Structure. 2010;18(10):1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Lee LW, Mapp AK. Transcriptional switches: Chemical approaches to gene regulation. J Biol Chem. 2010;285(15):11033–11038. doi: 10.1074/jbc.R109.075044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koehler AN. A complex task? Direct modulation of transcription factors with small molecules. Curr Opin Chem Biol. 2010;14(3):331–340. doi: 10.1016/j.cbpa.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravindranathan P, et al. Peptidomimetic targeting of critical androgen receptor-coregulator interactions in prostate cancer. Nat Commun. 2013;4:1923. doi: 10.1038/ncomms2912. [DOI] [PubMed] [Google Scholar]
- 30.Shaginian A, et al. Design, synthesis, and evaluation of an alpha-helix mimetic library targeting protein-protein interactions. J Am Chem Soc. 2009;131(15):5564–5572. doi: 10.1021/ja810025g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tosovská P, Arora PS. Oligooxopiperazines as nonpeptidic alpha-helix mimetics. Org Lett. 2010;12(7):1588–1591. doi: 10.1021/ol1003143. [DOI] [PubMed] [Google Scholar]
- 32.Patchett AA, Nargund RP. Privileged structures—An update. Annu Rep Med Chem. 2000;35:289–298. [Google Scholar]
- 33.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci STKE. 2004;2004(219):pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 34.Dial R, Sun ZY, Freedman SJ. Three conformational states of the p300 CH1 domain define its functional properties. Biochemistry. 2003;42(33):9937–9945. doi: 10.1021/bi034989o. [DOI] [PubMed] [Google Scholar]
- 35.De Guzman RN, Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding. Biochemistry. 2005;44(2):490–497. doi: 10.1021/bi048161t. [DOI] [PubMed] [Google Scholar]
- 36.Shibata T, Akiyama N, Noda M, Sasai K, Hiraoka M. Enhancement of gene expression under hypoxic conditions using fragments of the human vascular endothelial growth factor and the erythropoietin genes. Int J Radiat Oncol Biol Phys. 1998;42(4):913–916. doi: 10.1016/s0360-3016(98)00298-3. [DOI] [PubMed] [Google Scholar]
- 37.Fang Y, Sullivan R, Graham CH. Confluence-dependent resistance to doxorubicin in human MDA-MB-231 breast carcinoma cells requires hypoxia-inducible factor-1 activity. Exp Cell Res. 2007;313(5):867–877. doi: 10.1016/j.yexcr.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Ryan HE, Lo J, Johnson RS. HIF-1 α is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17(11):3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276(17):13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 40.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 41.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 42.Benita Y, et al. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009;37(14):4587–4602. doi: 10.1093/nar/gkp425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tuominen VJ, Ruotoistenmäki S, Viitanen A, Jumppanen M, Isola J. ImmunoRatio: A publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 2010;12(4):R56. doi: 10.1186/bcr2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raj M, Bullock BN, Arora PS. Plucking the high hanging fruit: A systematic approach for targeting protein-protein interactions. Bioorg Med Chem. 2013;21(14):4051–4057. doi: 10.1016/j.bmc.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kung AL, et al. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell. 2004;6(1):33–43. doi: 10.1016/j.ccr.2004.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.