Significance
Many chemotherapeutics function by inducing cellular apoptosis, or programmed cell death, in tumor cells. Here, using a unique enzymatically driven technology, we find that numerous protein fragments are released into the bloodstream from apoptotic cells within hours after chemotherapy. These circulating signatures of cell death, if confidently assigned to tumor cells, may form the foundation for entirely novel, rapid, and inexpensive biomarkers of chemotherapeutic efficacy. Diagnostic tests to rapidly assess cell death after treatment could serve an important role in management for many cancers.
Abstract
It is known that many chemotherapeutics induce cellular apoptosis over hours to days. During apoptosis, numerous cellular proteases are activated, most canonically the caspases. We speculated that detection of proteolytic fragments released from apoptotic cells into the peripheral blood may serve as a unique indicator of chemotherapy-induced cell death. Here we used an enzymatic labeling process to positively enrich free peptide α-amines in the plasma of hematologic malignancy patients soon after beginning treatment. This N-terminomic approach largely avoids interference by high-abundance proteins that complicate traditional plasma proteomic analyses. Significantly, by mass spectrometry methods, we found strong biological signatures of apoptosis directly in the postchemotherapy plasma, including numerous caspase-cleaved peptides as well as relevant peptides from apoptotic and cell-stress proteins second mitochondria-derived activator of caspases, HtrA serine peptidase 2, and activating transcription factor 6. We also treated hematologic cancer cell lines with clinically relevant chemotherapeutics and monitored proteolytic fragments released into the media. Remarkably, many of these peptides coincided with those found in patient samples. Overall, we identified 153 proteolytic peptides in postchemotherapy patient plasma as potential indicators of cellular apoptosis. Through targeted quantitative proteomics, we verified that many of these peptides were indeed increased post- vs. prechemotherapy in additional patients. Our findings reveal that numerous proteolytic fragments are released from dying tumor cells. Monitoring posttreatment proteolysis may lead to a novel class of inexpensive, rapid biomarkers of cell death.
It is known that the majority of small-molecule chemotherapeutics function by inducing apoptosis in cancer cells (1). Both in vitro and in vivo, apoptosis occurs rapidly, typically within 6–72 h after exposure to a chemotherapeutic (2, 3). Many proteases are activated during apoptosis (4), although the key effectors are the caspases, cysteine proteases that cleave intracellular elements and lead to cell death (5). Using imaging approaches, caspase activity in tumors has been identified postchemotherapy (6). Furthermore, through an unknown mechanism, some intracellular protein contents are released from apoptotic tumor cells into the bloodstream, including histones, cytochrome c, and a caspase-cleaved fragment of the intermediate filament protein cytokeratin 18 (7–9). However, these few existing markers unfortunately do not show sufficient sensitivity and specificity for use as clinical diagnostics of chemotherapeutic efficacy (10). The discovery of broad signatures of apoptosis, beyond the handful of markers already known, may offer sufficient diagnostic power to clinically monitor therapeutic response and greatly benefit cancer management.
We hypothesized that many more intracellular contents are likely released into the bloodstream after chemotherapy-induced apoptosis, yet they cannot be readily identified with existing technologies. Our laboratory has recently developed a unique, single-step labeling technology, using the enzyme subtiligase, to positively enrich free protein N termini generated by proteolysis, followed by mass spectrometry (MS)-based identification (11). This approach has particular advantages in blood plasma analysis, because it largely avoids interference from high-abundance proteins such as albumin, allowing for the identification of low-abundance species that could not be found through traditional plasma proteomics (12). Here we use this N-terminomic method to identify proteolytic fragments in the peripheral blood of hematologic malignancy patients within 24 h of chemotherapy induction. In these plasma samples, we first identified a number of caspase-cleaved and other proteolytically generated fragments not previously found in normal plasma. Remarkably, many of these same proteolytic products were also released from hematologic malignancy cells in culture treated with clinically relevant chemotherapeutics. Ultimately, using a combination of unbiased and targeted liquid chromatography-tandem MS (LC-MS/MS) approaches, we identified over 150 N-terminal fragments in postchemotherapy plasma derived from proteins not found in normal blood. These findings greatly expand the known library of proteolytic products released from dying cells. Furthermore, we developed a quantitative MS assay and found that many of these N termini are indeed increased in abundance post- vs. prechemotherapy in a larger cohort of hematologic malignancy patients. Overall, our results provide initial evidence that unbiased monitoring of proteolysis is a promising strategy to rapidly assess chemotherapeutic efficacy in cancer patients.
Results
Pipeline-Based Approach to Proteolytic Biomarker Identification in Plasma.
We hypothesized that tumor cells undergoing apoptosis in response to cytotoxic chemotherapy would release proteolytic peptides to the extracellular space over a time course of hours to days (Fig. 1A). In complex biological samples, we use the engineered enzyme subtiligase to biotin-tag free protein N termini and isolate them on streptavidin-coated beads. After trypsinization and elution, LC-MS methods are used to either identify or quantify the N-terminal peptides in the sample (13) (Fig. 1A). Of note, the N termini of 80–90% of native eukaryotic proteins are acetylated (14) and therefore do not react with subtiligase. In addition, this approach can be used even in the setting of high-abundance albumin without further depletion or chromatography steps (15). Thus, our approach allows for high sensitivity and specificity for proteolyic fragments.
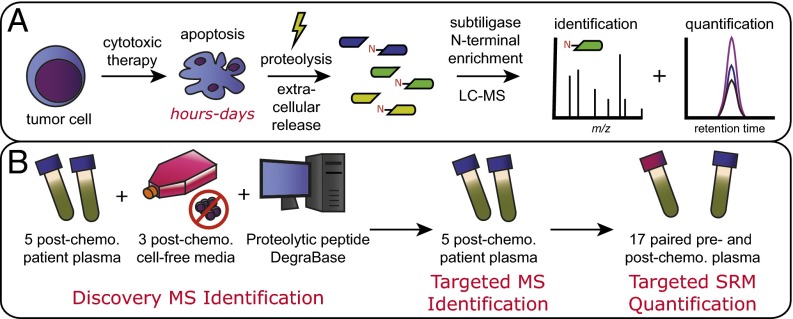
Fig. 1.
Approach to discovery of proteolytic biomarkers of cell death. (A) General strategy for apoptotic biomarker discovery. Tumor cells rapidly undergo apoptosis in response to chemotherapeutic treatment. Proteolysis is activated during apoptosis, and proteolytic fragments are released into the blood. Enzymatic labeling of free protein N termini combined with identification and quantification mass spectrometry approaches identifies potential biomarkers of cell death. (B) Pipeline for biomarker investigation. An initial discovery set of biomarkers is derived from MS experimentation on a set of high-yield patient plasma samples with significant decreases in circulating malignant cells after chemotherapy, studies in cell culture examining free N termini released from cells into the media after chemotherapy, and an extensive database of intracellular proteolytic events during apoptosis. This discovery dataset is used to generate a targeted MS method to more sensitively detect intracellular content release into the plasma in high-yield patients. Finally, an additional patient cohort was collected for quantitative SRM MS to determine relative changes in proteolytic biomarkers before and after chemotherapy. Peptides reproducibly increased posttherapy serve as the most promising biomarkers of cell death for further clinical validation.
In combination with subtiligase labeling, we used a pipeline-based strategy (Fig. 1B) modeled on a previously described approach to identify potential blood-based biomarkers (16, 17). This strategy first uses a cohort of “high-yield” samples to discover proteomic changes associated with a given condition. Here, using unbiased MS approaches on a quadrupole time-of-flight (QqTOF) instrument, we looked for proteolytic fragments released from patient tumor and cultured cells postchemotherapy. We combined these experimental data with an extensive database of proteolytic peptides found during cellular apoptosis, the DegraBase (18), to develop a targeted “inclusion list” for MS identification on an Orbitrap instrument. This approach allowed us to further expand our list of proteolytic fragments found in patient samples postchemotherapy. Finally, we used targeted, quantitative selected reaction monitoring (SRM) methods on a triple-quadrupole instrument (19) to measure changes in proteolytic N-terminal peptides pre- vs. postchemotherapy in a larger cohort of patients.
Unbiased Discovery MS Combined with N-Terminal Labeling Reveals Numerous Apoptosis-Related Peptides in Patient Plasma Postchemotherapy.
For our discovery samples, we sought a patient cohort with the highest probability of demonstrating proteolytic fragments in the blood postchemotherapy. We therefore identified hematologic malignancy patients with circulating malignant cells prechemotherapy and a significant drop in these cells (decrease of ≥7 × 106 cells per mL blood by hematopathology analysis) within 24 h of initiation of chemotherapy.
Although patients with these clinical characteristics are relatively rare, we were able to obtain 1.5-mL cell-free plasma samples from five patients [two acute myeloid leukemia (AML), one non-Hodgkin lymphoma (NHL) of diffuse large B-cell lymphoma subtype, one B-acute lymphoblastic leukemia (ALL), and one multiple myeloma evolved to plasma cell leukemia (PCL)] (Fig. 2A; additional clinical details are available in Table S1). We performed N-terminal labeling and reverse-phase high-pH fractionation into 10 fractions per sample, and evaluated each fraction in data-dependent acquisition mode on a QqTOF MS instrument.
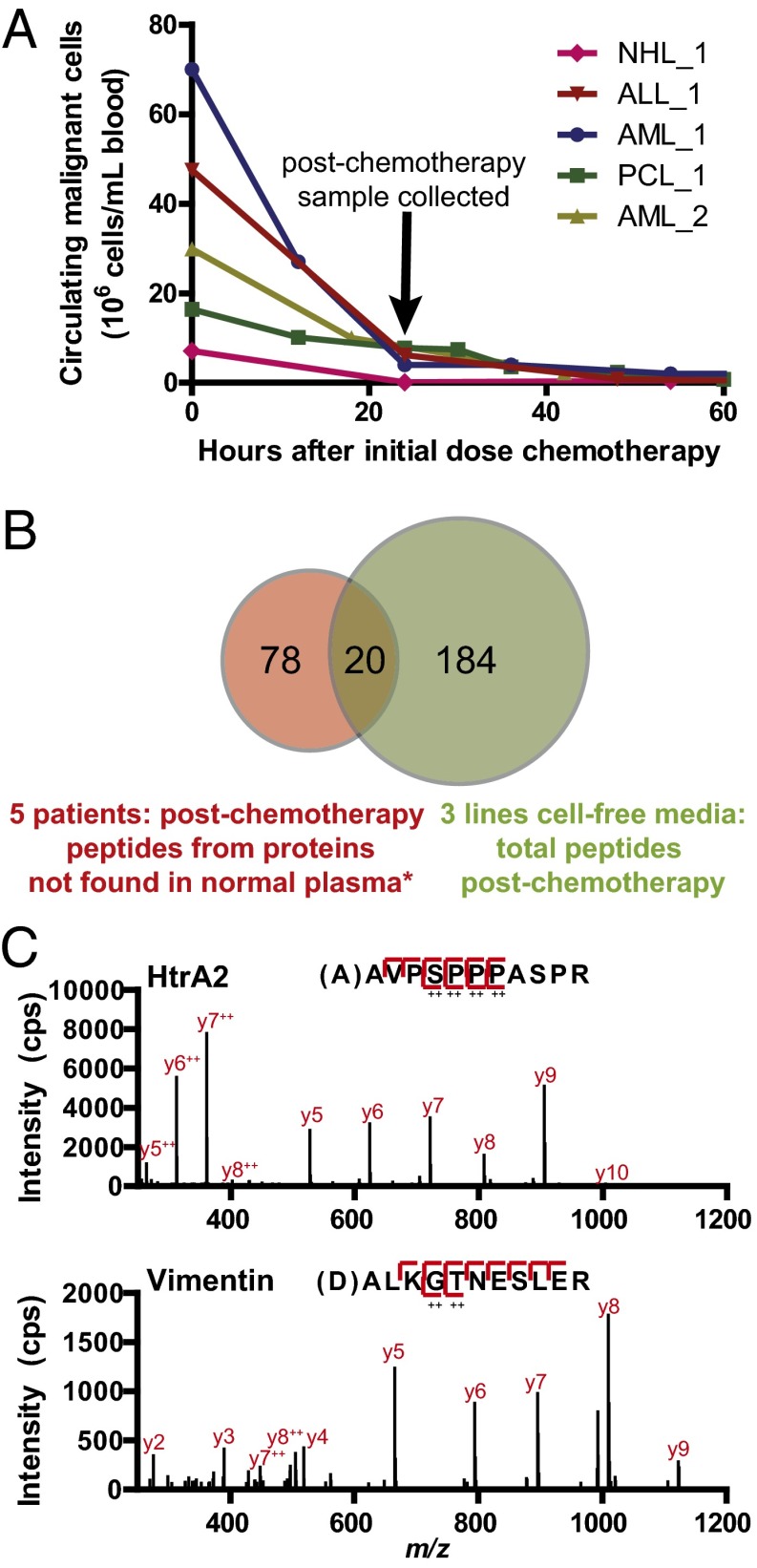
Fig. 2.
Identification of proteolytic fragments released postchemotherapy in discovery and targeted MS. (A) High-yield hematologic malignancy patient cohort for initial discovery experiments all show large decreases in circulating malignant cells postchemotherapy, suggesting extensive apoptosis directly in the peripheral blood. (B) From initial discovery MS experiments, we found 98 unique N-terminal peptides in high-yield patient postchemotherapy plasma derived from proteins not found in normal plasma [*as listed in Wildes and Wells (12)]. We found a number of overlapping peptides between proteolytic fragments released from apoptotic blood cancer cells in culture, further suggesting that the fragments in blood are generated during cell death. (C) Sample mass spectra for two biologically relevant markers of apoptosis using targeted MS on the LTQ Orbitrap Velos for patient NHL_1.
In the postchemotherapy samples, we sought to identify proteolytic fragments derived from proteins not found previously in normal blood plasma and serum. We hypothesized that these new N termini would be the strongest indicators of release of cleaved intracellular contents into the extracellular medium. Positive results would provide an initial indication of whether our approach for apoptotic biomarker identification was feasible. For this comparison with postchemotherapy samples, we used both a normal plasma sample analyzed here as well as an extensive database of >700 normal blood proteolytic N-terminal peptides previously identified by subtiligase labeling (12).
In each patient, we identified 195 (AML_1), 177 (NHL_1), 104 (ALL_1), 124 (PCL_1), and 110 (AML_2) unique N-terminal peptides. Significantly, in each of the five high-yield patient samples, we indeed identified between 5 and 60 N-terminal peptides derived from proteins not found in normal blood, for a total of 98 new peptides across all samples. Remarkably, these new peptides demonstrated strong cellular signatures of apoptosis, suggesting that they directly result from chemotherapy effects (a full list of peptides can be found in Dataset S1). For example, these signatures include the mature, processed N termini from Smac and HtrA2, which are released from mitochondria to promote caspase activation during apoptosis (20). We also identified the biologically active form of ATF-6, a transcription factor cleaved during cell stress such as that induced by chemotherapy (21). In addition, we found numerous peptides with an aspartic acid residue inferred at the P1 position, immediately N-terminal to the identified cleavage site, typical of caspase-cleavage events (5, 11, 18). Of particular note, we found multiple cleaved peptides from the intermediate filament protein vimentin with an inferred aspartic acid at P1. Vimentin, expressed in mesenchymally derived cells such as leukocytes (22), is analogous to cytokeratin 18 in epithelial cells. In aggregate, in these initial experiments, we found 23 high-confidence, but still candidate, caspase-cleaved fragments in the blood, whereas only one, derived from cytokeratin 18 (9), was known before. These results provide strong evidence that proteolytically cleaved peptides are directly released into the plasma after chemotherapy and can be identified using our N-terminal labeling method.
Peptides Released from Cultured Hematologic Malignancy Cells Coincide with Those Found in Postchemotherapy Plasma.
Our previous cellular work had focused on identifying caspase-cleaved peptides present in whole-cell lysates after induction of apoptosis (11, 23, 24). Here, as a complement to our experiments in patient samples, we instead chose to study proteolytic products released from cultured cells into the media after chemotherapy. We reasoned that this system would more closely resemble the physiology of intracellular content release to the plasma in patients treated for blood cancers.
We evaluated three cell lines treated with different drugs: (i) MM1.S, derived from multiple myeloma and treated with the proteasome inhibitor bortezomib, (ii) MOLM-13, derived from acute myeloid leukemia and treated with the nucleoside analog cytarabine, and (iii) SU-DHL-8, derived from diffuse large B-cell lymphoma and treated with the DNA-damaging agent doxorubicin. All of these conditions reflect the diagnoses of patients in our discovery cohort combined with clinically used chemotherapeutics. Under each condition, we either treated with drug or mock-treated the cells for at least 21 h. Treated cells demonstrated at least 50% apoptosis (Fig. S1). After removing whole cells, proteins in the media were precipitated with trichloroacetic acid and then subjected to N-terminal labeling by subtiligase. FBS-free media were used in these experiments to avoid contamination from normal bovine plasma proteins.
MS analysis on a QqTOF instrument demonstrated that in all cell types the number of proteolytic fragments in the media postchemotherapy increased compared with the control samples (Table S2 and Dataset S2). Released contents from MM1.S and SU-DHL-8 lines in particular showed strong signatures of apoptosis. For example, the number of released proteolytic fragments with Asp at P1 sites increased from 3 in the control to 28 posttreatment for MM1.S, and from 1 to 23 in SU-DHL-8. Across the three cell lines, we identified 204 unique N-terminal peptides released into the media postchemotherapy. Importantly, 20 of these peptides from cell-culture experiments were identical to those found in discovery experiments on patient plasma (Fig. 2B and Dataset S2). This remarkable degree of overlap further suggests that the proteolytic fragments in patient plasma are a direct result of intracellular content release after chemotherapy. Notably, the overlapping peptides found in both cultured cells and patient samples included fragments of Smac, HtrA2, and multiple caspase-cleaved vimentin peptides. These results again support that monitoring proteolytic fragments holds promise as an indicator of cell death postchemotherapy.
Targeted Inclusion List Enables Sensitive Detection of Proteolytic Peptides in Postchemotherapy Plasma.
We next sought to interrogate our high-yield patient plasma samples for additional proteolytic peptides not initially found by previous unbiased discovery MS of normal blood (12). An inclusion list approach on an Orbitrap instrument allows for increased sensitivity of detection for targeted peptides (16, 17, 25). In this approach, peptides falling within a narrow mass window around those on the inclusion list are preferentially selected for sequencing. To build our targeted inclusion list, we used (i) all peptides found in unbiased discovery experiments on plasma samples, (ii) all peptides found released from cultured hematologic malignancy cells postchemotherapy, and (iii) a selection of proteolytic peptides derived from a database of apoptotic samples, the DegraBase (18). These peptides from the DegraBase included those derived from proteins relevant to apoptosis or cell stress, peptides found to be rapidly cleaved during apoptosis by quantitative MS experiments (26), and peptides derived from relatively high abundance substrates (24). This strategy, ultimately including 672 peptides (listed in Dataset S3), aimed to both confirm peptides already found in plasma as well as identify additional biologically relevant peptides in plasma that were not found earlier.
We implemented this inclusion list strategy on an Orbitrap-based MS instrument to analyze the same five patient samples as used in unbiased discovery experiments. In each of the patient samples, we identified between 5 and 94 proteolytic peptides deriving from proteins not found in normal plasma (12), with a total of 140 unique peptides in all (Dataset S3). Notably, the targeted inclusion list strategy identified 54 peptides not found in the unbiased discovery experiments. In addition, we previously identified in normal plasma (12) a single caspase-cleaved protein fragment. This was derived from gelsolin, an actin-binding protein located at high abundance both intracellularly and in the blood (27). We also included this fragment for further study, because it was identified in all postchemotherapy samples.
Combining the results from the targeted and discovery experiments, we have identified 153 proteolytic peptides that represent an initial library of proteolytic biomarkers of cell death for further evaluation (Table S3). Forty-seven of these peptides (30.7%) demonstrated a D-at-P1 motif, suggestive of caspase cleavage. This percentage is very similar to the proportion of D-at-P1 peptides found in typical studies of apoptotic whole-cell lysate (18). In addition, based on protein expression data in the PaxDb (28), 142 (92.8%) of these peptides are derived from proteins that are typically present intracellularly rather than in the blood. Our methods could sensitively detect in the cell-free plasma many proteins typically present at <10 ppm intracellularly (Table S1). These cumulative results further support the notion that our methods are detecting the release of intracellular contents postchemotherapy.
A Quantitative Proteomic Assay Demonstrates Increases in Proteolytic Fragment Abundance Post- vs. Prechemotherapy.
If these markers of proteolysis are to be useful in a diagnostic context, they must distinguish relative increases in proteolytic fragments after chemotherapy compared with before. We therefore used targeted SRM methods on a triple-quadrupole instrument to quantitatively measure these fragments. SRM allows for highly sensitive, label-free quantification of peptides by monitoring the intensity and LC coelution of targeted parent ion–fragment ion pairs (“transitions”; see Fig. 3A for sample data) (19). To develop accurate SRM assays, we first sequenced by LC-MS/MS crude spot-synthesized peptides for 121 of the 153 targets in our library. The remaining peptides either could not be synthesized by this method or were not detected by LC-MS/MS. One hundred and seventeen (96.6%) of these synthetic peptides demonstrated similar MS/MS spectra and LC retention times to those identified in plasma, suggesting a high rate of true positive identification in plasma experiments. Importantly, as others have previously shown (29), these synthetic peptides allowed us to develop higher-quality SRM assays: For each well-characterized peptide, we can obtain the most intense fragment ions and LC retention time directly on the triple-quadrupole instrument. For the remaining peptides, we developed SRM assays by selecting coeluting peptide transitions in either plasma or cell-culture samples, similar to that done previously (24, 30). Overall, SRM assays were successfully developed for 140 of the 153 peptides of interest.
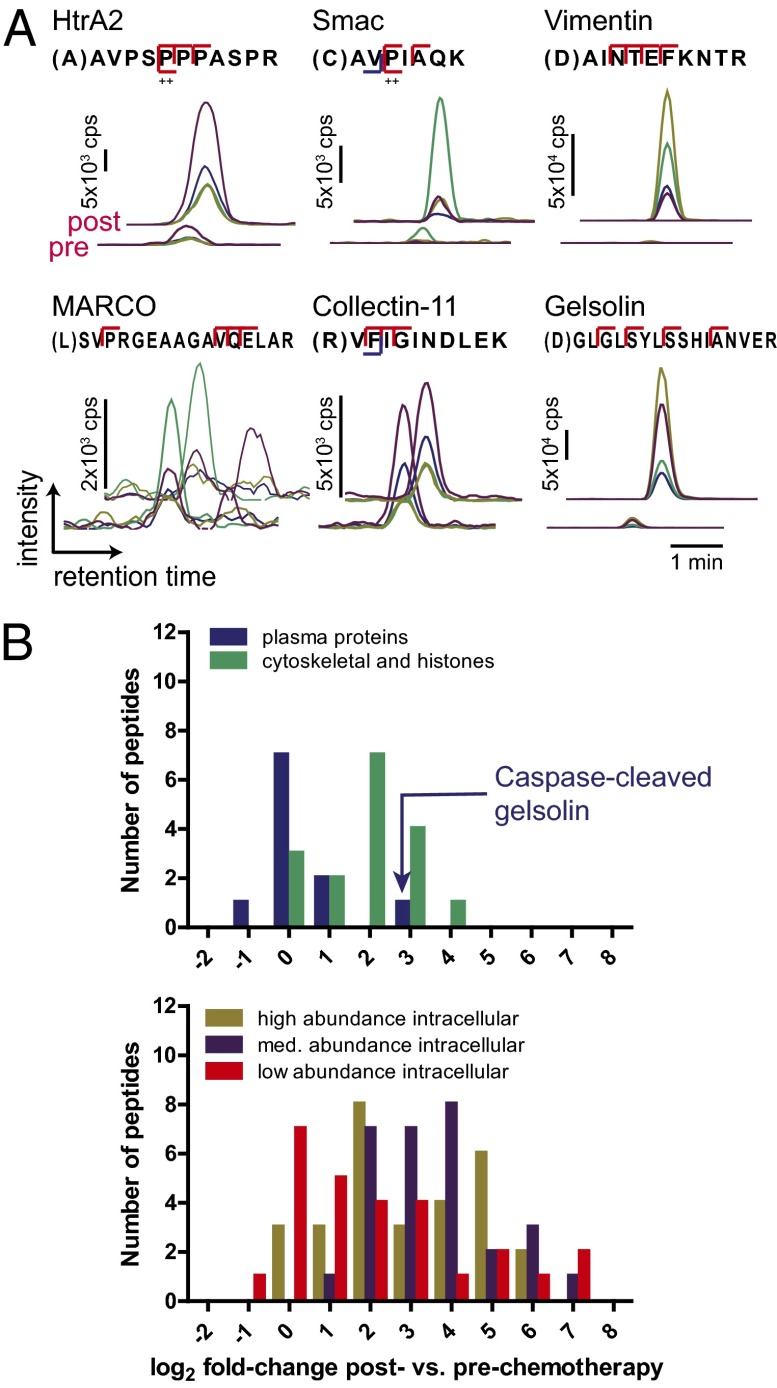
Fig. 3.
Quantitative increases in proteolytic fragments after chemotherapy using a targeted SRM assay. (A) Sample raw data from 140 peptide SRM assay for patient AML_1. For each target peptide, four parent ion–fragment ion pairs (or transitions) are monitored for coelution to confirm identification. Each trace represents the intensity of a single transition. Total peptide intensity is the sum of the area under the curve for all transitions. Three peptides (Upper), from typical intracellular proteins, are greatly increased in the plasma postchemotherapy. Of three peptides from typical plasma proteins (Lower), only the caspase-cleaved fragment of gelsolin is greatly increased postchemotherapy. (B) Log2 fold changes in peptide abundance post- vs. prechemotherapy in patient AML_1. In contrast to plasma proteins, peptides derived from typical intracellular proteins show large changes in the blood postchemotherapy, some increased over 50-fold. Peptides included in each category are listed in Dataset S4.
We next applied this completed SRM method to hematologic malignancy patient samples. As an initial case, we studied the only high-yield postchemotherapy patient sample that also had a paired pretreatment sample (AML_1 in Fig. 2A). We applied N-terminal enrichment to 500 µL of plasma at each time point and analyzed the unfractionated peptides by SRM in duplicate with intensity normalization by spike-in protein standards (Materials and Methods). In the postchemotherapy sample, we detected 100 of the 140 peptides (71.4%) with intensity signal above baseline noise. More importantly, by total peak area intensity, 90 of these peptides were increased in abundance post- vs. prechemotherapy, with 77 showing at least a twofold increase (Dataset S4). Fragments from typically intracellular proteins highly increased postchemotherapy included the N termini of Smac and HtrA2 as well as caspase-cleaved fragments of vimentin (Fig. 3A). In contrast, of the 10 detected peptides derived from proteins typically found at high abundance in normal plasma (from PaxDb analysis), 9 showed little change in abundance (Fig. 3A, MARCO and Collectin-11, and Fig. 3B, Upper). The only exception was the caspase-cleaved fragment of gelsolin, which showed an 8.7-fold increase after treatment. In contrast to these results from typical plasma proteins, peptides arising from intracellular proteins showed a wide range of abundance increases postchemotherapy, some over 50-fold (Fig. 3B, Lower). These results firmly demonstrate that the appearance of proteolytic fragments in the plasma is indicative of postchemotherapy apoptosis.
We collected pre- and postchemotherapy plasma samples from another 16 hematologic malignancy patients for additional quantitative validation. Postchemotherapy samples were collected between 12 and 96 h after initiation of treatment. Patients ranged in diagnosis, severity of disease, treatment regimen, and degree of response (clinical details are in Table S4). We applied our SRM method to these patient samples and examined changes pre- and postchemotherapy. In Fig. 4A, we display 16 peptides increased postchemotherapy across multiple patients, with examples of approximately twofold increases from two patients in Fig. 4B. Overall, these peptides represent the most promising targets for further exploration in clinical development as biomarkers of chemotherapeutic efficacy.
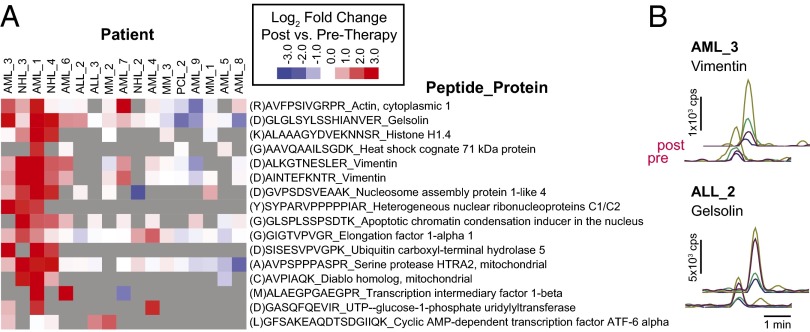
Fig. 4.
Quantitative increases in proteolytic peptides across a larger patient cohort. (A) Heat map displaying a subset of peptides with post- vs. prechemotherapy increases across 17 patients (log2 fold change in SRM peak area intensity; samples were measured in duplicate; heat map value represents mean fold change). Intriguingly, there is marked patient-to-patient variability, with some showing many increased peptides postchemotherapy and others showing little change. D-at-P1 cleavage position [“(D)” before peptide name] indicates a putative caspase proteolytic event. Gray indicates peptides not detected in the SRM assay. (B) Sample SRM data for peptides showing approximately twofold intensity increases in additional patients.
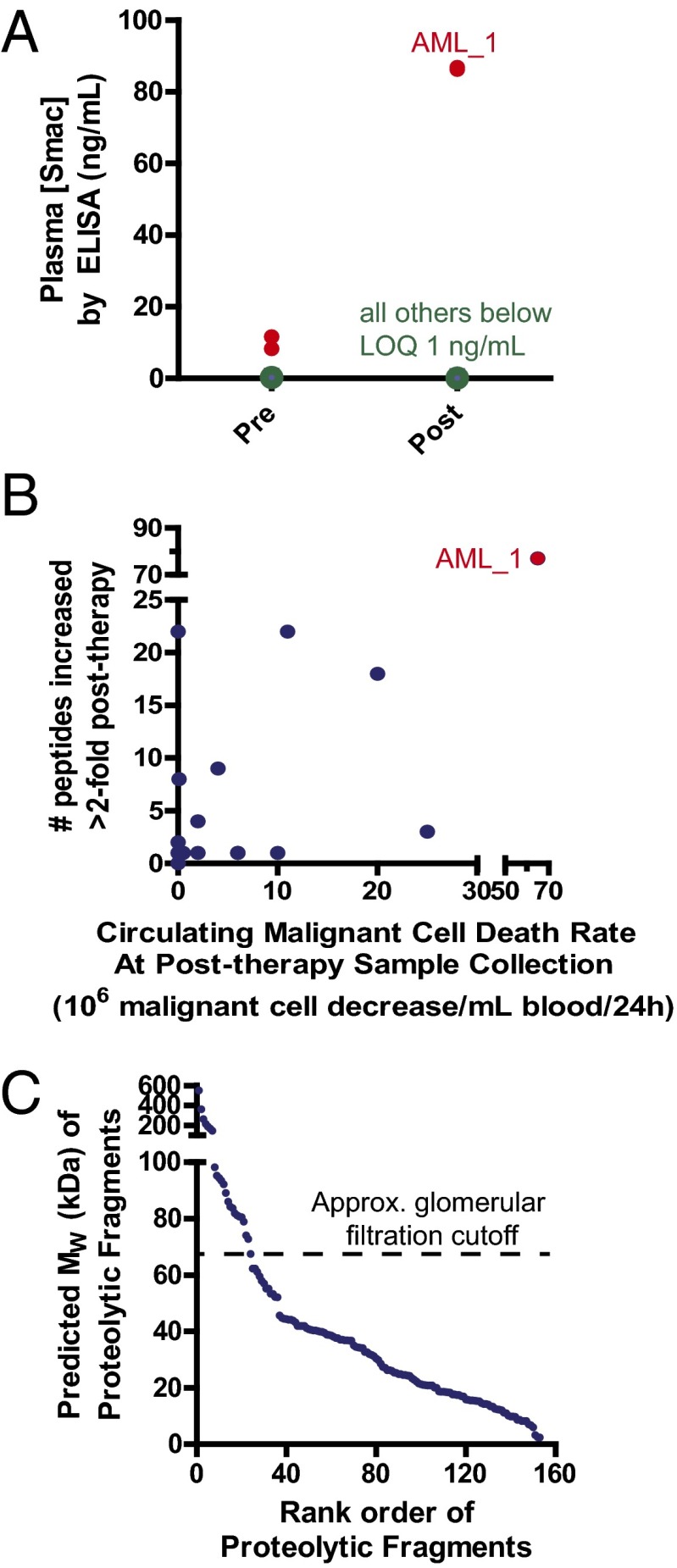
We sought to confirm our quantitative proteomic results by an independent method. Although specific antibodies are not available for the endoproteolytic fragments we found to be increased postchemotherapy, we were able to use a sandwich ELISA toward the protein Smac. In this protein, we monitored the N terminus of the intact, mature protein without any further endoproteolysis. Whereas for most patients the levels of Smac fell below the ELISA limit of quantification, for the patient AML_1 we positively identified Smac in both the pre- and postchemotherapy samples. Notably, the measured abundance increase, from 10 ng/mL pre- to 86 ng/mL posttreatment (Fig. 5A), is directly in-line with the 8.5-fold increase measured by SRM.
Fig. 5.
Confirmation and further analysis of monitoring proteolysis postchemotherapy. (A) An ELISA for full-length Smac protein confirms an ∼8.5-fold increase postchemotherapy, as found by SRM assay, for patient AML_1. The additional 12 patients tested did not have blood concentrations either pre- or posttreatment above the limit of quantification (LOQ) in this assay (1 ng/mL), and so changes could not be determined. (B) Although there is a statistically significant positive correlation (Pearson R = 0.86, P < 0.0001), there is a large variability between the decrease in circulating malignant cells postchemotherapy at time of collection and the number of proteolytic peptides increased by SRM. Note the split axes to display the results of patient AML_1. (C) The majority of proteolytic fragments released into the blood, based on the single cleavage site identified extended to the protein C terminus, have molecular weights below that of serum albumin. Therefore, they may be rapidly filtered into the urine, leaving a short time window for detection between induction of apoptosis and clearance. Proteolytic fragments are ranked by predicted molecular weight.
Discussion
Here we have demonstrated that specific enzymatic labeling of protein N termini, integrated with a combination of unbiased and targeted MS approaches, reveals that many more proteolytic fragments are released from dying cells than were previously known. Our experimental approach allowed us to identify these fragments, which could not be detected by typical plasma proteomic methods. We further used quantitative MS approaches to show that many of these proteolytic N termini are increased within days of chemotherapy initiation across multiple blood cancer patients. These results may ultimately lead toward a strategy for validating novel, rapid, and inexpensive protein-based biomarkers of chemotherapeutic efficacy.
Through targeted quantitative proteomics, we found that a greater rate of malignant cell decrease in the peripheral blood postchemotherapy correlates with a greater number of increased proteolytic fragments (Fig. 5B). This finding, along with the overlap in results between cultured tumor cells and patient samples, suggests that the proteolytic fragments we identified correspond to death of tumor cells. However, we cannot rule out the possibility that many of these peptides result from the death of normal somatic cells, particularly hematopoietic cells in the bone marrow, which are sensitive to many forms of chemotherapy.
One of the surprising findings in our study is the high degree of patient-to-patient variability in proteolytic peptides identified postchemotherapy. In our high-yield samples, we identified 5- to 10-fold more peptides postchemotherapy from the patients AML_1 and NHL_1 (Table S1) compared with the other three patients, even though the other three (ALL_1, PCL_1, and AML_2) also demonstrated large, rapid decreases in circulating malignant cell count (Fig. 2A). Large variability was also observed in our quantitative SRM assay (Fig. 4A).
There are many potential reasons for this observation. It is possible that decreases in circulating malignant cells do not always reflect apoptosis occurring directly in the blood. Instead, it could be a reflection of tumor cells partitioning away from the blood and toward the bone marrow or lymph nodes but without death. Alternatively, the mechanism by which intracellular contents are released is still unknown. Release may only occur when normal phagocytic functions of macrophages, which typically sequester cellular fragments generated during apoptosis, become overwhelmed. Therefore, there may be patient-to-patient variation in the threshold where intracellular content release occurs. Furthermore, these patients had different diagnoses, different disease burdens, and were treated with different drugs. There may be disease- or drug-specific tumor effects that govern the release of these contents.
Another important issue that likely governs proteolytic fragment detection is renal clearance. It is well-known that proteins with molecular weight below that of serum albumin (69.4 kDa) are rapidly filtered through the renal glomeruli (31). For the 153 proteolytic fragments studied here, extending from the identified cleavage site to the protein C terminus, the large majority are predicted to be below this size cutoff (Fig. 5C). Therefore, there may only be a short time window between the induction of apoptosis in a tumor and the renal excretion of these proteolytic fragments.
We are encouraged that we were able to detect many putative caspase-derived peptides postchemotherapy when to date the only validated product is the caspase-cleaved peptide from cytokeratin 18 (9). The abundance and biological function of these caspase substrates appear similar to those found intracellularly during apoptosis (18). We also identified numerous biologically relevant noncaspase N-terminal peptides directly in the blood after treatment. These findings highlight the ability of our N-terminal enrichment technology to identify proteolytic fragments not previously found by traditional plasma proteomics.
To demonstrate the feasibility of our methods, we initially studied hematologic cancers, where cell death occurs directly in communication with the blood compartment. In these patients specifically selected for this proof-of-principle study, we reveal that numerous proteolytic peptides are released from dying cells. Many of these proteolytic fragments may have diagnostic utility, but measuring their clinical performance characteristics will require further study in larger-scale human studies. It is also possible that many more such fragments exist but have not been identified in this limited patient cohort. Further improvements in mass spectrometer sensitivity may also allow us to identify specific proteolytically cleaved fragments in the blood of patients treated for solid tumors. Important end points, which we could not evaluate rigorously in our heterogeneous sample, include whether increases in proteolytic fragments correlate with other measures of therapeutic efficacy (bone marrow biopsy, positron emission tomography/computed tomography scans, etc.) as well as patient overall survival. These studies will also reveal whether the markers we identify are truly the result of tumor cell death versus normal somatic tissues, and whether the variability we see is due to differences in tumor type, burden, or somatic toxicity, which are all important clinical parameters to monitor. Such larger-scale trials will require the development of medium- to high-throughput assays using antibodies specific for the proteolytic fragment of interest, as the current N-terminomic method is not suited to evaluation of hundreds of samples from dozens of patients. Of note, our SRM results (Fig. 4A) and Smac ELISA experiments indicate that antibody-based assays may have to be highly sensitive and specific to detect potentially small (<1 ng/mL) changes in protein levels posttreatment.
Overall, our results demonstrate the promise of monitoring proteolysis as a strategy to rapidly determine the cell-death response after chemotherapy. Our findings greatly expand the potential repertoire of circulating markers of apoptosis beyond the few already known. If demonstrated to correlate with treatment efficacy, these markers could be of great use in early-stage studies of new anticancer compounds or other therapeutics that lead to apoptotic cell death. Alternatively, proteolytic fragments found to be specific for death of normal bone marrow or gastrointestinal tissues could serve as new markers of toxicity for an array of drug treatments. By applying similar methods to other tumor types and in larger patient cohorts, it may be possible to identify an entirely new class of general, cancer-type, or drug-specific biomarkers of therapeutic efficacy. Such diagnostic tests would represent an important advance toward the goal of personalized therapeutic regimens.
Materials and Methods
Patient Samples and N-Terminal Labeling.
All patient samples were obtained through human subjects and informed consent protocols approved by the University of California, San Francisco Committee on Human Research. Whole blood was centrifuged after collection at 5,000 × g for 5 min and plasma (citrate or EDTA anticoagulant) was stored at −80 °C until processing for experiments. For discovery MS, 1.5 mL of plasma was used; 0.25 or 0.5 mL was used for SRM experiments. N-terminal labeling was performed similar to that previously described (12). Isolated N-terminal peptides for discovery were fractionated using reverse-phase high-pH chromatography before MS analysis (24).
Cell-Culture Studies.
The MM1.S and SU-DHL-8 lines were obtained from the American Type Culture Collection; MOLM-13 was from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). Cell lines were grown in RPMI-1640 media without FBS for 24 h before drug treatment at the indicated doses (details are available in SI Materials and Methods). After treatment, cells and debris were separated from media by low-speed (800 × g, 5 min) followed by high-speed (24,000 × g, 1 h) centrifugation. Total protein in the media was precipitated with trichloroacetic acid, resuspended in 8 M guanidine⋅HCl, and then subjected to N-terminal labeling as described (30).
Mass Spectrometry.
Unbiased discovery experiments and targeted discovery were analyzed on an AB SCIEX QSTAR Elite QqTOF instrument and a Thermo Scientific LTQ Orbitrap Velos instrument, respectively, with in-line reverse-phase low-pH chromatography (see SI Materials and Methods for details of MS parameters). Crude synthetic peptides matching proteolytic N-terminal peptides found in the discovery experiments were purchased from JPT Peptide Technologies. SRM methods were developed as described previously (30) and applied to unfractionated samples on an AB SCIEX QTRAP 5500 triple-quadrupole instrument. Data were analyzed with Skyline (32), and intensity normalization between pre- and postchemotherapy samples was performed using spike-in protein standards.
Smac ELISA.
ELISA testing was typically performed at 1:2 plasma dilution in assay buffer using the manufacturer’s protocol (RayBiotech).
Supplementary Material
Acknowledgments
We thank Dr. Scott Kogan, Dr. Anne Deucher, Kathleen Kushner, residents, and staff from the University of California, San Francisco (UCSF) Department of Laboratory Medicine for assistance in obtaining patient samples. MOLM-13 cells were a kind gift of Dr. Scott Kogan. SU-DHL-8 cells were a kind gift of Dr. Peter Park (Immunogen). We thank Dr. Kazutaka Shimbo for assistance in MS method development. We thank the staff of the UCSF Bio-Organic Biomedical Mass Spectrometry Resource, directed by Dr. Alma Burlingame and supported by National Institutes of Health–National Institute of General Medical Sciences Grant 8P41GM103481 and the Howard Hughes Medical Institute, for providing access. This work was supported by National Institutes of Health Grant CA154802, a Discovery grant from The Rogers Foundation, and The Stephen and Nancy Grand Multiple Myeloma Translational Initiative (to J.A.W.). A.P.W. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG 111-12).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405987111/-/DCSupplemental.
References
- 1.Kaufmann SH, Earnshaw WC. Induction of apoptosis by cancer chemotherapy. Exp Cell Res. 2000;256(1):42–49. doi: 10.1006/excr.2000.4838. [DOI] [PubMed] [Google Scholar]
- 2.Blankenberg FG. In vivo imaging of apoptosis. Cancer Biol Ther. 2008;7(10):1525–1532. doi: 10.4161/cbt.7.10.6934. [DOI] [PubMed] [Google Scholar]
- 3.Renz A, et al. Rapid extracellular release of cytochrome c is specific for apoptosis and marks cell death in vivo. Blood. 2001;98(5):1542–1548. doi: 10.1182/blood.v98.5.1542. [DOI] [PubMed] [Google Scholar]
- 4.Moffitt KL, Martin SL, Walker B. Proteases implicated in apoptosis: Old and new. J Pharm Pharmacol. 2010;62(5):563–576. doi: 10.1211/jpp.62.05.0002. [DOI] [PubMed] [Google Scholar]
- 5.Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–1087. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- 6.Yang TJ, Haimovitz-Friedman A, Verheij M. Anticancer therapy and apoptosis imaging. Exp Oncol. 2012;34(3):269–276. [PubMed] [Google Scholar]
- 7.Beachy SH, Repasky EA. Using extracellular biomarkers for monitoring efficacy of therapeutics in cancer patients: An update. Cancer Immunol Immunother. 2008;57(6):759–775. doi: 10.1007/s00262-007-0445-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greystoke A, et al. Assessment of circulating biomarkers for potential pharmacodynamic utility in patients with lymphoma. Br J Cancer. 2011;104(4):719–725. doi: 10.1038/sj.bjc.6606082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olofsson MH, et al. Cytokeratin-18 is a useful serum biomarker for early determination of response of breast carcinomas to chemotherapy. Clin Cancer Res. 2007;13(11):3198–3206. doi: 10.1158/1078-0432.CCR-07-0009. [DOI] [PubMed] [Google Scholar]
- 10.Dean E, Greystoke A, Ranson M, Dive C. Biomarkers of cell death applicable to early clinical trials. Exp Cell Res. 2012;318(11):1252–1259. doi: 10.1016/j.yexcr.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 11.Mahrus S, et al. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134(5):866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wildes D, Wells JA. Sampling the N-terminal proteome of human blood. Proc Natl Acad Sci USA. 2010;107(10):4561–4566. doi: 10.1073/pnas.0914495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiita AP, Seaman JE, Wells JA. Global analysis of cellular proteolysis by selective enzymatic labeling of protein N-termini. Methods Enzymol. doi: 10.1016/B978-0-12-417158-9.00013-3. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polevoda B, Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol. 2003;325(4):595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- 15.Gerszten RE, et al. Challenges in translating plasma proteomics from bench to bedside: Update from the NHLBI Clinical Proteomics Programs. Am J Physiol Lung Cell Mol Physiol. 2008;295(1):L16–L22. doi: 10.1152/ajplung.00044.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Addona TA, et al. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nat Biotechnol. 2011;29(7):635–643. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiteaker JR, et al. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nat Biotechnol. 2011;29(7):625–634. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawford ED, et al. The DegraBase: A database of proteolysis in healthy and apoptotic human cells. Mol Cell Proteomics. 2013;12(3):813–824. doi: 10.1074/mcp.O112.024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: Workflows, potential, pitfalls and future directions. Nat Methods. 2012;9(6):555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 20.Saelens X, et al. Toxic proteins released from mitochondria in cell death. Oncogene. 2004;23(16):2861–2874. doi: 10.1038/sj.onc.1207523. [DOI] [PubMed] [Google Scholar]
- 21.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10(11):3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68(18):3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crawford ED, et al. Conservation of caspase substrates across metazoans suggests hierarchical importance of signaling pathways over specific targets and cleavage site motifs in apoptosis. Cell Death Differ. 2012;19(12):2040–2048. doi: 10.1038/cdd.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimbo K, et al. Quantitative profiling of caspase-cleaved substrates reveals different drug-induced and cell-type patterns in apoptosis. Proc Natl Acad Sci USA. 2012;109(31):12432–12437. doi: 10.1073/pnas.1208616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaffe JD, et al. Accurate inclusion mass screening: A bridge from unbiased discovery to targeted assay development for biomarker verification. Mol Cell Proteomics. 2008;7(10):1952–1962. doi: 10.1074/mcp.M800218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agard NJ, et al. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc Natl Acad Sci USA. 2012;109(6):1913–1918. doi: 10.1073/pnas.1117158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bucki R, Levental I, Kulakowska A, Janmey PA. Plasma gelsolin: Function, prognostic value, and potential therapeutic use. Curr Protein Pept Sci. 2008;9(6):541–551. doi: 10.2174/138920308786733912. [DOI] [PubMed] [Google Scholar]
- 28.Wang M, et al. PaxDb, a database of protein abundance averages across all three domains of life. Mol Cell Proteomics. 2012;11(8):492–500. doi: 10.1074/mcp.O111.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picotti P, et al. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat Methods. 2010;7(1):43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 30.Wiita AP, et al. Global cellular response to chemotherapy-induced apoptosis. Elife. 2013;2:e01236. doi: 10.7554/eLife.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lote CJ. Principles of Renal Physiology. 5th Ed. New York: Springer; 2012. [Google Scholar]
- 32.MacLean B, et al. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.