Significance
Photosynthesis is one of the most fundamental biological processes on Earth. To date, species capable of performing (bacterio)chlorophyll-based phototrophy have been reported in six bacterial phyla. Here we report a phototrophic bacterium belonging to the rare and understudied phylum Gemmatimonadetes. This strain, isolated from a freshwater lake in the Gobi Desert, contains fully functional photosynthetic reaction centers. Its photosynthesis genes appear to originate from an ancient horizontal gene transfer from a purple phototrophic bacterium. Our findings not only demonstrate that Gemmatimonadetes represents a new phototrophic bacterial phylum, but also present, to our knowledge, the first evidence that genes for (bacterio)chlorophyll-based phototrophy can be transferred between distant bacterial phyla.
Keywords: anoxygenic photosynthesis, horizontal gene transfer, bacterial pigments, aerobic photoheterotroph, fluorescence imaging system
Abstract
Photosynthetic bacteria emerged on Earth more than 3 Gyr ago. To date, despite a long evolutionary history, species containing (bacterio)chlorophyll-based reaction centers have been reported in only 6 out of more than 30 formally described bacterial phyla: Cyanobacteria, Proteobacteria, Chlorobi, Chloroflexi, Firmicutes, and Acidobacteria. Here we describe a bacteriochlorophyll a-producing isolate AP64 that belongs to the poorly characterized phylum Gemmatimonadetes. This red-pigmented semiaerobic strain was isolated from a freshwater lake in the western Gobi Desert. It contains fully functional type 2 (pheophytin-quinone) photosynthetic reaction centers but does not assimilate inorganic carbon, suggesting that it performs a photoheterotrophic lifestyle. Full genome sequencing revealed the presence of a 42.3-kb–long photosynthesis gene cluster (PGC) in its genome. The organization and phylogeny of its photosynthesis genes suggests an ancient acquisition of PGC via horizontal transfer from purple phototrophic bacteria. The data presented here document that Gemmatimonadetes is the seventh bacterial phylum containing (bacterio)chlorophyll-based phototrophic species. To our knowledge, these data provide the first evidence that (bacterio)chlorophyll-based phototrophy can be transferred between distant bacterial phyla, providing new insights into the evolution of bacterial photosynthesis.
Photosynthesis is one of the most ancient and fundamental biological processes (1, 2). Phototrophic organisms transform solar radiation into metabolic energy, which fuels most of the Earth’s ecosystems (3). It is generally assumed that the earliest phototrophs were anaerobic (i.e., living in the absence of free oxygen) anoxygenic (i.e., not producing oxygen) prokaryotes (2). After the rise of oxygenic Cyanobacteria ∼2.7 Gyr ago, the Earth’s atmosphere started to become gradually oxygenated, reaching the present oxygen concentration roughly 0.6 Gyr ago (4, 5). Thus, early anaerobic phototrophs were forced either to adapt to the new oxic conditions or to retreat to anoxic habitats, leading to present phylogenetically and physiologically diverse phototrophic lineages.
To date, species using (bacterio)chlorophyll-based photosynthetic reaction centers (chlorophototrophs) have been reported in six bacterial phyla: Cyanobacteria, Proteobacteria, Chlorobi, Chloroflexi, Firmicutes, and Acidobacteria (6). Each of these lineages contains a unique apparatus for solar energy conversion differing in light-harvesting complex architecture, pigment composition, and function of reaction centers (7). In general, photosynthetic reaction centers can be divided into two main groups. FeS-based (type 1) reaction centers are used by green sulfur bacteria (classified into Chlorobi), heliobacteria (phototrophic Firmicutes), and phototrophic Acidobacteria. Pheophytin-quinone (type 2) reaction centers are present in green nonsulfur bacteria (Chloroflexi) and purple bacteria (phototrophic Proteobacteria). Oxygenic Cyanobacteria contain both type 1 and type 2 reaction centers.
Although Cyanobacteria, green sulfur bacteria, and purple bacteria were discovered more than 100 y ago (8), green nonsulfur bacteria and heliobacteria were not described until the second half of the 20th century (9, 10). The most recently identified organism representing a novel phylum containing chlorophototrophs is Candidatus Chloracidobacterium thermophilum, described in 2007 (11). These six phyla account for only a small portion of described phyla within the domain Bacteria (12, 13). Given that (bacterio)chlorophyll-based phototrophy represents an ancient process, we hypothesized that there remain chlorophototrophic lineages awaiting discovery, and, based on this notion, conducted a large isolate-screening project in various freshwater lakes with the aim of discovering novel phototrophic lineages.
Results and Discussion
Strain Isolation and Characterization.
To facilitate the screening process, we assembled an infrared fluorescence imaging system for detection of bacteriochlorophyll (BChl)-containing organisms. Principle and system components are listed in SI Appendix, Fig. S1. We screened more than 5,000 bacterial colonies obtained from various aquatic systems. Along with the more than 100 various phototrophic proteobacterial strains detected, we isolated an infrared fluorescence-positive colony originating from freshwater Swan Lake in the western Gobi Desert (SI Appendix, Fig. S2). This red-pigmented colony was transferred several times onto R2A agar plates, and the obtained isolate was named AP64. The AP64 strain exhibited the best growth rate (2–3 wk) on agar media under semiaerobic conditions (SI Appendix, Table S1).
In the AP64 cells, which appear as 1∼6-μm–long rods, the presence of BChl was confirmed by infrared epifluorescence microscopy (Fig. 1A). Unexpectedly, AP64’s 16S rRNA sequence was found to be 96% identical to that of Gemmatimonas aurantiaca T-27T, the sole type strain of the recently described phylum Gemmatimonadetes (14). In addition, phylogenetic analysis of 16S rRNA genes clearly placed AP64 within Gemmatimonadetes (Fig. 1B). Gemmatimonadetes is a sister phylum to Fibrobacteres, but is not closely related to any phyla known to contain chlorophototrophs (Fig. 1B). To date, none of the cultured strains of this phylum (15, 16) demonstrated phototrophic capability.
Fig. 1.
Microscopy images of the phototrophic Gemmatimonadetes strain AP64 and 16S rRNA gene phylogeny showing the position of the Gemmatimonadetes phylum among the Bacteria domain. (A, Left) Infrared epifluorescence microscopy image (false colors). (Center) Scanning electron microscopy image. (Right) Transmission electron microscopy image of an AP64 cell. (B) phylogenetic tree of 16S rRNA genes based on the data from the All Species Living Tree project (www.arb-silva.de/). Orange wedges indicate the bacterial phyla that contain chlorophototrophic species. Scale bar represents changes per position.
Genome Sequencing and Identification of the Photosynthesis Gene Cluster.
To gain further insight into the phylogeny and metabolism of AP64, we sequenced its genome by combining Illumina and 454 pyrosequencing techniques. The genomic sequence thus obtained consisted of seven contigs with a total of 4,706,869 bases, with a GC content of 64.4% (SI Appendix, Table S2). It contained a complete rRNA operon consisting of 16S, 23S, and 5S rRNA genes located at a single 347,817-bp–long contig (Contig_6). All three rRNA genes showed the closest matches with rRNA genes of G. aurantiaca T-27, with sequence identities of 96% for 16S, 96% for 23S, and 97% for 5S (SI Appendix, Table S3). Similarly, an additional analysis of 40 housekeeping genes in AP64 showed that 39 of these genes had the highest sequence identities to their counterparts in G. aurantiaca T-27 (SI Appendix, Table S3), further supporting the assignment of AP64 as a member of Gemmatimonadetes.
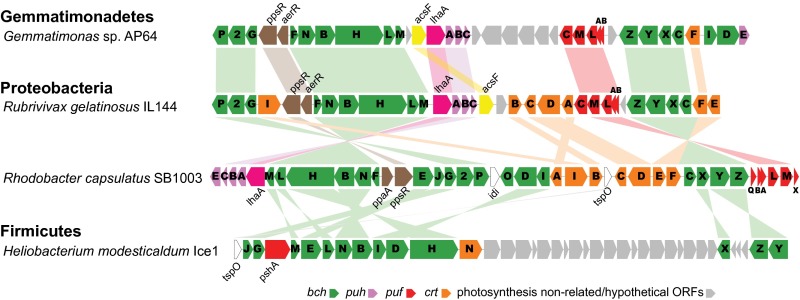
A 42.3-kb–long photosynthesis gene cluster (PGC) was identified on the 620,776-bp–long Contig_5 (Fig. 2 and SI Appendix, Fig. S3). The clustering of photosynthesis genes into a PGC was first described in Rhodobacter capsulatus (previously Rhodopseudomonas capsulata) (17–19). The PGCs are a typical feature in most purple phototrophic bacteria (20–22) and heliobacteria (23), but are not present in other chlorophototrophic lineages.
Fig. 2.
Gene organization of the PGC of AP64 in comparison with Proteobacteria and Firmicutes. Genome data were referenced for the PGC composition: R. capsulatus SB1003 (GenBank accession no. CP001312), R. gelatinosus IL144 (GenBank accession no. AP012320), and H. modesticaldum Ice1 (GenBank accession no. CP000930). bch (green), bacteriochlorophyll biosynthesis genes; puh (pink), genes encoding reaction center assembly proteins; puf (red), genes encoding reaction center proteins; crt (brownish yellow), carotenoid biosynthesis genes; gray, photosynthesis nonrelated genes or hypothetical ORFs.
Interestingly, the AP64 PGC arrangement closely resembles that of Proteobacteria (Fig. 2). The PGC in AP64 contains the same conserved gene order, crtF-bchCXYZ followed by the pufBALM operon, as was previously identified in Proteobacteria (21, 22). Similarly, the gene arrangement bchFNBHLM-acsF-lhaA-puhABC resembles the gene organization found in Rubrivivax gelatinosus IL144 (24), differing only in the position of acsF, and in R. capsulatus SB1003 (20, 25), where acsF was absent (Fig. 2). The presence of puf genes encoding bacterial reaction center subunits suggests the presence of type 2 photosynthetic reaction centers. In addition, the genome contains a complete gene inventory of the bacteriochlorophyll biosynthesis pathway (SI Appendix, Tables S4 and S5). The AP64 genome lacks the crucial genes of any known carbon fixation pathway (26). Similar to pure heterotrophs, the genome of AP64 contains all of the essential genes for the citric acid cycle and glycolysis, suggesting that this strain most likely carries out a photoheterotrophic lifestyle.
Expression and Functionality of the Photosynthetic Apparatus.
The expression of photosynthetic reaction centers in AP64 cells was demonstrated by its in vivo absorption spectrum, which displayed two infrared BChl a bands at 819 nm and 866 nm (Fig. 3A), resembling the absorbance of inner and peripheral light-harvesting complexes in some aerobic anoxygenic phototrophic bacteria (27). Carotenoids were responsible for most of the light absorption between 400 and 600 nm, with main absorption peaks at 478, 507, and 542 nm (Fig. 3A). The AP64 cells contained 3.5 ± 1.1 mg (mean ± SD; n = 5) of BChl a g−1 protein, approximately one order of magnitude less than typical anoxygenic photoautotrophs but similar to the levels reported in aerobic anoxygenic phototrophs (28). Further analysis of the pigment composition of AP64 by HPLC identified two forms of BChl a, esterified either with geranyl-geranyl or with phytol side chains (peaks 7 and 9 in Fig. 3C).
Fig. 3.
Photosystem functionality and pigment characterization of strain AP64. (A) In vivo absorption spectrum. (Inset) The 77K fluorescence emission spectrum. (B) Fluorescence induction and relaxation kinetics recorded by infrared fluorometry. (C) HPLC elution profile of pigment extracts from strains Gemmatimonas sp. AP64 and G. aurantiaca T-27 at 480 nm for carotenoids and 770 nm for BChl a. Note that traces are shifted vertically. Numbers above peaks indicate main pigments: 1 and 2, putative (2S,2’S)-oscillol 2,2’-di-(α-l-rhamnoside) (29); 3–6, unknown carotenoids; 7, BChl aGG (geranlylgeranyl); 8, spirilloxanthin; 9, BChl aP (phytol); 10, bacteriophaeophytin aP.
Based on the chromatography data, we determined that there were 62.1 ± 5.3 BChl a molecules per reaction center (mean ± SD; n = 4). In addition, AP64 contains several main carotenoids, mostly various derivatives of spirilloxanthin and oscillol (29) (Fig. 3C). As judged from its absorption spectrum and the high carotenoid-to-BChl a ratio (5.5 ± 1.7 mol:mol), most of the carotenoids probably do not play a role in light harvesting, but do serve in photoprotection. This idea is supported by the presence of abundant polar carotenoids (peaks 1–4) in the nonphototrophic relative G. aurantiaca T-27 as well (Fig. 3C). In line with the presence of these carotenoids, we have identified six carotenogenesis genes in the AP64 genome (SI Appendix, Table S6) and propose a putative biosynthesis pathway for spirilloxanthin and oscillol series carotenoid biosynthesis (SI Appendix, Fig. S4), based on available genetic information and previously reported data (29, 30).
We tested the functionality of the photosynthetic apparatus with infrared kinetic fluorometry (Fig. 3B). The recorded fluorescence transients resembled those of purple nonsulfur bacteria and aerobic anoxygenic phototrophs (28). The recorded FV/FM ratio of 0.71 ± 0.02 and turnover rate of 60.9 ± 18.8 s−1 (mean ± SD; n = 4) confirm that AP64 contains fully functional type 2 photosynthetic reaction centers connected to an efficient electron transfer chain. Despite the presence of the light-harvesting apparatus, AP64 is not an obligate phototroph, because it also grows well in the dark (SI Appendix, Table S1). It requires a supply of organic substrates for growth and metabolism, and thus is a facultative photoheterotroph. Indeed, exposure to light caused a 37.5 ± 6.9% reduction in AP64 respiration (SI Appendix, Fig. S5), demonstrating that photophosphorylation driven by light energy supplements oxidative phosphorylation using organic carbon substrates.
We tested the capacity of AP64 to assimilate CO2 using radioactive assays and found only a minimum amount of incorporated radioactivity and no significant difference between light and dark treatments (SI Appendix, Fig. S5). These findings suggest that this activity in AP64 derives solely from the anaplerotic carboxylation enzymes identified in its genome (SI Appendix, Table S7). Thus, AP64 represents a photoheterotrophic organism, whose ability to harvest light may provide additional energy for its metabolism and improve the economy of carbon utilization, as has been demonstrated in marine photoheterotrophic Proteobacteria (31).
Distribution of Phototrophic Gemmatimonadetes in Nature.
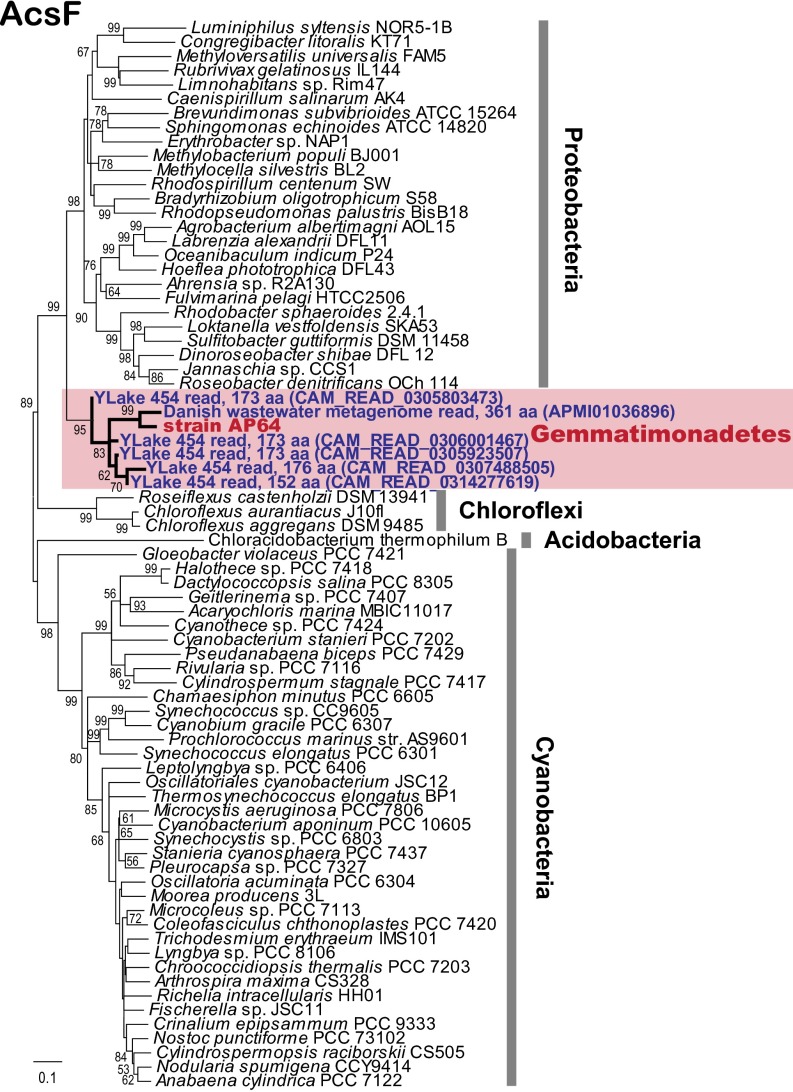
The ecology of phototrophic Gemmatimonadetes is unknown. 16S rRNA sequences related to Gemmatimonadetes have been found in soil and marine and lake sediments (32) (SI Appendix, Fig. S6); however, whether they originate from heterotrophic or phototrophic species is unclear. To target specifically phototrophic Gemmatimonadetes, we ran a search of protein homologs of oxygen-dependent Mg-protoporphyrin monomethylester cyclase (AcsF), a recently developed phylogenetic marker for aerobic and semiaerobic chlorophototrophs (33). The survey of available metagenomes stored in the CAMERA database (34) and in the National Center for Biotechnology Information (NCBI) whole-genome shotgun database identified five partial homologs predicted from Yellowstone Lake metagenomes (129–173 predicted amino acid positions with 88∼94% identities) and one full-length homolog from a Danish wastewater metagenome (362 positions with 88% identity) (SI Appendix, Table S8).
The homologs thus obtained formed a tight cluster with AP64 on the AcsF phylogenetic tree, suggesting that these sequences originated from phototrophic Gemmatimonadetes (Fig. 4). The presence of these genes in geographically dispersed and nonequivalent environments suggests that phototrophy may be a common strategy in Gemmatimonadetes rather than an unusual event occurring only in a specific environment.
Fig. 4.
Phylogenetic analysis of Gemmatimonadetes AcsF-like protein sequences predicted from environmental metagenomes in relation to other phototrophic bacterial phyla. Environmental Gemmatimonadetes AcsF-like sequences were retrieved from the CAMERA or NCBI WGS databases using tblastn searching (SI Appendix, Table S8). Pairwise deletion was performed at the sites containing missing data or alignment gaps. The unrooted tree was inferred using a neighbor-joining algorithm with 1,000 resamplings. Bootstrap values >50% are shown. Ylake, Yellowstone Lake. The total number of amino acids of the partial Gemmatimonadetes AcsF-like ORFs are shown next to the names. CAMERA/GenBank accession numbers are in parentheses.
Phylogeny of Photosynthesis Genes in Gemmatimonadetes and Evolutionary Significance.
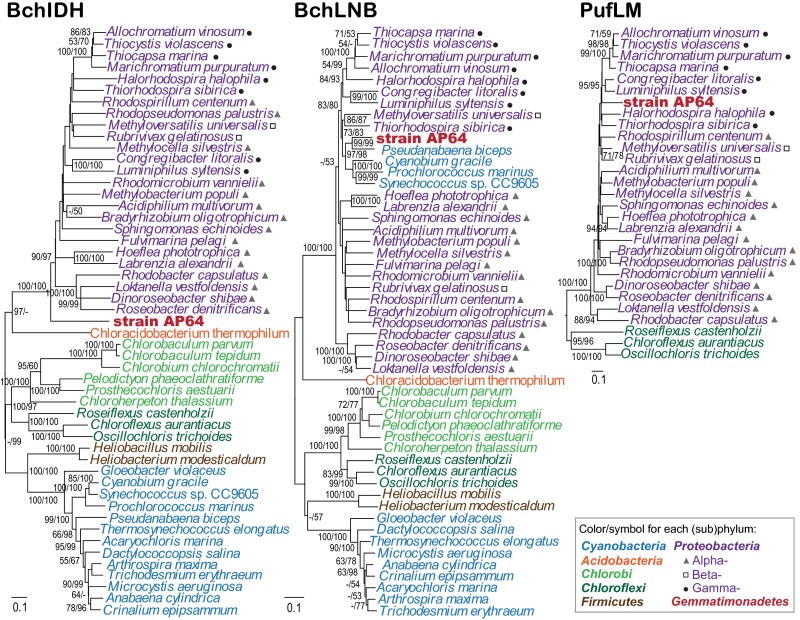
To elucidate the possible origin of phototrophic capacity in AP64, we studied the phylogeny of its photosynthesis genes in more detail using enzymes involved in the initial phase of the bacteriochlorophyll biosynthesis pathway, which exist only in chlorophototrophs and are shared by all known chlorophototrophic species (35). The enzymes that we used were magnesium chelatase encoded by bchIDH genes (36) and light-independent protochlorophyllide reductase encoded by bchLNB genes (37). The BchIDH and BchLNB trees separated main chlorophototrophic groups and placed Gemmatimonas sp. AP64 as an early-diverging member of the Proteobacterial clade on the BchIDH tree (Fig. 5A) while inside Proteobacteria on the BchLNB tree (Fig. 5B) together with a group of cyanobacteria for which horizontal gene transfer of BchIDH from Proteobacteria has been proposed previously (38). This finding clearly demonstrates a common origin of the bacteriochlorophyll biosynthesis pathway in AP64 and Proteobacteria.
Fig. 5.
Phylogenetic analyses of AP64 photosynthesis genes. (Left ) Analysis based on concatenated alignments of amino acid sequences of magnesium chelatase (BchIDH/ChlIDH; 1,810 common amino acid positions). (Center) Analysis based on concatenated alignments of amino acid sequences of light independent protochlorophyllide reductase (BchLNB/ChlLNB; 921 common amino acid positions). (Right) Analysis based on concatenated alignments of amino acid sequences of the photosynthetic reaction center subunits (PufLM; 507 common amino acid positions). Maximum likelihood (ML) and neighbor-joining (NJ) trees were inferred for strain AP64 and representative species from other six phototrophic phyla. The PufLM tree was rooted by Chroococcidiopsis thermalis (Cyanobacteria) D1/D2 sequences. ML/NJ bootstrap values >50% are shown on the trees. Scale bars represent changes per position.
Similarly, an analysis of reaction center subunit (PufLM) phylogeny placed AP64 onto a branch inside Proteobacterial sequences (Fig. 5C), confirming that AP64 contains a close homolog of Proteobacterial photosynthetic reaction centers. The similarity of photosynthetic apparatus between AP64 and Proteobacteria (Table 1) and their analogous PGC organization, as well as the close phylogenetic relationship of their photosynthesis genes, strongly suggest that phototrophy in Gemmatimonas spp. originated from an ancient horizontal transfer event of a complete set of photosynthesis genes from a purple photosynthetic bacterium.
Table 1.
Characteristics of phototrophic Gemmatimonadetes and six other bacterial phyla containing chlorophototrophic members
| Phylum | Common name* | Reaction center | Main pigments | Carbon fixation pathway† | Oxygen requirement | Discovery |
| Cyanobacteria | Type 1 | Chlorophylls, carotenoids, phycobilins | Reductive pentose phosphate (Calvin– Benson–Bassham) cycle | Aerobic | 19th century | |
| Type 2 | ||||||
| Proteobacteria | Purple bacteria | Type 2 | BChl a/b, carotenoids | Reductive pentose phosphate (Calvin– Benson–Bassham) cycle | Aerobic, semiaerobic, anaerobic | 19th century |
| Chlorobi | Green sulfur bacteria | Type 1 | BChl a/c/d/e, carotenoids | Reductive tricarboxylic acid (Arnon–Buchanan) cycle | Anaerobic | Early 20th century |
| Chloroflexi | Green nonsulfur bacteria | Type 2 | BChl a/c, carotenoids | 3-Hydroxypropionate (Fuchs–Holo) bi-cycle | Semiaerobic | 1974 (10) |
| Firmicutes | Heliobacteria | Type 1 | BChl g, carotenoids | Absent | Anaerobic | 1983 (11) |
| Acidobacteria | Type 1 | BChl a/c, carotenoids | Absent | Aerobic, semiaerobic | 2007 (12) | |
| Gemmatimonadetes | Type 2 | BChl a, carotenoids | Absent | Semiaerobic | This study |
Common or historical name used for phototrophic species in the particular phylum.
If present.
The question remains as to a suitable vector capable of distant interspecies transfer of a large-sized PGC. A complete PGC has been reported in a promiscuous self-transmissible R-prime plasmid of R. capsulatus (39) and in two plasmids of Roseobacter litoralis and Sulfitobacter guttiformis (40). Photosynthesis genes also can be packed into gene transfer agents (41, 42) or bacteriophages (43–45). Similar mobile elements might have facilitated the acquisition of purple bacterial photosynthetic reaction centers by an ancient counterpart of the heterotrophic Gemmatimonadetes strain T-27. The higher redox potential (∼0.5 V) of type 2 reaction centers compared with type 1 reaction centers might have allowed for easier incorporation into the respiratory electron transfer chain of this ancient Gemmatimonadetes bacterium. After the adoption of phototrophy as a means of energy production, phototrophic Gemmatimonadetes likely evolved independently, perhaps losing the acquired genes again in some lineages. Nevertheless, Gemmatimonas sp. AP64 represents, to our knowledge, the first known example of horizontal transfer of a complete photosynthesis gene package between distant bacterial phyla, providing new insights into the evolution of bacterial photosynthesis.
Materials and Methods
Bacterial Isolation and Colony Screening.
On December 15, 2011, 1 L of surface water (0.5 m) was collected approximately 3 m offshore in freshwater Swan Lake [in Chinese, 天鹅湖 (Tiān ér hú)]. This lake is located (latitude 42.005 N, longitude 101.585 E, ∼900 m above sea level) at the northern margin of the Badain Jaran Desert (∼49,000 km2 total area), part of the western Gobi Desert in the Inner Mongolia Autonomous Region of northern China (SI Appendix, Fig. S2). The lake has a surface area of approximately 1 km2, a maximum depth 0.65 m, a transparency of 0.65 m, salinity <0.1 ppt, pH 8.76, and conductivity of 1.70 mS cm−1. The catchment area of Swan Lake suffers from desertification, especially after the almost permanent cutoff of the Heihe River inflow in the past 30 y (46). Water sources are mainly groundwater, summer rainfalls, and sporadic runoff from small branches of the lower reaches of the Heihe River. In this region, the mean annual precipitation is <50 mm, and evaporation is roughly 2,000–2,500 mm/y (47–49).
In the laboratory, 1 µL of the original lake water was diluted into 100 µL of sterilized half-strength R2A medium, and the dilution was plated directly onto half-strength R2A agar plates (SI Appendix, Table S1). The plates were incubated at 25 °C under a 12-h light/12-h dark regimen until visible colonies formed (3–6 wk). Plates were scanned for BChl fluorescence emission using a modified PSI FluorCam 800MF fluorescence imaging system (SI Appendix, Fig. S1). In brief, a standard Petri dish (90 mm diameter) with colonies was illuminated with a bank of blue light-emitting diodes (470 nm). Infrared fluorescence from BChl a-containing colonies was registered by a monochrome CCD camera (Lumenera) protected by a long-pass >850-nm glass filter RG850 (Schott). The positive (IR fluorescent) colonies were resuspended in sterile medium and transferred onto new agar plates until pure cultures were obtained.
Cultivation of Strain AP64.
Strain AP64 was isolated from a 5-wk-old plate. We optimized the growth conditions by adjusting pH, temperature, oxygen level, and nutrient combinations (SI Appendix, Table S1). The best growth was achieved on modified R2A+ agar plates (medium composition shown in SI Appendix, Table S1). The plates were kept at 25∼28 °C in a reduced 5∼10% oxygen atmosphere under a 12-h light/12-h dark regimen. It takes 10∼14 d to obtain abundant round-shaped colonies (1.0∼2.0 mm diameter). Agar plates inoculated with the same amount of AP64 cells but kept in darkness showed similar growth patterns (SI Appendix, Table S1). We did not obtain satisfactory growth yields in the corresponding liquid medium, because AP64 seemed to prefer an attachment-to-solid surface lifestyle.
Microscopy, Spectroscopy, and Fluorometry.
Observation of cell morphology and monitoring of culture purity was performed by infrared epifluorescence microscopy (50). In vivo absorption spectra were recorded using a Shimadzu UV-3000 dual-beam spectrometer. The cells harvested directly from agar plates were resuspended into 70% glycerol to reduce light scattering. The same cells were also used to record 77K fluorescence emission spectra. Fluorescence was excited by a single cyan (505 nm) Luxeon Rebel light-emitting diode. The emission spectra were recorded by a PSI MCS 55/A high-resolution fiber optics spectrometer. BChl a fluorescence was recorded using a modified PSI FL200/PS laboratory fluorometer (28).
Pigment Analyses.
Approximately 1010 cells were harvested and extracted with 1 mL of 100% methanol for 5 min. The BChl a concentration was determined spectroscopically in 100% methanol (51). BChl a content was normalized on a per cell or per total protein basis. In addition, the extracts were analyzed by HPLC-MS using an Agilent 1100 Series HPLC system equipped with an in-line Agilent 1100 series LC/MSD trap mass spectrometer (28). BChl a per reaction center stoichiometry was determined using extract acidification as described previously (28). All reported values are presented as mean ± SDs determined from at least four independently grown batches of cells.
Radioactive Assays and Respiration Assays.
The carboxylation activity of strain AP64 was determined with a radiolabeled bicarbonate incorporation assay. Assays were performed under both anoxic and semiaerobic conditions. Bacterial cultures (20 mL, 6.2 × 108 cells mL−1) were labeled with 40 μCi NaH14CO3 and divided into 2-mL plastic transparent vials. The vials were placed into a temperature-controlled incubator (30 °C) illuminated with 150 μmol quanta m−2 s−1. The incubation time was 2 h. The incubation was terminated with the addition of 35% HCl, followed by continuous shaking for 24 h. To determine the incorporated radiolabeled compounds, 10 mL of EcoLite(+) scintillation mixture was added, and counts were performed with a liquid scintillation analyzer (Perkin-Elmer Tri-Carb 2810 TR). For comparison, the carboxylation activities of three control strains (heterotrophic G. aurantiaca T-27 and photoautotrophic Rhodobacter sphaeroides DSM158 and Synechocystis sp. PCC6803) were determined using the same protocol.
For respiration measurement, AP64 cells were placed in a temperature-stabilized measuring chamber at 30 °C, and the rate of oxygen consumption was registered with the Clark electrode for 20 min. The chamber was illuminated using a halogen lamp providing 150 μmol photons m−2 s−1. T-27 cells received the same treatment and served as a heterotrophic control. Carboxylation activity and respiration rate were expressed as mol CO2 or O2 g protein-1 hr-1. Quantification of protein concentrations in cellular extracts was determined with a modification of the micro-Lowry technique, using the Sigma-Aldrich Total Protein Kit.
Genome Sequencing and Data Analyses.
Bacterial cells were harvested directly from agar plates. Total genome DNA was extracted and purified using a DNA purification column kit (Tiangen). Initially, 16S rRNA was PCR-amplified using the standard 27F and 1492R primers. The amplified products were directly sequenced to identify the taxonomic identity of the obtained strain. Later, whole-genome shotgun sequencing was performed using an Illumina HiSeq-2000 platform at Meiji Biotech (Shanghai, China) and a Roche 454 GS FLX+ System at Macrogen (Seoul, South Korea). Procedures for DNA shearing, library preparation and quality control, sample loading, and sequencer operation were performed according to those companies’ standard protocols.
For Illumina technology, raw 101-bp paired-end reads produced from a 300-bp paired-end library were first subjected to quality control and trimming on the Galaxy Web server (52). High-quality reads were de novo assembled into contigs using Velvet version 1.2.10 (53). For 454 technology, raw reads with an average length of 800 bp were assembled using GS de novo Assembler version 2.8 (454 Life Sciences, Roche). To further assemble the achieved contigs from both Illumina and 454, all contigs were first shredded into 1-kb fake reads using the Biopieces package (http://www.biopieces.org) and then reassembled through GS de novo Assembler. Seven contigs were finally assembled with the contig length threshold set at 1.5 kb. Annotation of contigs was performed with the RAST (54) and BASys (55) servers with further manual corrections. Whole-genome alignments were performed and visualized in Mauve (56).
Phylogenetic Analyses.
For the overview tree of bacterial phyla, the aligned 16S rRNA gene sequences (ARB file, release LTPs111) were downloaded from the All Species Living Tree project (http://www.arb-silva.de) (13) and supplemented with cyanobacterial sequences retrieved from the SILVA database (57). AP64’s 16S rRNA gene sequence together with references from available Gemmatimonadetes pure cultures were added to the existing alignments and then to the ARB tree using the Add Species function of ARB (58). Reference sequences (environmental Gemmatimonadetes 16S rRNA gene sequences and BchIDH, BchLNB, PufLM, and AcsF amino acid sequences) were retrieved from the NCBI Web site. Multiple alignments were performed with Geneious version R6 (Biomatters) using the imbedded MUSCLE program. For BchIDH, BchLNB, and PufLM trees, sequence alignments of each peptide were concatenated, and the sites containing gaps were stripped from the final alignment before tree inference. Phylogenetic trees were calculated with MEGA version 5.2 (59) using both maximum likelihood and neighbor-joining algorithms with 1,000 times tree resampling. The Jones–Taylor–Thornton amino acid substitution model was used.
Supplementary Material
Acknowledgments
We thank Yapeng Liu for help during field sampling, Petra Kučerová for help with the HPLC-MS analyses, and Peter Nixon for providing Synechocystis PCC6803 WT strain. This work was supported by Czech Projects GAČR P501/10/0221, Algatech CZ.1.05/2.1.00/03.0110, and Algain CZ.1.07/2.3.00/30.0059. The early stage of the AP64 genome sequencing project was partially supported by National Natural Science Foundation of China (NSFC) Grant 30900045 (to Y.Z.). Field work was supported by NSFC Grant 30860015 (to F.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.E.B. is a guest editor invited by the Editorial Board.
Data deposition: The final genome assembly data of strain AP64 have been submitted to GenBank under BioProject PRJNA213561 (accession no. AUXF00000000). Raw Illumina and 454 reads were deposited at the NCBI Sequence Read Archive under BioSample SAMN02296694 (accession nos. SRR94398–SRR943988). The associated 16S rRNA gene sequence from the assembled AP64 genome was deposited in GenBank (accession no. KF481682).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400295111/-/DCSupplemental.
References
- 1.Canfield DE, Rosing MT, Bjerrum C. Early anaerobic metabolisms. Philos Trans R Soc Lond B Biol Sci. 2006;361(1474):1819–1834. doi: 10.1098/rstb.2006.1906. discussion 1835–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hohmann-Marriott MF, Blankenship RE. Evolution of photosynthesis. Annu Rev Plant Biol. 2011;62:515–548. doi: 10.1146/annurev-arplant-042110-103811. [DOI] [PubMed] [Google Scholar]
- 3.Falkowski PG, Raven JA. Aquatic Photosynthesis. 2nd Ed. Princeton, NJ: Princeton Univ Press; 2007. p. 484. [Google Scholar]
- 4.Nisbet EG, Sleep NH. The habitat and nature of early life. Nature. 2001;409(6823):1083–1091. doi: 10.1038/35059210. [DOI] [PubMed] [Google Scholar]
- 5.Kasting JF, Siefert JL. Life and the evolution of Earth’s atmosphere. Science. 2002;296(5570):1066–1068. doi: 10.1126/science.1071184. [DOI] [PubMed] [Google Scholar]
- 6.Raymond J. Coloring in the tree of life. Trends Microbiol. 2008;16(2):41–43. doi: 10.1016/j.tim.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Overmann J, Garcia-Pichel F. The phototrohic way of life. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes: Prokaryotic Communities and Ecophysiology. 4th Ed. Berlin: Springer; 2013. pp. 203–257. [Google Scholar]
- 8.Gest H, Blankenship RE. Time line of discoveries: Anoxygenic bacterial photosynthesis. Photosynth Res. 2004;80(1-3):59–70. doi: 10.1023/B:PRES.0000030448.24695.ec. [DOI] [PubMed] [Google Scholar]
- 9.Pierson BK, Castenholz RW. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch Microbiol. 1974;100(1):5–24. doi: 10.1007/BF00446302. [DOI] [PubMed] [Google Scholar]
- 10.Gest H, Favinger JL. Heliobacterium chlorum, an anoxygenic brownish-green photosynthetic bacterium containing a “new” form of bacteriochlorophyll. Arch Microbiol. 1983;136:11–16. [Google Scholar]
- 11.Bryant DA, et al. Candidatus Chloracidobacterium thermophilum: An aerobic phototrophic Acidobacterium. Science. 2007;317(5837):523–526. doi: 10.1126/science.1143236. [DOI] [PubMed] [Google Scholar]
- 12.Söhngen C, Bunk B, Podstawka A, Gleim D, Overmann J. BacDive—the Bacterial Diversity Metadatabase. Nucleic Acids Res. 2014;42(Database issue) D1:D592–D599. doi: 10.1093/nar/gkt1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yarza P, et al. The All-Species Living Tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31(4):241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, et al. Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int J Syst Evol Microbiol. 2003;53(Pt 4):1155–1163. doi: 10.1099/ijs.0.02520-0. [DOI] [PubMed] [Google Scholar]
- 15.Davis KER, Joseph SJ, Janssen PH. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl Environ Microbiol. 2005;71(2):826–834. doi: 10.1128/AEM.71.2.826-834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joseph SJ, Hugenholtz P, Sangwan P, Osborne CA, Janssen PH. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl Environ Microbiol. 2003;69(12):7210–7215. doi: 10.1128/AEM.69.12.7210-7215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yen HC, Marrs B. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J Bacteriol. 1976;126(2):619–629. doi: 10.1128/jb.126.2.619-629.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Youvan DC, Bylina EJ, Alberti M, Begusch H, Hearst JE. Nucleotide and deduced polypeptide sequences of the photosynthetic reaction-center, B870 antenna, and flanking polypeptides from R. capsulata. Cell. 1984;37(3):949–957. doi: 10.1016/0092-8674(84)90429-x. [DOI] [PubMed] [Google Scholar]
- 19.Zsebo KM, Hearst JE. Genetic-physical mapping of a photosynthetic gene cluster from R. capsulata. Cell. 1984;37(3):937–947. doi: 10.1016/0092-8674(84)90428-8. [DOI] [PubMed] [Google Scholar]
- 20.Haselkorn R, et al. The Rhodobacter capsulatus genome. Photosynth Res. 2001;70(1):43–52. doi: 10.1023/A:1013883807771. [DOI] [PubMed] [Google Scholar]
- 21.Swingley WD, Blankenship RE, Raymond J. Evolutionary relationships among purple photosynthetic bacteria and the origin of proteobacterial photosynthetic systems. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT, editors. Advances in Photosynthesis and Respiration, The Purple Phototrophic Bacteria. Vol 28. Dordrecht, The Netherlands: Springer; 2009. pp. 17–29. [Google Scholar]
- 22.Zheng Q, et al. Diverse arrangement of photosynthetic gene clusters in aerobic anoxygenic phototrophic bacteria. PLoS ONE. 2011;6(9):e25050. doi: 10.1371/journal.pone.0025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong J, Inoue K, Bauer CE. Tracking molecular evolution of photosynthesis by characterization of a major photosynthesis gene cluster from Heliobacillus mobilis. Proc Natl Acad Sci USA. 1998;95(25):14851–14856. doi: 10.1073/pnas.95.25.14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagashima S, et al. Complete genome sequence of phototrophic betaproteobacterium Rubrivivax gelatinosus IL144. J Bacteriol. 2012;194(13):3541–3542. doi: 10.1128/JB.00511-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strnad H, et al. Complete genome sequence of the photosynthetic purple nonsulfur bacterium Rhodobacter capsulatus SB 1003. J Bacteriol. 2010;192(13):3545–3546. doi: 10.1128/JB.00366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77(6):1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yurkov VV, Beatty JT. Aerobic anoxygenic phototrophic bacteria. Microbiol Mol Biol Rev. 1998;62(3):695–724. doi: 10.1128/mmbr.62.3.695-724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koblížek M, Mlcousková J, Kolber Z, Kopecký J. On the photosynthetic properties of marine bacterium COL2P belonging to Roseobacter clade. Arch Microbiol. 2010;192(1):41–49. doi: 10.1007/s00203-009-0529-0. [DOI] [PubMed] [Google Scholar]
- 29.Takaichi S, Maoka T, Takasaki K, Hanada S. Carotenoids of Gemmatimonas aurantiaca (Gemmatimonadetes): Identification of a novel carotenoid, deoxyoscillol 2-rhamnoside, and proposed biosynthetic pathway of oscillol 2,2'-dirhamnoside. Microbiol-Sgm. 2010;156:757–763. doi: 10.1099/mic.0.034249-0. [DOI] [PubMed] [Google Scholar]
- 30.Takaichi S. Distribution and biosynthesis of carotenoids. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT, editors. Advances in Photosynthesis and Respiration, The Purple Phototrophic Bacteria. Vol 28. Dordrecht, The Netherlands: Springer; 2009. pp. 97–117. [Google Scholar]
- 31.Hauruseu D, Koblížek M. Influence of light on carbon utilization in aerobic anoxygenic phototrophs. Appl Environ Microbiol. 2012;78(20):7414–7419. doi: 10.1128/AEM.01747-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeBruyn JM, Nixon LT, Fawaz MN, Johnson AM, Radosevich M. Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. Appl Environ Microbiol. 2011;77(17):6295–6300. doi: 10.1128/AEM.05005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boldareva-Nuianzina EN, Bláhová Z, Sobotka R, Koblížek M. Distribution and origin of oxygen-dependent and oxygen-independent forms of Mg-protoporphyrin monomethylester cyclase among phototrophic proteobacteria. Appl Environ Microbiol. 2013;79(8):2596–2604. doi: 10.1128/AEM.00104-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun SL, et al. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: The CAMERA resource. Nucleic Acids Res. 2011;39(Database issue):D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raymond J, Zhaxybayeva O, Gogarten JP, Gerdes SY, Blankenship RE. Whole-genome analysis of photosynthetic prokaryotes. Science. 2002;298(5598):1616–1620. doi: 10.1126/science.1075558. [DOI] [PubMed] [Google Scholar]
- 36.Gibson LCD, Willows RD, Kannangara CG, von Wettstein D, Hunter CN. Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: Reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc Natl Acad Sci USA. 1995;92(6):1941–1944. doi: 10.1073/pnas.92.6.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujita Y, Matsumoto H, Takahashi Y, Matsubara H. Identification of a nifDK-like gene (ORF467) involved in the biosynthesis of chlorophyll in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 1993;34(2):305–314. [PubMed] [Google Scholar]
- 38.Bryant DA, et al. Comparative and functional genomics of anoxygenic green bacteria from the taxa Chlorobi, Chloroflexi, and Acidobacteria. In: Burnap RL, Vermaas W, editors. Advances in Photosynthesis and Respiration, Functional Genomics and Evolution of Photosynthetic Systems. Vol 35. Dordrecht, The Netherlands: Springer; 2012. pp. 47–102. [Google Scholar]
- 39.Marrs B. Mobilization of the genes for photosynthesis from Rhodopseudomonas capsulata by a promiscuous plasmid. J Bacteriol. 1981;146(3):1003–1012. doi: 10.1128/jb.146.3.1003-1012.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen J, et al. Think pink: Photosynthesis, plasmids and the Roseobacter clade. Environ Microbiol. 2012;14(10):2661–2672. doi: 10.1111/j.1462-2920.2012.02806.x. [DOI] [PubMed] [Google Scholar]
- 41.Marrs B. Genetic recombination in Rhodopseudomonas capsulata. Proc Natl Acad Sci USA. 1974;71(3):971–973. doi: 10.1073/pnas.71.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lang AS, Beatty JT. Genetic analysis of a bacterial genetic exchange element: The gene transfer agent of Rhodobacter capsulatus. Proc Natl Acad Sci USA. 2000;97(2):859–864. doi: 10.1073/pnas.97.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann NH, Cook A, Millard A, Bailey S, Clokie M. Marine ecosystems: Bacterial photosynthesis genes in a virus. Nature. 2003;424(6950):741. doi: 10.1038/424741a. [DOI] [PubMed] [Google Scholar]
- 44.Lindell D, et al. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci USA. 2004;101(30):11013–11018. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharon I, et al. Photosystem I gene cassettes are present in marine virus genomes. Nature. 2009;461(7261):258–262. doi: 10.1038/nature08284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li ZW, Wang ZY, Liu L. Characteristics of meanders and oxbow lakes in Qinghai-Tibet plateau. In: Murillo Munoz RE, editor. River Flow 2012. Vol 1. Boca Raton, FL: CRC Press; 2012. pp. 727–734. [Google Scholar]
- 47.Mischke S, Demske D, Schudack ME. Hydrologic and climatic implications of a multidisciplinary study of the mid to late Holocene Lake Eastern Juyanze. Chin Sci Bull. 2003;48(14):1411–1417. [Google Scholar]
- 48.Qi SZ, Luo F. Environmental degradation problems in the Heihe River Basin, northwest China. Water Environ J. 2007;21(2):142–148. [Google Scholar]
- 49.Hartmann K, Wunnemann B. Hydrological changes and Holocene climate variations in NW China, inferred from lake sediments of Juyanze palaeolake by factor analyses. Quat Int. 2009;194:28–44. [Google Scholar]
- 50.Atamna-Ismaeel N, et al. Bacterial anoxygenic photosynthesis on plant leaf surfaces. Environ Microbiol Rep. 2012;4(2):209–216. doi: 10.1111/j.1758-2229.2011.00323.x. [DOI] [PubMed] [Google Scholar]
- 51.Permentier HP, et al. Composition and optical properties of reaction centre core complexes from the green sulfur bacteria Prosthecochloris aestuarii and Chlorobium tepidum. Photosynth Res. 2000;64(1):27–39. doi: 10.1023/A:1026515027824. [DOI] [PubMed] [Google Scholar]
- 52.Goecks J, Nekrutenko A, Taylor J, Team G. Galaxy Team Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aziz RK, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Domselaar GH, et al. BASys: A Web server for automated bacterial genome annotation. Nucleic Acids Res. 2005;33(Web Server issue):W455-9. doi: 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue) D1:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32(4):1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.