Significance
During cell division mitochondria and their genomes need to be transmitted to the daughter cells. In the parasitic protozoa Trypansoma brucei we find a unique situation. It has a single mitochondrion with a single-unit genome that is physically connected to the flagellum. Here we identify the β-barrel mitochondrial outer membrane protein TAC40 that localizes to this connection. TAC40 is essential and defines a novel subclass of mitochondrial porins that are specialized in mitochondrial genome inheritance. A comparative analysis reveals a conserved concept of a mitochondrial DNA inheritance mechanism in trypanosomes and yeast that depends on a physical linkage between mitochondrial DNA and the cytoskeleton, which is organized around a β-barrel protein of the mitochondrial porin family.
Keywords: organelle, parasitic protozoa
Abstract
Mitochondria cannot form de novo but require mechanisms allowing their inheritance to daughter cells. In contrast to most other eukaryotes Trypanosoma brucei has a single mitochondrion whose single-unit genome is physically connected to the flagellum. Here we identify a β-barrel mitochondrial outer membrane protein, termed tripartite attachment complex 40 (TAC40), that localizes to this connection. TAC40 is essential for mitochondrial DNA inheritance and belongs to the mitochondrial porin protein family. However, it is not specifically related to any of the three subclasses of mitochondrial porins represented by the metabolite transporter voltage-dependent anion channel (VDAC), the protein translocator of the outer membrane 40 (TOM40), or the fungi-specific MDM10, a component of the endoplasmic reticulum–mitochondria encounter structure (ERMES). MDM10 and TAC40 mediate cellular architecture and participate in transmembrane complexes that are essential for mitochondrial DNA inheritance. In yeast MDM10, in the context of the ERMES, is postulated to connect the mitochondrial genomes to actin filaments, whereas in trypanosomes TAC40 mediates the linkage of the mitochondrial DNA to the basal body of the flagellum. However, TAC40 does not colocalize with trypanosomal orthologs of ERMES components and, unlike MDM10, it regulates neither mitochondrial morphology nor the assembly of the protein translocase. TAC40 therefore defines a novel subclass of mitochondrial porins that is distinct from VDAC, TOM40, and MDM10. However, whereas the architecture of the TAC40-containing complex in trypanosomes and the MDM10-containing ERMES in yeast is very different, both are organized around a β-barrel protein of the mitochondrial porin family that mediates a DNA–cytoskeleton linkage that is essential for mitochondrial DNA inheritance.
Mitochondria are a hallmark of all eukaroytic cells. They derive from an endosymbiontic event between a free-living bacterium and a presumably prokaryotic host cell. More than 1.5 billion years of evolution resulted in a great diversification of mitochondria. As a consequence, the shape and number of organelles per cell as well as size, content, copy number, and organization of their genomes vary greatly between different taxons (1). However, all eukaryotes must be able to faithfully transmit mitochondria to their offspring (2, 3).
Unlike most other eukaryotes, the parasitic protozoa Trypanosoma brucei has a single mitochondrion throughout its life and its cell cycle. Due to the single-unit nature of the mitochondrion, its duplication must be coordinated with the duplication of the nucleus (4). The mitochondrial genome of T. brucei, termed kinetoplast DNA (kDNA), is essential for growth of both the procyclic insect stage and the bloodstream form of the parasite (5). It consists of a disk-shaped single-unit kDNA network that localizes to a distinct region within the mitochondrial matrix (6). The kDNA is physically connected with the cytosolic basal body, the organizing center of the eukaryotic flagellum, via a high-order transmembrane structure termed tripartite attachment complex (TAC) (7) of which only few components have been identified (8–10). Replication of the kDNA network occurs at a defined stage of the cell cycle shortly before the onset of the nuclear S phase. After replication, the kDNA networks need to be correctly positioned so that during cell and mitochondrial division each daughter cell receives a single organelle with a single kDNA network. This process requires an intact TAC and is mediated by the movement of the basal body: one kDNA network remains connected to the basal body of the old flagellum whereas the other one segregates with the basal body of the new flagellum (7, 11).
Unlike trypanosomes, Saccharomyces cerevisiae propagates by budding and contains highly dynamic mitochondria that constantly divide and fuse (12, 13). Mitochondrial inheritance in budding yeast therefore requires a mechanism to move mitochondria and their genomes from the mother cell into the growing bud. The protein-associated mitochondrial genomes of S. cerevisiae, termed nucleoids, localize to dozens of globular foci that are distributed all over the organelles. Most actively replicating nucleoids are associated with a protein complex that includes the outer membrane (OM) protein MDM10 as a central unit, as well as the proteins MDM12, MDM34, and MMM1 (14–16). The protein complex forms the endoplasmic reticulum (ER)–mitochondria encounter structure (ERMES) tethering the ER to the mitochondrion (17). The ERMES has also been suggested to connect to cytosolic actin fibers that mediate the movement of mitochondria to the bud of dividing yeast cells (14, 18, 19). Besides its role in mitochondrial inheritance, the ERMES has been implicated in maintenance of mitochondrial morphology and in phospholipid and calcium exchange as well as in the assembly of the protein translocase of the mitochondrial OM (TOM) (20, 21). Some of the proposed ERMES functions are controversial and there is evidence that some of them might be due to secondary effects caused by the drastically altered mitochondrial morphology (22).
The central ERMES subunit, the β-barrel protein MDM10 belongs to the mitochondrial porin superfamily, which comprises the three members voltage-dependent anion channel (VDAC), Tom40, and MDM10. Whereas VDAC and Tom40 have so far been found in all eukaryotes, including T. brucei (23, 24), MDM10 is specific to the fungal clade.
In this study we identify a mitochondrial OM protein of T. brucei as a novel component of the TAC. We show that the protein defines a novel subclass of the mitochondrial porin superfamily that is specialized in mitochondrial DNA inheritance.
Results
A 40-kDa Protein Is a Component of the TAC.
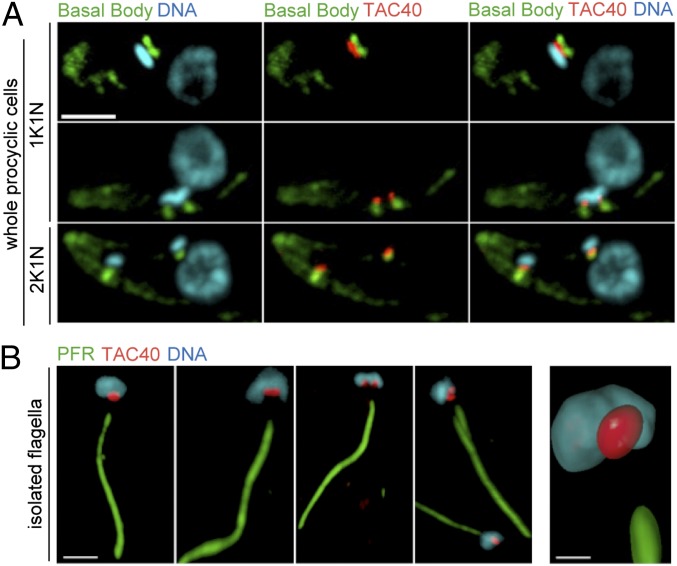
We have recently characterized the mitochondrial OM proteome of T. brucei (25). From this proteome we now have selected a 40-kDa protein, encoded by the ORF Tb927.4.1610, for further analysis. The protein has previously been identified as a VDAC-like mitochondrial OM protein in a genome-wide bioinformatic analysis (26). A cell line expressing a C-terminally hemagglutinin epitope (HA)-tagged version of the 40-kDa protein was analyzed by immunofluorescence (IF), using triple labeling with the DNA stain 4′,6-diamidino-2-phenylindol (DAPI), the monoclonal antibody YL1/2, and the anti-HA antibody. YL1/2 labels tyrosinated α-tubulin and in trypanosomes can be used as a marker for mature basal bodies (27). The results show that the tagged 40-kDa protein localizes between the basal body of the flagellum and the kDNA across the cell cycle in both the procyclic (Fig. 1A) and the bloodstream form of T. brucei (Fig. S1). The TAC resists extraction by nonionic detergents, which allows it to isolate flagella that are still attached to the kDNA (7). In Fig. 1B isolated flagella from the cell lines expressing the tagged 40-kDa protein were stained for the kDNA, for the HA tag, and for the main component of the paraflagellar rod (PFR), a complex structure that runs adjacent to the axoneme of the flagellum. The results show that the tagged 40-kDa protein localizes between the basal body and the kDNA in isolated flagella and therefore is a stable component of the TAC. We refer to it as TAC40 in the following.
Fig. 1.
TAC40 of procyclic cells localizes to the TAC. (A) Maximum-intensity projections of IF confocal microscopy images from whole procyclic T. brucei cells expressing C-terminally HA-tagged TAC40. Green, basal body region stained with the YL1/2 antibody; blue, DAPI-stained nuclear DNA and kDNA; red, HA-tagged TAC40. Three cell cycle stages containing one kDNA and one nucleus (1K1N, Top and Middle) or two kDNA and one nucleus (2K1N, Bottom) are shown. (Scale bar, 3 μm.) (B) The same as A but isolated flagella representing different cell cycle stages are shown (Left). Green, paraflagellar rod (PFR). (Scale bar, 3 μm.) (Right) three-dimensional volume reconstruction of the kDNA/TAC region from an isolated flagellum. (Scale bar, 1 μm.)
TAC40 Is a β-Barrel OM Protein.
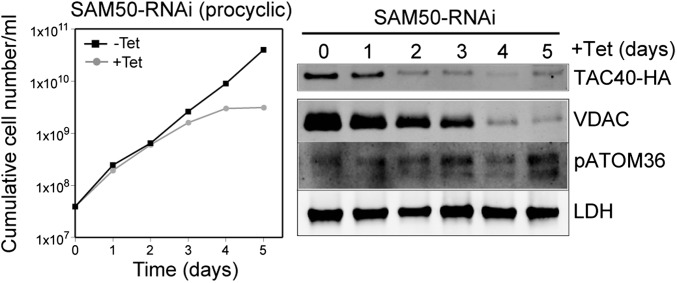
To verify that TAC40 is a mitochondrial β-barrel OM protein, we measured the steady-state levels of tagged TAC40 in a cell line ablated for 50 kDa subunit of the sorting and assembly machinery (SAM50) (Fig. 2) (28), the essential core component of the mitochondrial β-barrel protein insertion machinery, and we observed a rapid decrease of the protein level. The same effect is seen for the β-barrel protein VDAC but not for the abundant non–β-barrel OM membrane protein pATOM36 (28) and the matrix protein lipoamide dehydrogenase (LDH). We have previously shown that such a decrease indicates inhibition of protein import because mislocalized proteins get rapidly degraded (24). Thus, TAC40 is a mitochondrial OM protein with β-barrel structure that belongs to the mitochondrial porin superfamily. A BLAST search in the Uniprot database detected orthologs of TAC40 in essentially all trypanosomatids, but not in any other eukaryote. An alignment of TAC40 sequences with representative members of the VDAC, Tom40, and MDM10 families showed that these four proteins are about equally distant subfamilies of the mitochondrial porin superfamily (Fig. S2).
Fig. 2.
In vivo import-dependent accumulation of TAC40 depends on SAM50. (Left) Growth curve of uninduced (−Tet) and induced (+Tet) SAM50-RNAi cell line of procyclic T. brucei. (Right) Immunoblots of total cellular extracts from procyclic SAM50-RNAi cells that constitutively express HA-tagged TAC40 collected at the indicated times of induction. Immunoblots were probed for HA-tagged TAC40, the β-barrel protein VDAC, the non–β-barrel mitochondrial OM protein pATOM36, and LDH.
TAC40 Is Required for kDNA Maintenance.
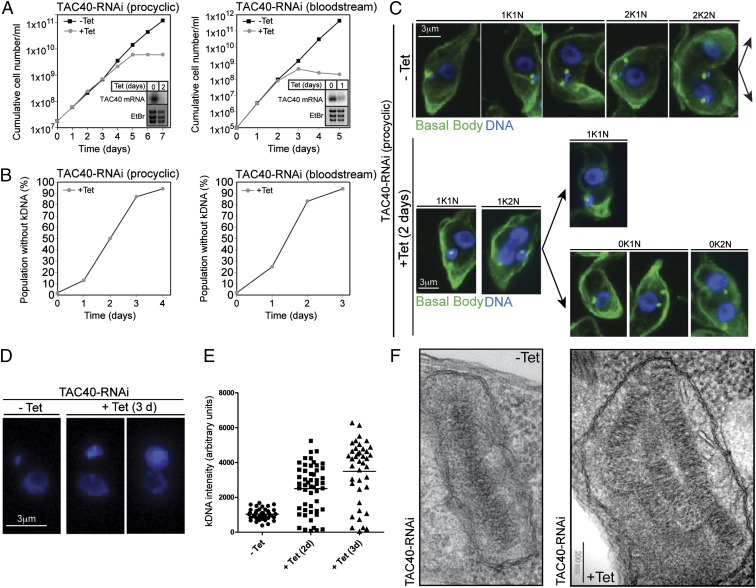
We set out to characterize cellular functions of TAC40. Inducible RNAi shows that TAC40 is essential for normal growth of procyclic and bloodstream forms of T. brucei (Fig. 3A). In the procyclic form analysis of DAPI-stained cells revealed a rapid loss of kDNA after induction of RNAi approaching 50% after 2 d (Fig. 3B), whereas the growth arrest is evident after 3–4 d only (Fig. 3A). In line with the shorter generation time the growth arrest and the loss of kDNA occur even earlier in bloodstream form cells (Fig. 3 A and B, Right). Hence, cells lacking kDNA networks are still able to divide at least once before growth ceases.
Fig. 3.
TAC40 is required for kDNA maintenance but not for kDNA replication. (A) Growth curve of uninduced (−Tet) and induced (+Tet) TAC40-RNAi cell lines of procyclic and bloodstream form T. brucei. (Insets) Northern blots demonstrating ablation of the TAC40 mRNA. Ethidium bromide-stained gel showing the rRNA region is used as a loading control. (B) Time course depicting the fraction of cells lacking kDNA during induction of TAC40-RNAi, as determined by microscopic examination of DAPI-stained cells. (C) IF images of uninduced (−Tet) and 2-d–induced (+Tet) TAC40-RNAi cells. Green, basal body region stained with the YL1/2 antibody; blue, DAPI-stained nuclear DNA and kDNA. For the uninduced population, five cell cycle stages consisting of cells containing one kDNA and one nucleus (1K1N), two kDNAs and one nucleus (2K1N), and two kDNAs and two nuclei (2K2N) are shown. The induced population in addition contains the abnormal cells containing no kDNA and one nucleus (0K1N) and cells lacking kDNA but having two nuclei (0K2N). (D) DAPI staining reveals large kDNA networks in the induced TAC40-RNAi cell line. (E) Quantification of the kDNA fluorescence intensity during induction of TAC40-RNAi in the kDNA-containing cell population. (F) Transmission electron micrographs showing the kDNA region of uninduced (−Tet) and 3-d–induced cells (+Tet).
A comparative IF analysis of different cell cycle stages of uninduced and induced procyclic RNAi cells (Fig. 3C) suggests that in the absence of TAC40 the newly formed basal bodies cannot form a complete TAC and therefore are unable to connect to the organellar DNA. Subsequent cell division results in two daughter cells: one in which the mitochondrion contains a highly enlarged kDNA network and another one that lacks kDNA altogether (Fig. 3 C–E). Electron micrographs show that the enlarged kDNA networks often consist of two or more kDNA discs with an undisturbed ultrastructure that are tightly packed (Fig. 3F). This suggests that the entire kDNA network is overreplicated in the induced RNAi cell line, indicating that TAC40 is required for segregation of kDNA networks but not for their replication. The kDNA segregation defect and the associated overreplication of the remaining kDNA are reminiscent of what has been observed in cells ablated for p166 and p197, two previously characterized TAC components (8, 10).
TAC40 Regulates Neither Mitochondrial Morphology nor Protein Import.
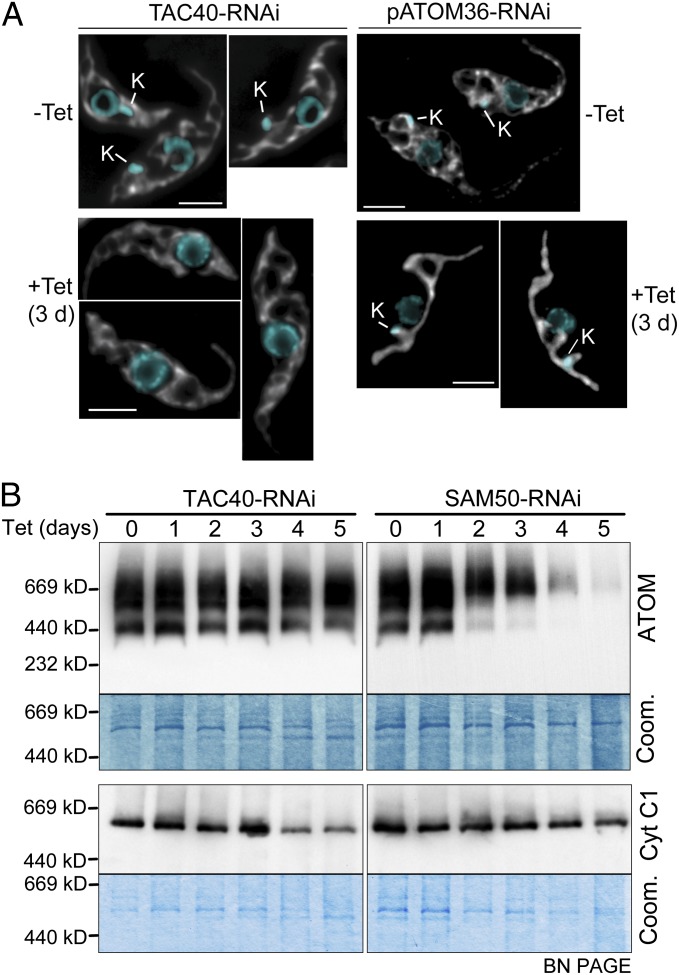
In yeast the lack of MDM10 leads to a dramatic collapse of mitochondrial morphology (15). Moreover, a fraction of the protein is associated with the SAM, where it mediates assembly of the TOM complex (21, 29). In T. brucei the shape of the mitochondrion shows marked differences between the procyclic and the bloodstream form. However, ablation of TAC40 does not affect mitochondrial morphology in either of the two forms as evidenced by IF using mitochondria-specific antisera (Fig. 4A and Fig. S3). Note that the successful ablation of TAC40 is demonstrated by the lack of kDNA in the induced cells. For comparison, RNAi cells for pATOM36 show an example of an OM protein whose ablation dramatically alters mitochondrial morphology (Fig. 4A, Right and Fig. S3). The blue native gel in Fig. 4B in which the ATOM complex was analyzed during induction of RNAi furthermore shows that TAC40 is not required for ATOM complex assembly. However, the bc1-respiratory complex that contains the mitochondrially encoded cytochrome b declines at late time points of induction, indicating the TAC40-induced loss of the kDNA. The SAM50 RNAi cell line serves as a positive control. It shows that due to lack of assembly of β-barrel proteins, the ATOM complex disappears, whereas the bc1 complex is only marginally affected at very late time points of induction.
Fig. 4.
TAC40 regulates neither mitochondrial morphology nor protein import. (A) Procyclic TAC40 and pATOM36 RNAi cell lines were tested for alterations of mitochondrial morphology. Maximum-intensity projections of IF confocal microscopy images of uninduced (−Tet) and induced (+Tet) cells stained with DAPI (blue) and with antibodies specific for the mitochondrial matrix protein HSP60 are shown. The position of the kDNA (K) is indicated. (Scale bars, 3 μm.) (B, Left) Duplicate immunoblots of digitonin-extracted crude mitochondrial fractions from the TAC40-RNAi cell line collected at the indicated times after induction of RNAi were separated on 4–13% blue native gradient gels (BN-PAGE) and probed with ATOM and cytochrome C1 (Cyt C1) antiserum detecting the ATOM and the bc1-complex, respectively. As a loading control a Coomassie-stained section of the same gels is shown. (Right) The same analysis as in Left, done for the SAM50 RNAi cell line.
Immunoprecipitations (IPs) using mitochondria containing tagged SAM50 combined with mass spectrometric analysis identified TAC40 as a SAM50-interacting protein (Fig. S4). Thus, reminiscent of MDM10 of yeast, a small fraction of TAC40 that might not be visible by IF is associated with SAM50 (29). TAC40 as a β-barrel protein requires SAM50 for assembly, and the small amount of TAC40 detected in SAM50 pull-down experiments therefore likely reflects transient binding during import and assembly.
TAC40 and Trypanosomal Orthologs of ERMES Components.
In fungi and animals mitochondria are physically linked to the ER. In fungi this interaction is mediated by the ERMES composed of the four proteins: MDM10, MDM12, MDM34, and MMM1 (17, 20). Like MDM10, all other ERMES components were thought to be specific for the fungal lineage. However, in a recent bioinformatic study putative ERMES orthologs were identified in a wide range of eukaryotes, including trypanosomatids (30). In T. brucei the ORFs Tb927.11.9230 and Tb927.8.4850 were identified as putative MDM12 and MDM34 orthologs, whereas an MMM1 ortholog and an MDM10 ortholog were not found. In the absence of an MDM10 ortholog, TAC40 could be assumed to be a binding partner of these putative trypanosomal MDM12 and MDM34 proteins. To test this idea, we produced cell lines allowing tetracycline-inducible expression of the two N- and C-terminally myc-tagged proteins. Subsequent experiments showed that all tagged versions of MDM12 and MDM34 could be extracted by low concentrations of digitonin (Fig. S5A) (24) and that in cell fractionations yielding enriched mitochondrial, ER, and cytosolic fractions MDM12 and MDM34 localize to the cytosol (Fig. S5B). These results are consistent with the IF analysis, which shows a cytosolic staining for both proteins (Fig. S5A). Tagged TAC40 in contrast specifically localizes to mitochondria in the same experiments. Moreover, analysis of cells shortly after tetracycline induction indicates that within approximately an order of magnitude the expression levels of the C-terminally tagged proteins do not influence the localization (Fig. S6). Furthermore, reciprocal IPs in cell lines expressing tagged MDM12 and MDM34 in combination with tagged TAC40 did not reveal interaction between the two proteins (Fig. S7). Finally, we produced inducible RNAi cell lines targeting MDM12 and MDM34, respectively. The results show that MDM12 but not MDM34 is essential for normal growth. Importantly, however, in contrast to the TAC40 RNAi cell line, there is no loss of the kDNA in either of the two cell lines (Fig. S8).
In summary, these results show that in agreement with the absence of an MDM10 ortholog in T. brucei (25) the putative MDM12 and MDM34 orthologs do not colocalize with the mitochondrion. These findings highlight the identification of TAC40 as an OM protein that is functionally distinct from MDM10.
kDNA Maintenance Is the Only Essential Function of TAC40.
TAC40 of trypanosomes is required for mitochondrial DNA maintenance but does not perform any of the other functions associated with yeast MDM10. Mitochondrial translation and thus the kDNA are essential for the survival of T. brucei throughout its life cycle (5, 31). To determine whether the growth arrest observed in induced TAC40-RNAi cells is linked to kDNA maintenance or to another essential process that has not explicitly been investigated in our study, we used an engineered T. brucei cell line, which as a bloodstream form is able to grow without kDNA due to a single compensatory mutation in the nuclear-encoded γ-subunit of the mitochondrial ATPase (31, 32). Fig. S9 shows that RNAi-mediated ablation of TAC40 in this cell line leads to a complete loss of kDNA without affecting growth. This firmly links the observed growth arrest caused by the absence of TAC40 to its role in kDNA maintenance and indicates that this is the only essential function of the protein.
Discussion
To date, five mitochondrial OM β-barrel proteins have been identified in the genome of T. brucei. These are the ortholog of Sam50, a member of the Omp85 family, and four β-barrel proteins from the general mitochondrial porin superfamily. One of these is the T. brucei VDAC, functioning as a metabolite transporter (23). A second one is the protein import pore ATOM, whose evolutionary origin is controversial, but shows at least some similarity to the mitochondrial porin family (24, 33, 34). Of the two remaining proteins, one is TAC40, analyzed in the present study.
Functional analysis of TAC40 does not allow grouping into any of the three mitochondrial porin subfamilies VDAC, TOM40, or MDM10. Bioinformatic analysis shows that the clade of TAC40 sequences is as distantly related to the other subfamilies as they are related to each other (Fig. S2). Instead, TAC40 is an integral part of the TAC that links the kDNA to the basal body of the flagellum and as such is essential for inheritance of the single-unit mitochondrial genome of T. brucei. The fact that it can be detected in the TAC of isolated flagella suggests that it plays a structural rather than a regulatory role in mitochondrial DNA maintenance. Based on these results, we conclude that TAC40 defines a novel as yet unknown subfamily of the mitochondrial porin superfamily.
Within this superfamily, TAC40 appears to be functionally more closely related to MDM10 than to the two other members in the sense that both proteins are required for cellular organization and mitochondrial DNA maintenance. However, unlike MDM10, ablation of TAC40 affects neither mitochondrial morphology nor protein import. Moreover, the highly specific localization of TAC40 suggests it is not involved in ER–mitochondria contact sites, because it is unlikely that such sites would be restricted to the TAC region only. Furthermore, whereas genes encoding orthologs of the ERMES components MDM12 and MDM34 have been found in the T. brucei genome (30), the corresponding proteins are cytosolic and unlike their counterparts in yeast are not required for mitochondrial DNA maintenance. Our results therefore also show that the presence of ERMES component orthologs in an organism does not necessarily indicate that they form an ERMES-like protein complex.
β-Barrel membrane proteins are exclusively found in the OMs of bacteria and bacteria-derived organelles (35). They were likely the only proteins present in the OM of the original endosymbiont from which all mitochondria derive. The fact that in such disparate eukaryotes as trypanosomatids and yeast a β-barrel protein is a core component of the mitochondrial DNA inheritance machinery therefore indicates that the mechanism for the process arose very early in evolution.
Despite the large evolutionary divergence of yeast and trypanosomatids (36) and despite the differences in the organization of their mitochondrial genomes, faithful mitochondrial DNA inheritance in both organisms requires a physical connection of the mitochondrial genomes with the cytoskeleton. The latter is tubulin based in T. brucei (7) and actin based in yeast (14). Thus, although the TAC in trypanosomes and the ERMES in yeast are very differently organized, they both contain a central β-barrel protein of the general mitochondrial porin superfamily (Fig. S10).
We can thus imagine two possible scenarios for the relation of MDM10 and TAC40. They may have evolved independently from precursors within the mitochondrial porin family. Alternatively, they may be descendants of a single common β-barrel protein precursor that, as part of a larger protein complex, was responsible for mitochondrial inheritance and organization before the divergence of yeast and trypanosomatids. In the latter scenario the postulated ancestral protein would have given rise to TAC40, which remained specialized in mitochondrial inheritance and to MDM10, which acquired additional functions such as tethering the ER to mitochondria.
In summary, our study shows that structural components of mitochondrial DNA inheritance machinery are more conserved than originally thought. Moreover, the single TAC connecting the single-unit kDNA to the single flagellum also provides an example that trypanosomal biology can reveal an extreme manifestation of a conserved biological concept.
Materials and Methods
Transgenic Cell Lines.
Transgenic procyclic cell lines are based on T. brucei 29-13 and were grown at 27 °C in SDM-79 supplemented with 10% (vol/vol) FCS. Transgenic bloodstream form cell lines are based on a single marker strain (37), in which one allele of the ATPase subunit gamma has been replaced by a mutant version of the protein in which L262 has been substituted by proline (32). Bloodstream form T. brucei were cultivated at 37 °C in HMI-9 medium containing 10% FCS. Transformation, cloning, and selection of transgenic cell lines were done as described in ref. 38. Procyclic RNAi cell lines for SAM50 have previously been described (39). RNAi of TAC40 was done by using a pLew-100–derived stem-loop construct containing the blasticidine resistance gene (40). As an insert, we used a 479-bp fragment (nucleotides 600–1,078) of the TAC40 ORF (Tb927.4.1610). To detect TAC40 in the 29-13 and the SAM50-RNAi cell lines one of its alleles was in situ tagged with a triple-HA tag as described in ref. 41. The putative MDM12 (Tb927.11.9230) and MDM34 (Tb927.8.4850) orthologs of T. brucei were N- and C-terminally tagged with triple–c-myc epitopes, using a pLew-100 derivative.
Immunofluorescence.
IF of procyclic 29-13 cell lines was essentially done as described in ref. 42. In short, cells were fixed in PBS containing 4% (wt/vol) paraformaldehyde for 10 min and subsequently permeabilized for 5 min with PBS containing 2% (wt/vol) Triton X-100. First and second antibodies were diluted in PBS and incubated for 1 h each at ambient temperature. Vectashield containing 1.5 μg/mL DAPI (Vector Laboratories) was used as a mounting medium. The following primary antisera were used: monoclonal anti-HA antibody HA11 produced in mouse (Covance Research Products) (dilution 1:1,000); monoclonal anti-tyrosinated tubulin antibody YL1/2 produced in rat (dilution 1:500), which stains the basal body region (27) (generous gift of K. Gull, Oxford University, Oxford); polyclonal anti-PFR antiserum produced in rat (dilution 1:1,000) (generous gift of T. Seebeck, University of Bern); and polyclonal anti-ATOM antiserum (dilution 1:1,000) produced in rabbits and a polyclonal anti–heat-shock protein 60 (Hsp60) antiserum produced in mouse (dilution 1:1,000) (generous gift of A. Hehl, University of Zurich, Zurich).
The images were acquired by conventional IF microscopy (Fig. 3 and Figs. S3, S5, and S6), using the Leica DMI6000 B microscope. FITC goat anti-rabbit (dilution 1:100) from Sigma, IRDye 680LT goat anti-mouse (dilution 1:100) from Li-Cor, and Alexa Fluor 488 goat anti-rabbit (dilution 1:750) and Alexa Fluor 594 goat anti-mouse (dilution 1:1,000) sera from Abcam were used as secondary antibodies. The images in Figs. 1 and 4 and Fig. S1 were acquired using a spinning-disk confocal microscope (Andromeda; Till photonics) equipped with a 488-nm diode laser (Toptica iBeam); an interline transfer CCD camera (Andor iXon) and a 60× oil immersion objective (NA 1.35; Olympus); or a deconvolution microscope (iMic digital microscope), equipped with a polychrome illumination system (Till Polychrome V), a 60× oil immersion objective (NA 1.35; Olympus), and a CCD camera (Hamamatsu ORCA-R2). The acquired images were processed using Imaris (Bitplane, version 7.6).
Relative Quantification of the Size of kDNA Networks.
Uninduced and induced TAC40-RNAi cell cultures were fixed, permeabilized, and mounted using Vectashield containing 1.5 μg/mL of DAPI as described above. Images were acquired using exposure times in which maximum fluorescence intensities did not lead to saturation. Using the AF6000 software, the kDNA regions were marked with circles. For each of the marked kDNAs three areas outside of but close to the kDNA region were also marked to determine the average background. The mean value from the three background regions was then subtracted from the integrated area of the marked kDNAs.
IPs and Mass Spectrometric Analysis.
Isotonically isolated mitochondria (43) containing C-terminally HA-tagged SAM50 were solubilized for 15 min on ice in 300 μL of lysis buffer and processed as described in ref. 28. Three biological replicates of the IP were performed. Results from the same IP analysis have previously been used as a negative control for the experiment shown in figure S4 of ref. 28.
Transmission Electron Microscopy.
Uninduced and induced TAC40-RNAi cells of T. brucei were fixed in culture for 1 h, using 2.5% glutaraldehyde or 2.5% glutaraldehyde in 0.15 M Hepes buffer. Samples were postfixed with 1% (wt/vol) osmium tetroxide, block stained with 1% (wt/vol) uranyl acetate, dehydrated in ascending concentrations of ethanol, and embedded in Epon812. Ultrathin sections were stained with uranyl acetate and lead citrate, using EMstain (Leica), and observed in a Philips CM12 electron microscope at 80 kV equipped with a Morada camera system, using iTEM software.
Sequence Homology Analysis.
For the sequence alignment, 9 Tom40, 10 MDM10, and 9 VDAC sequences from different organisms were selected from the Uniprot database. All annotated sequences from each family were clustered by similarity and one sequence was chosen per cluster. T. brucei VDAC (23) and T. brucei ATOM (24) were also included. Six TAC40 sequences with low relative redundancy from different trypanosomatids were identified by a BLAST search. All sequences were simultaneously aligned using Clustal Omega (44). The evolutionary tree was calculated based on Jukes–Cantor genetic distances (45) with the software Geneious.
Miscellaneous.
Isolation of flagella was done as described in ref. 46. For immunoblots the following primary antibodies were used: mouse monoclonal anti–EF-1a antibody (dilution 1:1,000) (Santa Cruz Biotechnology), rabbit polyclonal anti-VDAC antiserum (dilution 1:1,000), rabbit anti-LDH antiserum (dilution 1:1,000) (generous gift of L. Krauth-Siegel, University of Heidelberg, Heidelberg), and rabbit polyclonal ATOM antiserum (1:5,000). Northern blots and BN-PAGE were done as described in ref. 24.
Supplementary Material
Acknowledgments
We thank J. Wideman for communicating the accession numbers of trypanosomal MDM12 and MDM34 candidates to us. M.N. and A.H. gratefully acknowledge fellowships from the Peter und Traudl Engelhorn Foundation. Research in the groups of C.M. and B.W. was funded by grants from the Deutsche Forschungsgemeinschaft and the Excellence Initiative of the German Federal and State Governments (Exzellenzcluster 294 BIOSS Centre for Biological Signalling Studies). Research in the laboratory of A.S. was supported by Grant 138355 of the Swiss National Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404854111/-/DCSupplemental.
References
- 1.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4(9):a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott SV, Cassidy-Stone A, Meeusen SL, Nunnari J. Staying in aerobic shape: How the structural integrity of mitochondria and mitochondrial DNA is maintained. Curr Opin Cell Biol. 2003;15(4):482–488. doi: 10.1016/s0955-0674(03)00070-x. [DOI] [PubMed] [Google Scholar]
- 3.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6(11):815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 4.McKean PG. Coordination of cell cycle and cytokinesis in Trypanosoma brucei. Curr Opin Microbiol. 2003;6(6):600–607. doi: 10.1016/j.mib.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Cristodero M, Seebeck T, Schneider A. Mitochondrial translation is essential in bloodstream forms of Trypanosoma brucei. Mol Microbiol. 2010;78(3):757–769. doi: 10.1111/j.1365-2958.2010.07368.x. [DOI] [PubMed] [Google Scholar]
- 6.Jensen RE, Englund PT. Network news: The replication of kinetoplast DNA. Annu Rev Microbiol. 2012;66:473–491. doi: 10.1146/annurev-micro-092611-150057. [DOI] [PubMed] [Google Scholar]
- 7.Ogbadoyi EO, Robinson DR, Gull K. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol Biol Cell. 2003;14(5):1769–1779. doi: 10.1091/mbc.E02-08-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Z, Lindsay ME, Roy Chowdhury A, Robinson DR, Englund PT. p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation. EMBO J. 2008;27(1):143–154. doi: 10.1038/sj.emboj.7601956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochsenreiter T, Anderson S, Wood ZA, Hajduk SL. Alternative RNA editing produces a novel protein involved in mitochondrial DNA maintenance in trypanosomes. Mol Cell Biol. 2008;28(18):5595–5604. doi: 10.1128/MCB.00637-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gheiratmand L, Brasseur A, Zhou Q, He CY. Biochemical characterization of the bi-lobe reveals a continuous structural network linking the bi-lobe to other single-copied organelles in Trypanosoma brucei. J Biol Chem. 2013;288(5):3489–3499. doi: 10.1074/jbc.M112.417428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson DR, Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 1991;352(6337):731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- 12.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11(12):872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 13.Shaw JM, Nunnari J. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 2002;12(4):178–184. doi: 10.1016/s0962-8924(01)02246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boldogh IR, et al. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell. 2003;14(11):4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126(6):1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Youngman MJ, Hobbs AE, Burgess SM, Srinivasan M, Jensen RE. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J Cell Biol. 2004;164(5):677–688. doi: 10.1083/jcb.200308012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325(5939):477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meeusen S, Nunnari J. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J Cell Biol. 2003;163(3):503–510. doi: 10.1083/jcb.200304040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimmer KS, Jakobs S, Vogel F, Altmann K, Westermann B. Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast. J Cell Biol. 2005;168(1):103–115. doi: 10.1083/jcb.200410030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornmann B, Walter P. ERMES-mediated ER-mitochondria contacts: Molecular hubs for the regulation of mitochondrial biology. J Cell Sci. 2010;123(Pt 9):1389–1393. doi: 10.1242/jcs.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wideman JG, et al. Roles of the Mdm10, Tom7, Mdm12, and Mmm1 proteins in the assembly of mitochondrial outer membrane proteins in Neurospora crassa. Mol Biol Cell. 2010;21(10):1725–1736. doi: 10.1091/mbc.E09-10-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen TT, et al. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic. 2012;13(6):880–890. doi: 10.1111/j.1600-0854.2012.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pusnik M, et al. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol Biol Evol. 2009;26(3):671–680. doi: 10.1093/molbev/msn288. [DOI] [PubMed] [Google Scholar]
- 24.Pusnik M, et al. Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr Biol. 2011;21(20):1738–1743. doi: 10.1016/j.cub.2011.08.060. [DOI] [PubMed] [Google Scholar]
- 25.Niemann M, et al. Mitochondrial outer membrane proteome of Trypanosoma brucei reveals novel factors required to maintain mitochondrial morphology. Mol Cell Proteomics. 2013;12(2):515–528. doi: 10.1074/mcp.M112.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flinner N, Schleiff E, Mirus O. Identification of two voltage-dependent anion channel-like protein sequences conserved in Kinetoplastida. Biol Lett. 2012;8(3):446–449. doi: 10.1098/rsbl.2011.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephan A, Vaughan S, Shaw MK, Gull K, McKean PG. An essential quality control mechanism at the eukaryotic basal body prior to intraflagellar transport. Traffic. 2007;8(10):1323–1330. doi: 10.1111/j.1600-0854.2007.00611.x. [DOI] [PubMed] [Google Scholar]
- 28.Pusnik M, et al. An essential novel component of the noncanonical mitochondrial outer membrane protein import system of trypanosomatids. Mol Biol Cell. 2012;23(17):3420–3428. doi: 10.1091/mbc.E12-02-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meisinger C, et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell. 2004;7(1):61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 30.Wideman JG, Gawryluk RM, Gray MW, Dacks JB. The ancient and widespread nature of the ER-mitochondria encounter structure. Mol Biol Evol. 2013;30(9):2044–2049. doi: 10.1093/molbev/mst120. [DOI] [PubMed] [Google Scholar]
- 31.Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24(23):4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dean S, Gould MK, Dewar CE, Schnaufer AC. Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc Natl Acad Sci USA. 2013;110(36):14741–14746. doi: 10.1073/pnas.1305404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zarsky V, Tachezy J, Dolezal P. Tom40 is likely common to all mitochondria. Curr Biol. 2012;22(12):R479–R481, author reply R481–R482. doi: 10.1016/j.cub.2012.03.057. [DOI] [PubMed] [Google Scholar]
- 34.Harsman A, et al. Bacterial origin of a mitochondrial outer membrane protein translocase: New perspectives from comparative single channel electrophysiology. J Biol Chem. 2012;287(37):31437–31445. doi: 10.1074/jbc.M112.392118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bay DC, Hafez M, Young MJ, Court DA. Phylogenetic and coevolutionary analysis of the β-barrel protein family comprised of mitochondrial porin (VDAC) and Tom40. Biochim Biophys Acta. 2012;1818(6):1502–1519. doi: 10.1016/j.bbamem.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 36.Dacks JB, Walker G, Field MC. Implications of the new eukaryotic systematics for parasitologists. Parasitol Int. 2008;57(2):97–104. doi: 10.1016/j.parint.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99(1):89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 38.McCulloch R, Vassella E, Burton P, Boshart M, Barry JD. Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol Biol. 2004;262:53–86. doi: 10.1385/1-59259-761-0:053. [DOI] [PubMed] [Google Scholar]
- 39.Tschopp F, Charrière F, Schneider A. In vivo study in Trypanosoma brucei links mitochondrial transfer RNA import to mitochondrial protein import. EMBO Rep. 2011;12(8):825–832. doi: 10.1038/embor.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bochud-Allemann N, Schneider A. Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J Biol Chem. 2002;277(36):32849–32854. doi: 10.1074/jbc.M205776200. [DOI] [PubMed] [Google Scholar]
- 41.Oberholzer M, Morand S, Kunz S, Seebeck T. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol Biochem Parasitol. 2006;145(1):117–120. doi: 10.1016/j.molbiopara.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Schneider A, et al. Subpellicular and flagellar microtubules of Trypanosoma brucei brucei contain the same alpha-tubulin isoforms. J Cell Biol. 1987;104(3):431–438. doi: 10.1083/jcb.104.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider A, Charrière F, Pusnik M, Horn EK. Isolation of mitochondria from procyclic Trypanosoma brucei. Methods Mol Biol. 2007;372:67–80. doi: 10.1007/978-1-59745-365-3_5. [DOI] [PubMed] [Google Scholar]
- 44.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jukes TH, Cantor CR. Evolution of Protein Molecules. New York: Academic; 1969. [Google Scholar]
- 46.Sasse R, Gull K. Tubulin post-translational modifications and the construction of microtubular organelles in Trypanosoma brucei. J Cell Sci. 1988;90(Pt 4):577–589. doi: 10.1242/jcs.90.4.577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.