Significance
Different molecules act in sensing cytosolic nucleic acids derived from viruses, depending on the cell type and the virus. Epithelial cells and fibroblasts recognize viral invasion through cytosolic nucleic acid sensors and initiate antiviral immune responses by secreting cytokines. Retinoic acid-inducible gene 1 (RIG-I), a member of the DNA/RNA helicase family, plays a significant role as such a sensor. We identified DEAH (Asp-Glu-Ala-His) box polypeptide 29 (DHX29), another member of the family, as a unique cytosolic nucleic acid cosensor in human airway epithelial cells and fibroblasts. DHX29 directly bound nucleic acids, interacted with RIG-I, and triggered downstream signaling. DHX29 may be the optimal target for drug and vaccine design to control viral infections and viral-induced pathology in the airway.
Keywords: cytosolic sensor, airway infection
Abstract
The recognition of cytoplasmic nucleic acid is critical for innate immune responses against microbial infection and is responsible for autoimmunity induced by dead cells. Here, we report the identification of a unique cytosolic nucleic acid cosensor in human airway epithelial cells and fibroblasts: DEAH (Asp-Glu-Ala-His) box polypeptide 29 (DHX29), a member of the DExD/H (Asp-Glu-x-Asp/His)-box helicase family. Knocking down DHX29 by siRNA attenuated the ability of cells to mount type I IFN and IL-6 in response to cytosolic nucleic acids and various viruses by blocking the activation of interferon regulatory factor 3 and NF-κB-p65. The cytosolic nucleic acid sensing by DHX29 in human epithelial cells and fibroblasts is independent of stimulator of interferon genes but is dependent on retinoic acid-inducible gene 1 (RIG-I) and mitochondrial antiviral signaling protein (MAVS). DHX29 binds directly to nucleic acids and interacts with RIG-I and MAVS through its helicase 1 domain, activating the RIG-I–MAVS-dependent cytosolic nucleic acid response. These results suggest that DHX29 is a cytosolic nucleic acid cosensor that triggers RIG-I/MAVS-dependent signaling pathways. This study will have important implications in drug and vaccine design for control of viral infections and viral-induced pathology in the airway.
Human airway epithelia interface with the outside air environment. Viruses, bacteria, and other airborne microorganisms frequently cause mild to serious infections in humans, which may cause or exacerbate many human diseases, including pneumonia, asthma, and chronic obstructive pulmonary disease (1). Recent studies demonstrated that in addition to providing a physical barrier, epithelia can sense viral infection. This ability is critical to subsequent activation of antiviral innate and adaptive immunity (2–8).
In the past decade, various cytosolic nucleic acid sensors and their mechanisms of action have been uncovered. Cytosolic double-stranded RNA (dsRNA) polyinosinic:polycytidylic acid (poly I:C) and 5′-ppp-dsRNA, mimicking the virus-derived RNA, are sensed by RIG-I–like receptors (9–11), which signal through the adaptor protein mitochondrial antiviral signaling protein (MAVS) (also known as CARDIF, IPS-1, or VISA) (12–15). For double-stranded DNA (dsDNA), many sensors have been reported, including DAI, AIM2, RNA pol III, IFI16, DDX41, Mre11, DNA-PKcs/Ku70/Ku80, cGAS, LRRFIP1, HMGB1, LSm14A, and NLRP3 (16, 17). Some of these molecules depend on stimulator of interferon genes (STING) (also known as ERIS, MITA, or MYPS) as the signaling adaptor (18–22). Furthermore, in addition to RIG-I–like receptors, many other DExD/H helicase family members, such as DHX9, DHX36, DDX1, DDX21, DHX33, DDX3, and DDX60, have been reported to function in nucleic acid sensing (23–29). These reports suggest that, depending on the virus species, cell type, types of ligands, types of responses, and the response phase, cytosolic nucleic acids are sensed by various sensing molecules that lead to different downstream signaling (16, 17). Different cytosolic nucleic acid sensors also have been implicated in the sensing of viral infection in human airway epithelial and subepithelial cells (2–7).
In this study, we systemically analyzed the function of 59 members of the DExD/H (Asp-Glu-x-Asp/His) helicase family in sensing nucleic acids in human airway-derived epithelial cells and fibroblasts. We report that in the human airway system, DHX29 is engaged in cytosolic nucleic acid and virus sensing as a cosensor of the RIG-I/MAVS pathway, independently of STING.
Results
Human Epithelial Cells and Fibroblasts Sense Cytosolic DNA and RNA.
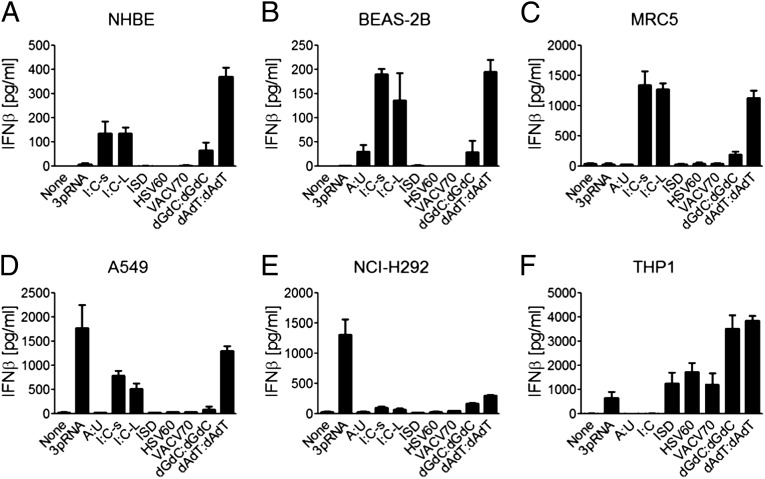
To obtain general insights in cytosolic RNA and DNA sensing by airway-derived cells, various known cytosolic nucleic acid ligands were transfected to various cells originating from epithelial or subepithelial tissue. Results from normal human bronchial/tracheal epithelial cells, transformed primary airway epithelial cells (BEAS-2B), two airway epithelial cell lines (A549 and NCI-H292), airway-derived primary fibroblast MRC5 cells, and THP-1 cell line-derived macrophages are shown in Fig. 1. The following interesting observations were obtained: (i) all epithelial cells and fibroblasts showed IFN-β responses to poly I:C and poly(deoxyadenylic-thymidylic):poly(deoxyadenylic-thymidylic) acid (poly dAdT:dAdT); (ii) only A549 and NCI-H292 cells made robust IFN-β responses to 5′-ppp-dsRNA (3pRNA), suggesting that although RIG-I is required for sensing both 3pRNA and short poly I:C and poly dAdT:dAdT, RIG-I may depend on different components to sense 3pRNA than to sense short poly I:C and poly dAdT:dAdT; and (iii) only THP1-derived macrophages but not airway-derived cells showed significant response to immunostimulatory DNA, HSV-60mer dsDNA, and VACV-70mer dsDNA, suggesting that the airway epithelial cells and fibroblasts have a defect in the STING-dependent sensing pathway (18, 21, 22, 30). These results support the concept that different cells may use different cytosolic nucleic acid sensors and the existence of a complex cytosolic nucleic acid-sensing system that cannot be fully explained by the current knowledge of nucleic acid sensing.
Fig. 1.
Human airway epithelial cells and fibroblasts sense cytosolic DNA and RNA. (A) Normal human bronchial/tracheal epithelial (NHBE) cells, (B) BEAS-2B cells, (C) primary airway-derived fibroblast MRC5 cells, (D and E) two airway epithelial cell lines (A549 and NCI-H292), and (F) THP1 cell line-derived macrophages were transfected with 10 µg/mL of various nucleic acids by using Lipofectamine 2000: 3pRNA, poly A:U (A:U), conventional poly I:C (I:C), short poly I:C (I:C-s), long poly I:C (I:C-L), immunostimulatory dsDNA (ISD), HSV-60mer dsDNA (HSV60), VACV 60mer dsDNA (VACV70), poly dGdC:dGdC, and poly dAdT:dAdT. Culture supernatants were collected 18 h later to measure IFN-β by ELISA. Values are mean ± SD of at least three independent experiments.
DHX29 Is Critical for Virus and Cytosolic Nucleic Acid Response in Human Airway Epithelial Cells and Fibroblasts.
RIG-I–like receptors, including RIG-I, MDA-5, and LGP-2, constitute a small subfamily of the DExD/H helicase superfamily (23). Previous studies showed that the DExD/H helicase superfamily, which contains at least 59 members, plays a much broader role in sensing nucleic acids. DHX9, DHX36, DDX1, DDX21, DDX41, DHX33, DDX3, and DDX60 were found to play important roles in nucleic acid sensing in various human and mouse cells (22–29).
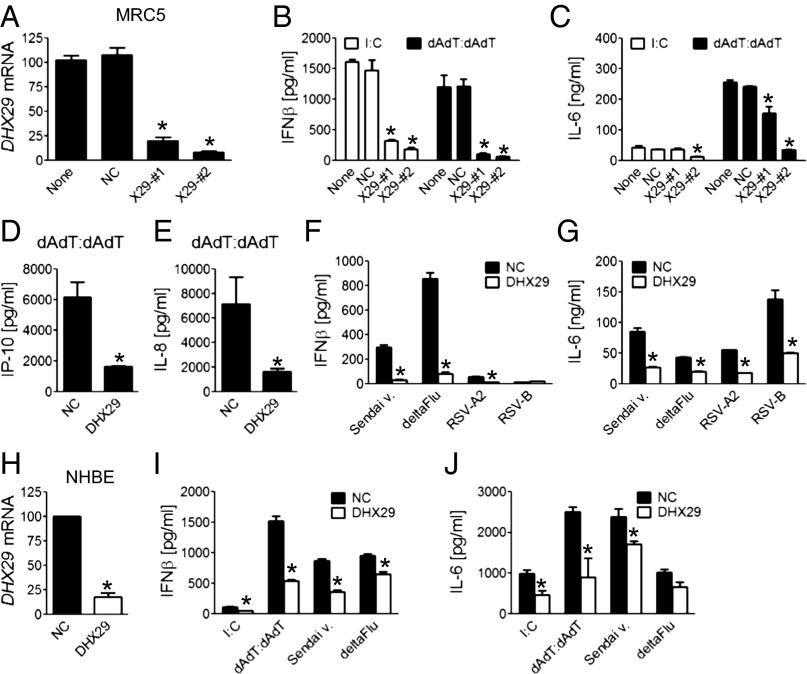
To obtain knowledge of nucleic acid sensing in human airway epithelial cells and fibroblasts, we systemically screened the function of 59 helicases in MRC5 cells and BEAS-2B cells in response to poly I:C or poly dAdT:dAdT using siRNA. The outcome was assessed by production of IFN-β and IL-6. We found that the knockdown of DHX29 or RIG-I had the most dramatic and consistent inhibitory effect on the IFN-β and IL-6 production induced by poly I:C or by poly dAdT:dAdT. Subsequently, the knockdown effect of DHX29 was confirmed using four individual sequence siRNAs for DHX29. Two different sequence siRNAs with more than 75% knockdown efficiency were able to reproduce the screening results with their effects correlating with the knockdown efficiency (Fig. 2 A–C). The production of IP-10 and IL-8 also was significantly reduced by the knockdown of DHX29 (Fig. 2 D and E). Similar results were obtained with airway epithelial cell lines BEAS-2B and A549 (Fig. S1 A–D). Cytokine production induced by the live virus was measured as well, showing a similarly marked reduction upon the knockdown of DHX29 (Fig. 2 F and G). Finally, the effect of knocking down DHX 29 expression was assessed in normal human bronchial/tracheal epithelial cells. Although the knockdown efficiency was not as good as in MRC5 cells, siRNA treatment of DHX29 resulted in a significant decrease in DHX29 expression (Fig. 2H) and in reduced production of IFN-β and IL-6 upon cytosolic nucleic acid and virus stimulation (Fig. 2 I and J). These results strongly support that DHX29 may represent a critical molecule in cytosolic nucleic acid sensing of human airway epithelial cells and fibroblasts.
Fig. 2.
DHX29 is critical for virus and cytosolic nucleic acid response in human airway epithelial cells and fibroblasts. MRC5 cells were treated with one of three siRNAs [nonsilencing negative control (NC), DHX29-#1, or DHX29-#2] or none (A–C). Two siRNAs (NC and DHX29-#2) were used in D–G by using Lipofectamine RNAiMax. Forty hours later, cells were either collected and lysed for quantitative RT-PCR (qPCR) to measure knockdown levels (A) or stimulated with poly I:C (B and C), poly dAdT:dAdT (B–E), or Sendai virus (Sendai v.), NS1 region-deleted influenza virus (deltaFlu), respiratory syncytial virus type A2 (RSV-A2), or RSV type B (RSV-B) (F and G). NHBE cells were treated as above with one of two siRNAs (NC or DHX29-#2) and were collected for qPCR (H) or stimulated with poly dAdT:dAdT, poly I:C, Sendai v., or deltaFlu (I and J). Eighteen hours after stimulation, cytokine concentrations in supernatants were measured by ELISA. Values are mean ± SD of at least three independent experiments (*P < 0.05 vs. NC).
DHX29 Is Specifically Expressed by Epithelial Cells and Fibroblasts in a Constitutive Fashion.
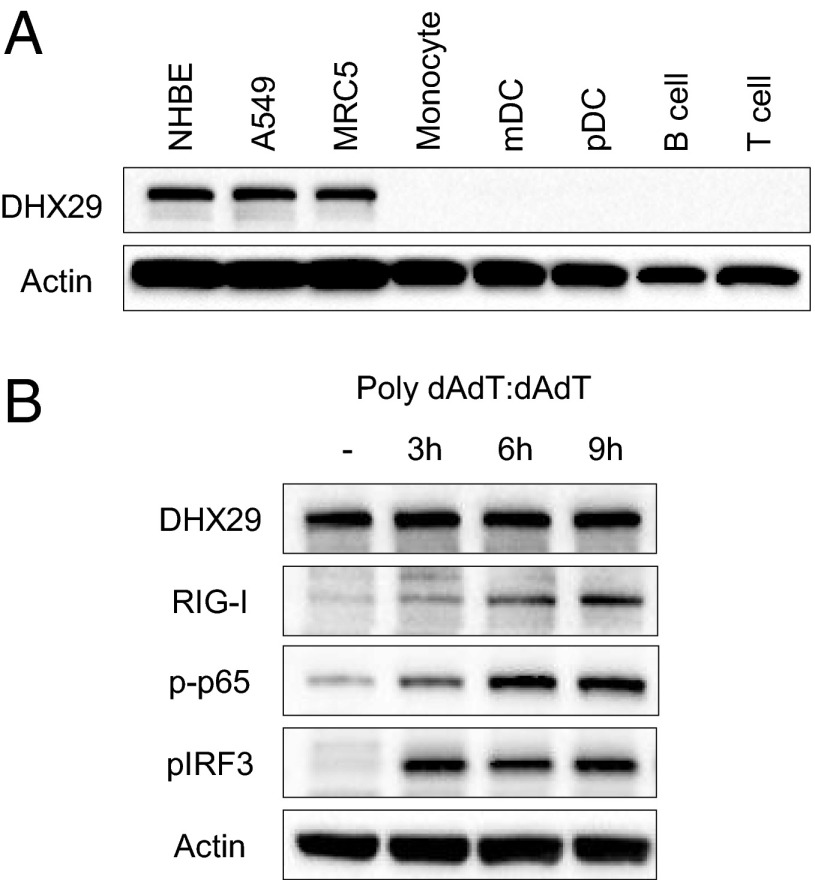
To assess the specificity of DHX29 expression in different cell types of the airway, DHX29 protein expression in airway-derived epithelial cells and fibroblasts was compared with that in peripheral blood-derived mononuclear cell (PBMC)-derived monocytes, myeloid dendritic cells, plasmacytoid dendritic cells, B cells, and T cells by Western blotting (Fig. 3A). The result showed dramatically specific expression of DHX29 in airway-derived cells but not in other cell subsets. Thus, DHX29 seems to be the sensing molecule that is specific to epithelial cells and fibroblasts. As for whether DHX29 expression is constitutive, DHX29 did not show any difference upon poly I:C or poly dAdT:dAdT stimulation (Figs. 3B and 4).
Fig. 3.
DHX29 is specifically expressed by epithelial cells and fibroblasts in a constitutive fashion. (A) Normal human bronchial/tracheal epithelial (NHBE) cells, A549 cells, MRC5 cells, monocytes, myeloid dendritic cells (mDC), plasmacytoid dendritic cells (pDC), B cells, and T cells were lysed, and 30,000 cell equivalents were run on an SDS/PAGE gel. Western blot analyses were done using antibodies against actin and DHX29. (B) A549 cells were stimulated with poly dAdT:dAdT for 3, 6, or 9 h. They then were lysed and run on an SDS/PAGE gel. Western blot analyses were done using antibodies against actin, DHX29, RIG-I, phosphorylated NF-κB-p65 (p-p65), and phosphorylated IRF3 (pIRF3).
Fig. 4.
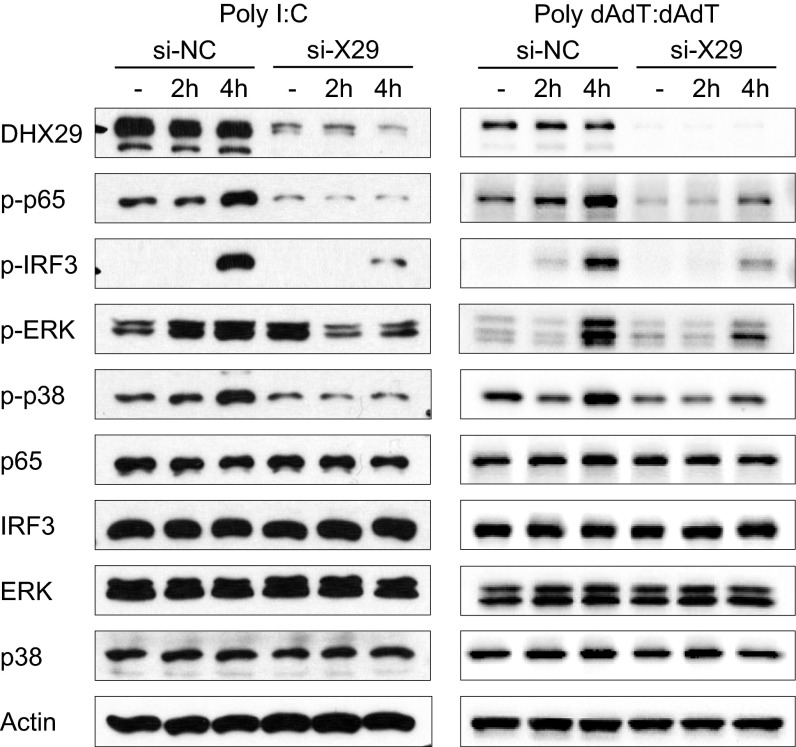
Knockdown of DHX29 results in impaired poly I:C- and poly dAdT:dAdT-induced signaling. MRC5 cells were transfected with nonsilencing negative control siRNA (si-NC) or DHX29 siRNA #2 (si-X29). Forty hours later, the cells were stimulated with poly I:C or poly dAdT:dAdT. Two or four hours following stimulation, the cells were collected, lysed, and run on SDS/PAGE gel. Western blot analyses were done using antibodies against actin, DHX29, and both the whole and phosphorylated forms of NF-κB-p65, IRF3, ERK, and p38-MAPK. Data are representative of at least three independent experiments.
Knockdown of DHX29 Results in Impaired Poly I:C- and Poly dAdT:dAdT-Induced Signaling.
To confirm the role of DHX29 in sensing nucleic acids in human epithelial cells and fibroblasts, poly I:C- or poly dAdT:dAdT-induced cell signaling was analyzed. MRC5 cells with DHX29 or nontarget control siRNA treatment were stimulated with cytosolic poly I:C or poly dAdT:dAdT. The cells then were collected, lysed, and applied to SDS/PAGE for Western blotting. As shown in Fig. 4, the phosphorylation of NF-κB-p65 and interferon regulatory factor 3 (IRF3) was reduced dramatically, and the phosphorylation of MAPK ERK and p38 was down-regulated moderately. The roles of both IRF3 and NF-κB-p65 in IFN-β production and NF-κB-p65 in IL-6 production were confirmed by knockdown experiments. On the other hand, ERK and p38 MAPK did not show any correlation with IFN-β and IL-6 production (Fig. S2). These results suggest that DHX29 may represent an upstream element of a nucleic acid-sensing pathway.
DHX29 Directly Binds to Poly I:C and Poly dAdT:dAdT.
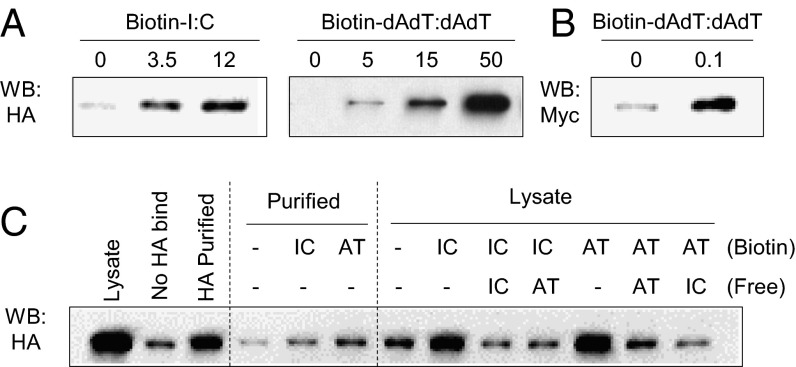
To investigate whether DHX29 plays a role as a sensor, binding of DHX29 to poly I:C and poly dAdT:dAdT was tested. HA-tagged DHX29 (HA-DHX29) overexpressed cell lysates were incubated with biotinylated nucleic acids. The pull-down of nucleic acids by using NeutrAvidin beads resulted in coprecipitation of DHX29, depending on the dose of poly I:C and poly dAdT:dAdT (Fig. 5A). This was confirmed with highly purified commercial recombinant Myc/DDK-DHX29 (Fig. 5B) and with HA column-purified HA-DHX29 (Fig. 5C). To clarify the specificity of these bindings, competition assays were done using nonlabeled free poly I:C and poly dAdT:dAdT. The results showed that both nonlabeled nucleic acids could complete the binding of either nucleic acid (Fig. 5C). Thus, DHX29 can bind both poly I:C and poly dAdT:dAdT and may function as a nucleic acid sensor.
Fig. 5.
DHX29 directly binds poly I:C and poly dAdT:dAdT. (A) 293T cells were overexpressed with HA-tagged DHX29 (HA-DHX29) and lysed. Then, the cell lysates were mixed with biotinylated poly I:C or poly dAdT:dAdT at various concentrations. After 2 h of incubation, NeutrAvidin beads were added to the mixture and incubated further. After another 2 h, the beads were washed thoroughly, and the proteins bound to the beads were eluted. The eluates were subjected to SDS/PAGE and Western blot analysis with anti-HA antibody. (B) Purified recombinant Myc-DDK–tagged DHX29 protein (100 ng) was mixed with 100 ng of biotinylated poly dAdT:dAdT and was subjected to the same procedures as in A. (C) HA-DHX29–overexpressed 293T cell lysates or HA-purified DHX29 were mixed with biotinylated poly I:C or poly dAdT:dAdT with or without nonbiotinylated free poly I:C or poly dAdT:dAdT, and were subjected to the same procedure as in A. Data are representative of at least three independent experiments.
DHX29 Interacts with RIG-I and MAVS.
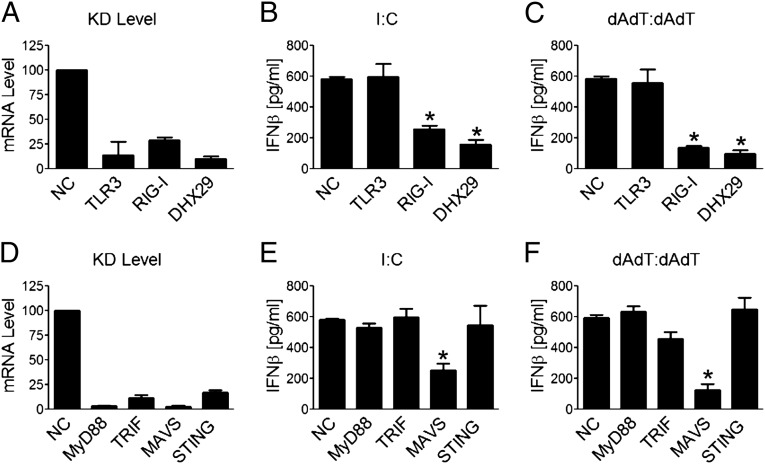
To assess which sensing and adaptor molecules are participating in sensing cytosolic nucleic acids in human epithelial cells and fibroblasts, the knockdown of a group of these molecules was carried out. For the sensing molecules, knockdown of RIG-I but not TLR3 resulted in a marked decrease in IFN-β and IL-6 responses (Fig. 6 A–C). For the adaptor molecules, knockdown of MAVS but not MyD88, TRIF, and STING resulted in a marked decrease in IFN-β and IL-6 responses (Fig. 6 D–F). These results show that the RIG-I/MAVS pathway mainly was involved in poly I:C and poly dAdT:dAdT responses in MRC5 cells.
Fig. 6.
RIG-I and MAVS are the indispensable molecules for cytosolic nucleic acid responses in MRC5 cells. Cells were transfected with siRNA as labeled. Forty hours later, cells were either collected and lysed for qPCR to check the knockdown (KD) levels (A and D) or stimulated with poly I:C (B and E) or with poly dAdT:dAdT (C and F). Eighteen hours following stimulation, culture supernatant IFN-β concentrations were measured by ELISA. Values are mean ± SD of at least three independent experiments (*P < 0.05 vs. NC).
To investigate whether DHX29 interacts with RIG-I and MAVS, endogenous coimmunoprecipitation (Co-IP) of these proteins was done. MRC5 cells with or without treatment with poly I:C or poly dAdT:dAdT were lysed and immunoprecipitated with anti-DHX29 antibody. Anti-DHX29 antibody could pull down both RIG-I and MAVS together with DHX29, especially after stimulation (Fig. 7A, Left). Co-IP with RIG-I resulted in pull down of DHX29 protein as well, which was not affected by RNase or DNase treatment (Fig. 7A, Center). The interaction of DHX29 and RIG-I also was confirmed with Co-IP by using purified proteins with or without poly I:C (Fig. 7A, Right). Furthermore, Co-IP of overexpressed HA-DHX29 and Myc-RIG-I in 293T cell lysates was performed using anti-HA agarose beads. Although full-length RIG-I showed very weak binding with DHX29, the C-terminal deletion mutant representing the active form of RIG-I (RIG-ΔC) (31) showed clear binding with DHX29 (Fig. 7B, Left). These results imply that although DHX29 can bind to RIG-I directly, it can bind strongly only when RIG-I goes through the conformational change induced by nucleic acid binding. Next, to visualize the localization of DHX29 with RIG-I, A549 cells were stimulated with poly dAdT:dAdT and observed by confocal microscopy. The endogenous staining of RIG-I and DHX29 showed that RIG-I was up-regulated strongly upon stimulation, and it showed clear colocalization or juxtaposition with DHX29 (Fig. 7E). Taken together, these data strongly suggest that DHX29 acts as a cosensor of cytosolic nucleic acids for RIG-I.
Fig. 7.
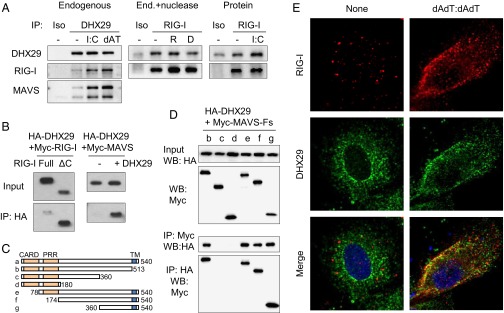
DHX29 interacts with RIG-I and MAVS. (A, Left) MRC5 cells stimulated with or without poly I:C or poly dAdT:dAdT (dAT) were lysed and mixed with anti-DHX29 antibody or with an isotype control and then incubated with Protein G agarose. Immunoprecipitated proteins were detected by Western blotting using anti-DHX29, anti–RIG-I, and anti-MAVS antibodies. (Center) Poly I:C-stimulated MRC5 cell lysates were treated with or without RNase (R) or DNase (D) before immunoprecipitation with anti–RIG-I antibody or with an isotype control. (Right) Highly purified DHX29 and RIG-I proteins with or without poly I:C were immunoprecipitated with anti-RIG-I antibody or with an isotype control. (B) Cell lysates from 293T cells overexpressed with Myc-tagged RIG-I (Myc–RIG-I; full-length or C-terminal deleted form) and HA-tagged DHX29 (Left), or with Myc-tagged MAVS (Myc-MAVS) with or without HA-DHX29 (Right), were incubated with anti-HA agarose beads. After 2 h, the beads were washed thoroughly and their binding proteins were eluted. The inputs and immunoprecipitated elutes (IP) were detected by Western blotting analysis using anti-Myc antibody. (C and D) Cell lysates from 293T cells overexpressed with HA-DHX29 together with various deletion mutants of Myc-MAVS (Myc-MAVS-Fs) were incubated with anti-HA or anti-Myc agarose beads. After 2 h, the beads were subjected to the same procedure as in B and were detected using anti-HA or anti-Myc antibodies. Data are representative of at least three independent experiments. (E) A549 cells were treated with or without poly dAdT:dAdT stimulation. After 6 h, the cells were fixed and stained with anti–RIG-I and anti-DHX29 antibodies and were observed with confocal microscopy. Red, RIG-I; green, DHX29; blue, DAPI.
As for MAVS, similar experiments were done using cell lysates from 293T cells overexpressed with Myc-MAVS with or without HA-DHX29. Immunoprecipitation with anti-HA beads showed strong Co-IP of MAVS only in the presence of DHX29 (Fig. 7B, Right). Then, MAVS domain mapping was done using various MAVS deletion mutation constructs (Fig. 7C). The Co-IP of HA-DHX29 (full) and Myc-MAVS (various fragments) defined a region of MAVS consisting of amino acids 360–510 as the site of interaction with DHX29. This was confirmed with Co-IP of both anti-Myc and anti-HA beads (Fig. 7D).
DHX29 Binds to Nucleic Acids, RIG-I, and MAVS via the Helicase 1 Domain.
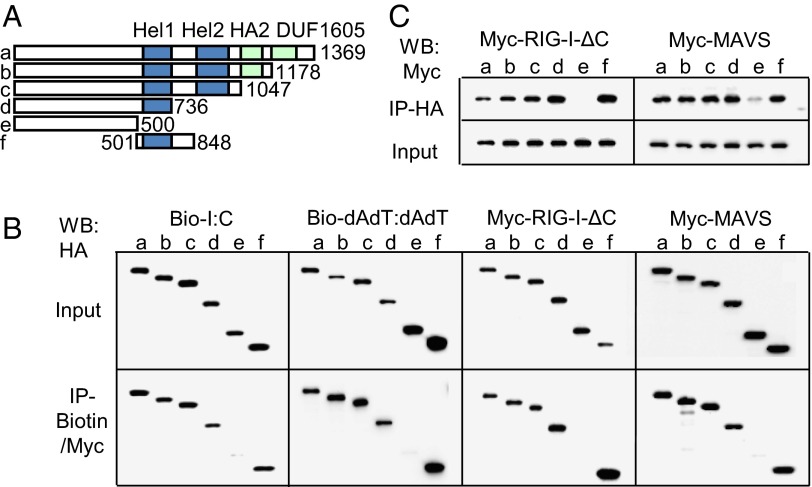
To understand the molecular interaction between DHX29 and nucleic acids, as well as DHX29 with RIG-I and MAVS, serial deletion and binding experiments with DHX29 were performed (Fig. 8A). First, each of the HA-DHX29 mutants was overexpressed in 293T cells. The lysates were mixed with biotinylated poly I:C or poly dAdT:dAdT and subsequently pulled down with NeutrAvidin beads. The results showed the defect of HA-DHX29 pull-down only when the helicase 1 domain was absent (Fig. 8B). As for RIG-I and MAVS, each HA-DHX29 mutant was co-overexpressed with Myc-RIG-I-dC or Myc-MAVS in 293T cells. Then, the cell lysates were subjected to Co-IP with anti-HA beads or anti-Myc beads. The Co-IP suggested that the helicase 1 domain of DHX29 was indispensable for binding with RIG-I and MAVS, and it was confirmed with both kinds of beads (Fig. 8 B and C). Thus, these results suggest that the helicase 1 domain is the crucial and indispensable domain for DHX29 to interact and form complexes with nucleic acids and RIG-I and MAVS.
Fig. 8.
DHX29 binds to nucleic acids, RIG-I, and MAVS via the helicase 1 domain. Cell lysates of 293T cells overexpressed with various deletion mutants of HA-DHX29 (A) mixed with biotinylated poly I:C or poly dAdT:dAdT were immunoprecipitated with NeutrAvidin beads (B). Cell lysates overexpressed with deletion mutants of HA-DHX29 with Myc–RIG-I-ΔC or with Myc-MAVS were immunoprecipitated with anti-Myc (B) or anti-HA beads (C). The immunoprecipitated elutes (IP) were detected by Western blot analysis with anti-HA (B) or anti-Myc (C) antibodies. Data are representative of at least three independent experiments.
Discussion
In this study, we made very interesting observations on nucleic acid sensing by human airway epithelial cells and fibroblasts. Airway-derived epithelial cells and fibroblasts have a defect in STING-dependent cytosolic DNA sensing, suggesting that STING-dependent DNA sensing is not universal in all cell types, and airway-derived cells and myeloid cells have different cytosolic nucleic acid sensors. Of the 59 helicases tested, DHX29 and RIG-I were found to play critical roles in type 1 IFN responses in airway-derived epithelial cells and fibroblasts following activation by poly I:C, poly dAdT:dAdT, or RNA virus. DHX29 is not expressed in other PBMC subsets and does not play a significant role in type 1 IFN responses in other cell types, including HEK293T cells, THP-1 cells, human primary monocytes, and the mouse dendritic cell line D2SC (22). Further biochemical analyses showed that DHX29 directly binds poly I:C and poly dAdT:dAdT as well as RIG-I and MAVS. These data suggest that DHX29 may function as a RIG-I coreceptor, and RIG-I may pair with different coreceptors in different cell types or for sensing different ligands, such as poly I:C, poly dAdT:dAdT, and 5′-ppp-dsRNA.
In terms of the binding of DHX29 to nucleic acids, it did not show a specificity for the binding site of poly I:C or poly dAdT:dAdT. Similar findings have been reported for RIG-I (32). These observations suggest that these helicases recognize the common structure in the two nucleic acids, which is not unusual for DExD/H helicase family members that do not have molecular specificity for DNA or RNA (33, 34). As for the binding domain in DHX29, only a helicase 1 domain defect showed impairment in forming a complex with other molecules. This may suggest that DHX29 forms a complex by binding directly and indirectly through this domain. Another possibility is that, as known for other helicase family members, there are multiple interacting domains. This is the case especially for interactions with nucleic acids (35–39); thus, only total depletion of such domains (represented as fragment “e”) showed total impairment of complex formation.
In terms of the types of response, the knockdown of DHX29 had more impact on IFN-β production than on IL-6. This may reflect the fact that MRC5 fibroblasts produce a great amount of IL-6 spontaneously upon senescence. Another possibility, as suggested by Choi et al. (32), is the presence of another sensing mechanism for the NF-κB pathway. This will be a future point of interest.
In conclusion, we could identify the DHX29–RIG-I–MAVS pathway as the cytosolic nucleic acid-sensing system in airway epithelial cells and fibroblasts. Furthermore, this study clearly shows that the innate immune system has significant diversity in the cytosolic nucleic acid-sensing system among different cells and response types, suggesting distinct selective pressure by various microorganisms. Many reported and unreported molecules, including helicase family members, might contribute to shaping each specific sensing pathway. Clarifying the whole picture of such pathways should enhance our understanding of various infections and immune pathologies, enabling us to design novel therapeutic strategies.
Materials and Methods
RNA Interference.
Transfections of siRNA were performed using Lipofectamine RNAiMAX (Invitrogen) with siRNA (Dharmacon) at a concentration of 5 pmol for 2 × 104 cells in 96-well plates or 25 pmol for 10 × 104 cells in 24-well plates according to the manufacturer’s instructions. Forty hours later, cells were used for further experiments.
Quantitative RT-PCR.
RPL13A was used as a housekeeping control to normalize the amounts of cDNA between each sample. Differences were calculated using the threshold cycle (Ct) and comparative Ct methods for relative quantification. Results were expressed as the relative expression of mRNA levels detected in control samples.
Plasmids.
cDNA plasmids were obtained from Open Biosystems. PCR assays were performed using Phusion Hot Start High-Fidelity DNA Polymerase (New England Biolabs) and primers designed to the ORFs of DHX29, DDX58 (RIG-I), or MAVS with terminal restriction enzyme sites to facilitate directional cloning. The amplicons were subcloned into pCMV-HA or pCMV-Myc vectors (Clontech) by using SfiI, NotI, KpnI, T4 polynucleotide kinase, and T4 DNA ligase (New England Biolabs). Based on pCMV-HA–full-length DHX29, pCMV-Myc–full-length RIG-I, and pCMV-Myc–full-length MAVS, truncated forms of HA-DHX29, Myc-RIG-I, and Myc-MAVS were generated by inverse PCR methods. Sequence fidelity of full-length and truncated cDNA clones was verified by DNA sequencing.
Statistical Analysis.
Data were analyzed for statistical significance by the two-tailed Student t test. A P value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Carson Harrod for critical reading. We thank Musheng Bao and Sandra Clayton for their technical support on confocal microscopy, and we thank Haruyuki Fujita for assistance with sorting cells. We thank all the colleagues in our laboratory. This work was supported by the US National Institutes of Health (R37 AI091947).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400139111/-/DCSupplemental.
References
- 1.Girard MP, Cherian T, Pervikov Y, Kieny MP. A review of vaccine research and development: Human acute respiratory infections. Vaccine. 2005;23(50):5708–5724. doi: 10.1016/j.vaccine.2005.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo Z, et al. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am J Respir Cell Mol Biol. 2007;36(3):263–269. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
- 3.Le Goffic R, et al. Cutting edge: Influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J Immunol. 2007;178(6):3368–3372. doi: 10.4049/jimmunol.178.6.3368. [DOI] [PubMed] [Google Scholar]
- 4.Liu P, et al. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J Virol. 2007;81(3):1401–1411. doi: 10.1128/JVI.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mibayashi M, et al. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81(2):514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Opitz B, et al. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 2007;9(4):930–938. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, et al. Role of double-stranded RNA pattern recognition receptors in rhinovirus-induced airway epithelial cell responses. J Immunol. 2009;183(11):6989–6997. doi: 10.4049/jimmunol.0901386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ioannidis I, Ye F, McNally B, Willette M, Flaño E. Toll-like receptor expression and induction of type I and type III interferons in primary airway epithelial cells. J Virol. 2013;87(6):3261–3270. doi: 10.1128/JVI.01956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 10.Broz P, Monack DM. Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol. 2013;13(8):551–565. doi: 10.1038/nri3479. [DOI] [PubMed] [Google Scholar]
- 11.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38(5):855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 13.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437(7062):1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 14.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19(6):727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38(5):870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unterholzner L. The interferon response to intracellular DNA: Why so many receptors? Immunobiology. 2013;218(11):1312–1321. doi: 10.1016/j.imbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun W, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA. 2009;106(21):8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong B, et al. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30(3):397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12(10):959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim T, et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107(34):15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34(6):866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Yuan B, Lu N, Facchinetti V, Liu YJ. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J Immunol. 2011;187(9):4501–4508. doi: 10.4049/jimmunol.1101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitoma H, et al. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39(1):123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, et al. The interaction between the helicase DHX33 and IPS-1 as a novel pathway to sense double-stranded RNA and RNA viruses in myeloid dendritic cells. Cell Mol Immunol. 2014;11(1):49–57. doi: 10.1038/cmi.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshiumi H, Sakai K, Matsumoto M, Seya T. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur J Immunol. 2010;40(4):940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- 29.Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol. 2011;31(18):3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478(7370):515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito T, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104(2):582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi MK, et al. A selective contribution of the RIG-I-like receptor pathway to type I interferon responses activated by cytosolic DNA. Proc Natl Acad Sci USA. 2009;106(42):17870–17875. doi: 10.1073/pnas.0909545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: Family matters. Curr Opin Struct Biol. 2010;20(3):313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linder P, Jankowsky E. From unwinding to clamping—the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12(8):505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 35.Dhote V, Sweeney TR, Kim N, Hellen CU, Pestova TV. Roles of individual domains in the function of DHX29, an essential factor required for translation of structured mammalian mRNAs. Proc Natl Acad Sci USA. 2012;109(46):E3150–E3159. doi: 10.1073/pnas.1208014109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang F, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479(7373):423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147(2):423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 38.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147(2):409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu B, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152(1-2):276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.