Abstract
Small noncoding RNAs (sncRNAs), such as microRNAs (miRNA), virus-derived sncRNAs, and more recently identified tRNA-derived RNA fragments, are critical to posttranscriptional control of genes. Upon viral infection, host cells alter their sncRNA expression as a defense mechanism, while viruses can circumvent host defenses and promote their own propagation by affecting host cellular sncRNA expression or by expressing viral sncRNAs. Therefore, characterizing sncRNA profiles in response to viral infection is an important tool for understanding host–virus interaction, and for antiviral strategy development. Human metapneumovirus (hMPV), a recently identified pathogen, is a major cause of lower respiratory tract infections in infants and children. To investigate whether sncRNAs play a role in hMPV infection, we analyzed the changes in sncRNA profiles of airway epithelial cells in response to hMPV infection using ultrahigh-throughput sequencing. Of the cloned sncRNAs, miRNA was dominant in A549 cells, with the percentage of miRNA increasing in a time-dependent manner after the infection. In addition, several hMPV-derived sncRNAs and corresponding ribonucleases for their biogenesis were identified. hMPV M2-2 protein was revealed to be a key viral protein regulating miRNA expression. In summary, this study revealed several novel aspects of hMPV-mediated sncRNA expression, providing a new perspective on hMPV–host interactions.
Keywords: human metapneumovirus, human metapneumovirus–derived snRNAs, miRNAs, sncRNA
Introduction
Human metapneumovirus (hMPV) is an important and recently identified member of Paramyxoviridae family that also includes respiratory syncytial virus (RSV) and parainfluenza virus.1 The genomic organization of hMPV is analogous to RSV, however hMPV lacks the nonstructural genes, NS1 and NS2, and the hMPV antisense RNA genome contains eight open reading frames in an order of 3′-N-P-M-F-M2-SH-G-L-5′. In terms of hMPV M2, it encodes two overlapping proteins: M2-1 and M2-2. Many clinical studies have shown that hMPV is a leading cause of lower respiratory infection in infants and children worldwide.2,3,4 Although many studies have been carried out, the host–hMPV interaction is still poorly understood.
The discovery that cells produce small noncoding RNAs (sncRNAs) that regulate gene expression in a sequence-specific fashion is a recent breakthrough in biology. MicroRNA (miRNA) is a class of highly conserved sncRNAs that usually suppresses gene expression by binding to the untranslated regions (UTR) of target mRNAs, representing a new mechanism for the posttranscriptional regulation of gene expression.5 The role of sncRNA in regulating antiviral immune responses has recently emerged.6,7 Other important classes of cellular sncRNAs include short interfering RNAs (siRNAs), small nucleolar RNAs (snoRNAs), and tRNA-derived RNA fragments (tRFs).8,9,10 Several viruses, majority of which are DNA viruses, also produce virus-encoded small ncRNAs to promote their replication or latency.11,12 Whether the profiles of sncRNAs, both host- and virus-encoded, are altered in response to hMPV infection, are unknown. In addition, it is not clear whether sncRNAs are functionally important in controlling cellular responses to hMPV infection.
High-throughput sequencing technologies, also known as deep sequencing or next-generation sequencing technologies, deliver fast, accurate, and large volume sequence data. The technology is frequently used for small RNA sequencing and discovery.13 We used this advanced technology and biological experiments, such as northern blot and real-time polymerase chain reaction (PCR), to investigate whether hMPV infection results in significant changes in the expression of sncRNAs. We found that hMPV infection led to significant changes in the expression profile of miRNAs in human airway epithelial cells. In airway epithelial cells, the changes in sncRNA profiles by hMPV infection were different from what we have recently reported for RSV infection.10 In response to RSV infection, the expression of tRFs is most significantly affected while there are minimal changes in overall miRNA expression. In contrast to RSV infection, hMPV infection did not induce tRFs but gradually enhanced the expression percentage of miRNAs in a time-dependent fashion. Expression levels of 174 miRNAs were significantly altered over a period of 15 hours. Among the altered miRNAs, 142 and 32 miRNAs were upregulated and downregulated, respectively, while 72 miRNAs, including miR-30a and miR-16, were not changed by hMPV infection. Interestingly, even though miR-30a and miR-16 were not affected by wild-type (WT) recombinant hMPV (WT-rhMPV), their expression was significantly induced by a mutant hMPV lacking M2-2 protein (ΔM2-2-rhMPV), demonstrating that the hMPV M2-2 protein antagonized the induction of miR-30a and miR-16. In addition, we identified several hMPV-derived sncRNAs.
Overall, these data suggest a new way in which paramyxoviruses regulate the host cell response to infection. Given the fundamental roles of sncRNAs in regulating the expression of genes, including those which are important in cellular signaling pathways in other viral infections,7 these findings will provide a foundation for future studies investigating the role of various miRNAs and its associated pathways in the cellular response to this important respiratory virus. The results also potentially provide sncRNA-centered information for designing antiviral drugs and live attenuated vaccines against hMPV infection.
Results
Global sncRNA profile in hMPV-infected A549 cells
Illumina ultrahigh-throughput sequencing was carried out in small RNAs from mock- or hMPV-infected A549 cells to characterize the global sncRNA expression profile. We obtained a total of 58,517,786 sequence reads. These data were processed similarly as described,10 and is also shown in Figure 1. After stripping adaptor sequences, individual sequences with cloning frequency (read numbers) ≥10 were mapped to the human genome (http://hgdownload.cse.ucsc.edu/downloads.html) to identify host-derived RNAs and also mapped to hMPV genome/antigenome to identify hMPV-derived sncRNAs. Sequences mapped to both human and hMPV genomes were considered as hMPV sequences when they align better to hMPV than to human genome. The small RNAs mapped to human genome were further identified as specific human sncRNA sequences by comparing them to the miRNA database (miRBase release 20; http://www.mirbase.org), the rRNA database (RDP; http://rdp.cme.msu.edu/), the tRNA database (GtRNAdb; http://gtrnadb.ucsc.edu/), the snoRNAs database (prepared after downloaded sno/miRNA track from the UCSC Table Browser), and the Exon-Intron Database (http://www.utoledo.edu/med/depts/bioinfo/database.html). The cloning frequency of sncRNAs from high-throughput sequencing provides a digital measure of their relative expression level. To investigate whether hMPV altered the expression profile of sncRNAs, we calculated the relative sequencing frequency of each sncRNA by dividing its raw read numbers by the total read numbers of human genome.
Figure 1.
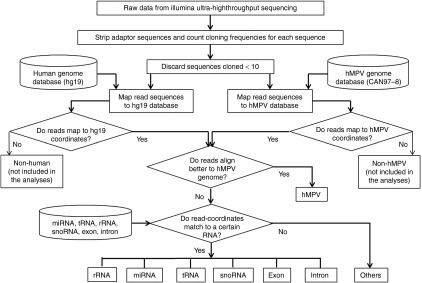
Pipeline of analyses of Illumina high-throughput sequencing data. Flowchart of the sequencing data analyses is depicted. hMPV, human metapneumovirus; miRNA, microRNA; rRNA, ribosomal RNA; snoRNA, small nucleolar RNA; tRNA, transfer RNA.
As shown in Figure 2a, sncRNA composition between mock- and hMPV-infected samples is different. The expression of miRNAs increased from 66.51% to more than 86% after 15 hours of hMPV infection, while small nucleolar RNAs (snoRNAs) were gradually altered from 17.91 to 1.49%. Minor difference was observed for other types of sncRNAs. Since the percentage of miRNAs clones increased in response to hMPV infection, we compare the miRNA profiling between the mock-infected cells and hMPV-infected cells at different time points postinfection As shown in Figure 2b, the expression level of 201 miRNAs was upregulated at least 1.5-fold, while 72 miRNAs was downregulated by ≤0.7-fold, over a period of 24 hours in infected cells, indicating that hMPV induces profound changes in global miRNA expression. At 6 hours postinfection, a number of miRNAs were already modified in response to the virus. In Supplementary Table S1, we have listed miRNAs with early change in their expressions and cloning frequency ≥500. At all time points postinfection, upregulated miRNAs were nearly four- to fivefold more than the downregulated miRNAs. There were also 82 miRNAs that were not affected by hMPV infection (Figure 2b). In addition to the change in miRNA expression, we also observed 22 hMPV-derived sncRNAs with cloning frequency >500 in hMPV-infected samples (Table 1). The Integrative Genomics Viewer was also used to demonstrate the distribution of hMPV-derived sncRNAs along the hMPV antigenome (Supplementary Figure S1). These hMPV-derived sncRNAs are unlikely to be random cleavage of viral RNAs because of the following reasons: (i) these hMPV-derived sncRNAs were restrictively antigenome derived, rather than genome derived, (ii) not all viral genes were cleaved to produce sncRNAs, and (iii) for those viral genes which produced sncRNAs, the corresponding regions account for a small portion of genes. For example, hMPV polymerase L is a gene with more 6,000 nt, however, only few regions have their respective RNA fragments. In addition, no quality issue was found, as evaluated by 5S rRNA northern hybridization (please see the northern blot controls for expression validation through the manuscript).
Figure 2.
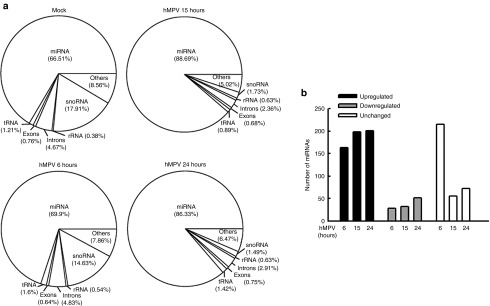
Kinetics of changes in sncRNA expression. (a) Human genome–derived small RNAs were sorted according to their origins, and their relative abundance was calculated by dividing their frequency numbers by total read numbers, and depicted in pie charts. (b) miRNAs, their abundance were changed with a fold change of ≥1.5 (upregulated) or ≤0.7 (downregulated), compared to baseline (uninfected), are represented at various times postinfection. Fold change is calculated on relative abundance for each time point compared to the baseline in mock infected cells. miRNAs whose expression was not changed are also represented. hMPV, human metapneumovirus; miRNA, microRNA; rRNA, ribosomal RNA; snoRNA, small nucleolar RNA; tRNA, transfer RNA.
Table 1. hMPV-encoded sncRNAs with cloning frequency ≥500.
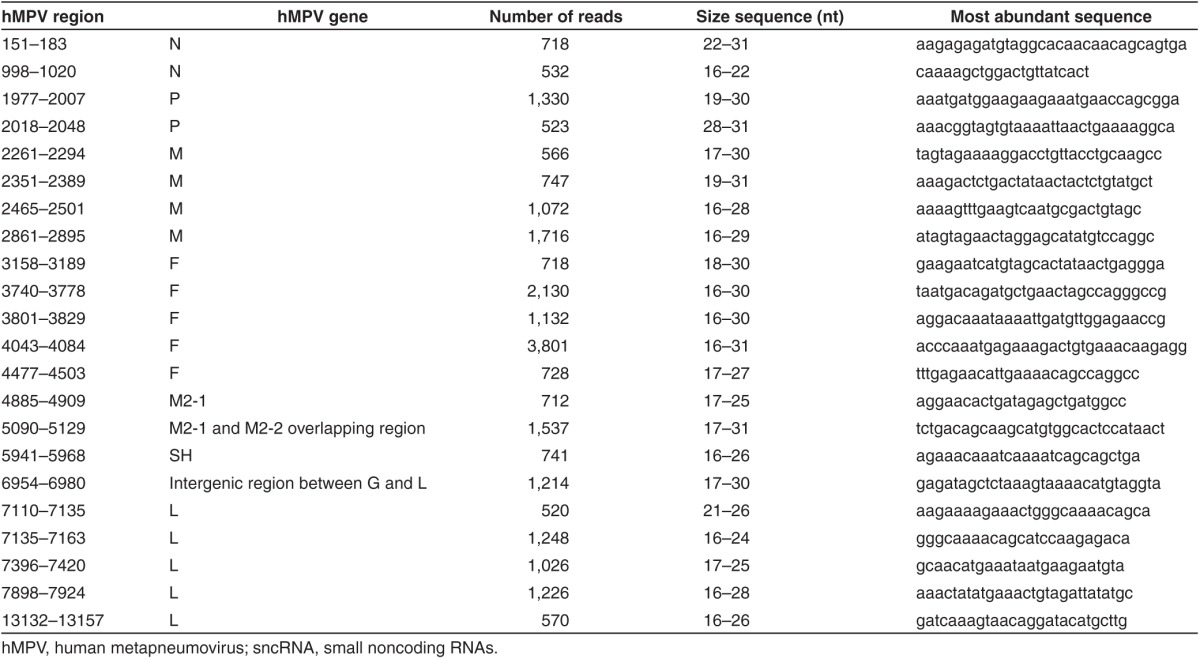
sncRNA expression validation
Among the altered miRNA, quantitative real-time PCR (qRT-PCR) was used to verify the expression of miRNAs which were reported to be affected by other viruses, but with functions incompletely characterized or uncharacterized. We used U6, a small nuclear RNA, as an internal control for qRT-PCR, as its expression was comparable in response to mock and hMPV infection (data not shown), although the overall expression of snoRNAs was changed by hMPV infection (Figure 2a). Let-7f has been reported to regulate viral replication or chemokine expression, and its expression is sensitive to the infection of RSV (a close family member of hMPV) and human immunodeficiency virus.14,15,16 Compared to other miRNAs in A549 cells, mock or hMPV-infected, let-7f was the second most abundant miRNAs at l5 hours postinfection and detectable by northern blot (Supplementary Figure S2). Consistent with our sequencing and northern blot data, qRT-PCR assay confirmed that hMPV infection significantly enhanced let-7f expression (Figure 3a). miR-452 is inducible by influenza H1N1.17 We found its expression was also significantly increased by hMPV (Figure 3a). Whether miR-452 plays a role in regulating host response to viral infection is currently unknown. We also picked up miR-374a* and miR-192 as the representative of downregulated miRNAs by hMPV for expression confirmation. Compared to other downregulated miRNAs, miR-192 and miR-374a* were relatively abundant with cloning frequency at mock-infected cells above 30,000. Both our sequencing data and qRT-PCR revealed that they were downregulated by hMPV infection. Similar downregulation of miR-192 occurs in human hepatoma cells in response to hepatitis C virus and hepatitis B virus infection18,19, and the involvement of miR-192 in enhancing replication of the hepatitis C virus has also been shown.20 Of note is the fact that the information on the regulation of miR-374a* by viral infection is very limited except their induction by hepatitis C virus.21 Our data demonstrate that miR-374a* was reduced by hMPV infection (Figure 3a). In this study, we also used qRT-PCR to confirm the expression of miR-30a and miR16, whose expression was not affected by hMPV infection.
Figure 3.
Experimental validation of sncRNA expression. (a) miRNA validation. Total RNA from indicated treatments in A549 cells was harvested at indicated time points postinfection, followed by reverse transcription using a TaqMan microRNA reverse transcription kit (Life Technologies). Quantitative real-time polymerase chain reaction was used to validate the expression of representative miRNAs. U6 snRNA was used as an internal control. All reactions were done in a 10-μl reaction volume in triplicate. Data are representative of two to three independent experiments. *P < 0.05, **P < 0.01, relative to the first bar in each group. (b). hMPV-derived sncRNA expression. A549 cells, mock- or hMPV-infected, were harvested at 15 hours postinfection, followed by total RNA extraction and loading to a denaturing polyacrylamide gel for northern hybridization using indicated probes. 5S rRNA is shown for equal loading. Data are representative of two to three independent experiments. hMPV, human metapneumovirus; miRNA, microRNA; rRNA, ribosomal RNA; snoRNA, small nucleolar RNA; tRNA, transfer RNA.
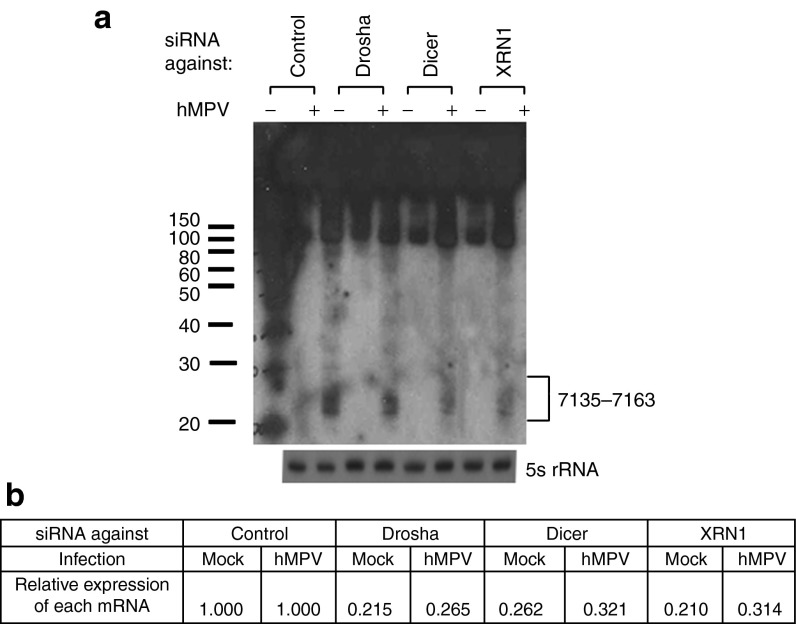
As shown in Table 1, hMPV infection resulted in the expression of hMPV-derived sncRNAs as well. It is commonly believed that negative-sense RNA paramyxoviruses do not produce viral miRNAs, as their replication does not occur in the nuclear compartment, where miRNA biogenesis is initialized.22,23 However, several other sncRNAs are known to be generated by alternative pathways, and some RNA viruses produce virus-derived sncRNAs.8,24,25 To confirm the sequencing data of hMPV-derived sncRNAs, we used northern blot to detect the two hMPV-derived sncRNAs. As shown in Figure 3b, two sncRNAs derived from the P and L, two genes of hMPV RNA-dependent RNA polymerase complex, were detectable by northern blot. The band corresponding to 2018–2048 was slightly higher than 7135–7163, which is consistent with their sequencing data showing that the overall length of 2018–2048 was longer than that of 7135–7163. In addition, we used 6- and10-day film exposure to acquire the bands corresponding to the regions of 7135–7163 and 2018–2048, respectively, which again is in a line with the sequencing data suggesting that there was more induced 7135–7163 than 2018–2048.
Function of let-7f in hMPV replication
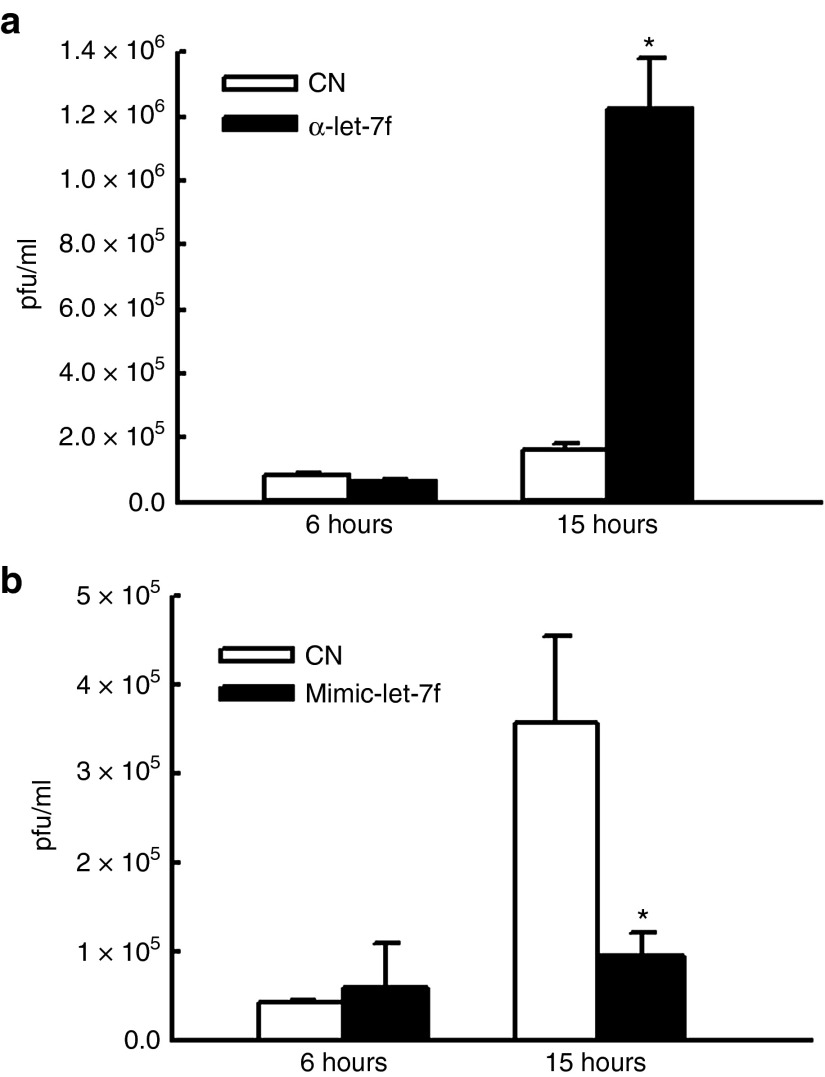
Among the differentially expressed miRNAs, we further explored let-7f function. Similar to hMPV infection, RSV infection has been shown to enhance the expression of let-7f expression as well, and the enhancement actually favors RSV replication.14 To determine whether hMPV-induced let-7f is functional, we first investigated whether hMPV replication is sensitive to the treatment of let-7f inhibitor. Commercially available miRNA inhibitors are usually chemically modified, single-stranded nucleic acids designed to specifically bind to and inhibit endogenous miRNA molecules. These ready-to-use inhibitors can be introduced into cells using transfection or electroporation parameters similar to those used for siRNAs and enable detailed study of miRNA biological effects. A549 cells were transfected with 150 nmol/l let-7f inhibitor or its control for 24 hours, followed by mock or hMPV infection at multiplicity of infection of 2. After 1 hour, the cells were replaced with fresh media and incubated for additional 15 hours. Cells were then scraped, followed by sonication and centrifugation to release the intracellular viral particles. The infectious hMPV particles were measured by immunostaining as we previously described and expressed as plaque-forming unit/ml (the number of infective particles within 1 ml of the sample).26,27,28 We found that cells transfected with let-7f inhibitor had a fold change of 6 in hMPV replication compared to control oligotransfected cells (Figure 4a). A real-time PCR for hMPV N gene copy numbers also showed similar trends (data not shown). miRNA mimics are small, chemically modified double-stranded RNAs that mimic endogenous miRNAs and enable miRNA functional analysis by upregulation of miRNA activity. In contrast to the effect of let-7f inhibitor on hMPV replication, lef-7f mimic significantly decreased hMPV replication 3.3-fold (Figure 4b). Transfection of let-7f-specific mimic or inhibitor did not result in more cytotoxicity in cells than control oligos as the trypan blue staining did not reveal the difference in dead cell numbers between let-7f-specific and control oligo-treated cells (data not shown). These data suggest that hMPV-modulated let-7f has an antiviral role.
Figure 4.
Let-7f inhibits human metapneumovirus (hMPV) replication. (a) Knockdown of let-7f inhibits hMPV replication. Let-7f inhibitor was transfected into A549 cells using lipofectamine 2000. At 24 hours post-transfection, cells were mock infected or infected with hMPV at multiplicity of infection (MOI) of 2. Cells were harvested at 6 or15 hours postinfection, and viral infectious particles were measured by immunostaining. (b) Let-7f mimic were transfected into A549 cell. At 2 hours post-transfection, cells were mock infected or infected with hMPV at MOI of 2. At 6 or 15 hours postinfection, viral titration was assayed. Data are representative of two to three independent experiments. *P < 0.05, relative to the white bar at each time point postinfection as indicated. pfu, plaque-forming unit.
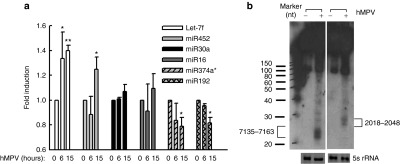
The role of hMPV M2-2 protein in miRNA expression
M2-2 is an hMPV protein that regulates viral RNA synthesis.28,29,30 Recently, we have discovered that hMPV M2-2 is a potent virulent factor that blocks mitochondrial antiviral signalling-mediated antiviral signaling leading to inhibited activity of two transcription factors: NF-κB and interferon regulatory factor (IRF)-3, and, subsequently reducing production of type I interferon (IFN).28 Since miRNAs are RNA molecules and its regulation has been shown to be regulated by transcription factors, such as NF-κB and IRF-3, and IFNs,31,32,33,34 we investigated whether M2-2 regulates the synthesis of miRNAs. In brief, A549 cells were infected with recombinant hMPV (rhMPV), either WT or a mutant lacking M2-2 (▵M2-2). At 15 hours postinfection, cells were harvested for RNA extraction followed by miRNA quantification by qRT-PCR. Interestingly, we did not observe any effect of M2-2 on the expression of let-7f and miR-452, two hMPV-induced miRNAs. M2-2 also did not affect the expression of miR-192 and miR-374a*, two hMPV-decreased miRNAs (data not shown). As mentioned above, M2-2 deletion significantly enhances IFN-α/β secretion and the activation of NF-κB and IRF-3 in response to hMPV infection.28 Comparable expression of let-7f, miR-452, miR-192, and miR-374a* in WT- or ▵M2-2-infected cells suggests that the effect of hMPV on the expression of these miRNAs does not depend on IFN signaling or the activation of NF-κB and IRF-3.
Notably, compared to WT infection, ▵M2-2 infection resulted in an increased expression of miR-16 and miR-30a in airway epithelial cells, although the expression of both miRNAs was not affected by the infection of naive hMPV or WT-rhMPV (Figures 3a and 5a). To confirm the role of M2-2 in modulation of miR-16 and miR-30a, the M2-2 protein was exogenously expressed. We found that M2-2 expression alleviated enhanced miR-16 and miR-30a expression following rhMPV-ΔM2-2 infection (Figure 5a), demonstrating a role of M2-2 in regulation of these two miRNAs. We also found that the inhibition of M2-2 on miR-16 induction was impaired in U4A cells, a cell line lacking IFN signaling because of JAK-1 knockout,35,36 suggesting that M2-2-mediated miR-16 expression depends on IFN signaling (Figure 5b). Comparable miR-16 expression in WT- and ΔM2-2-infected U4A cells also suggested that IRF-3 and NF-κB are not important for miR-16 induction, as M2-2 deletion resulted in more activated IRF-3 and NF-κB in U4A cells, akin to the regulatory role of M2-2 in the activation of IRF-3 and NF-κB in airway epithelial cells (data not shown). In contrast to miR-16, miR-30a seemed to be IFN independent. In the absence of JAK-1, ▵M2-2 induced more miR-30a than WT virus in U4A cells (Figure 5b). Besides direct inhibition of antiviral signaling pathway by M2-2, M2-2 also suppresses other viral gene expression to facilitate evasion of hMPV from host immunity.28 Therefore, it is possible that M2-2 regulates miR-30a through modulating other viral gene expression. However, our results show that M2-2 deletion does not change hMPV protein expression in U4A cells (Figure 5c), suggesting a direct regulatory function of M2-2 in miR-30a expression. We show, for the first time, that M2-2 regulates miRNA expression.
Figure 5.
M2-2 regulates microRNA (miRNA) expression. (a) A549 cells were transfected with plasmid expressing M2-2 or its control (0.1 µg/well). At 30 hours post-transfection, the cells were mock infected or infected with rhMPV, wild type (WT), or ΔM2-2, at multiplicity of infection (MOI) of 2. At 15 hours postinfection, total RNA was prepared followed by quantitative real-time polymerase chain reaction (qRT-PCR) to measure the expression of miR-30a and miR-16. (b) U4A cells were infected with rhMPV at MOI of 2 for 15 hours, followed by total RNA preparation and qRT-PCR to measure the expression of miR-30a and miR-16. U6 snRNA was used as an equal loading internal control. Data are representative of two to three independent experiments. *P < 0.05, **P < 0.01, relative to the first bar in each group. (c) The total protein from U4A cells was also subjected to Western blot to assess the viral gene expression. hMPV, human metapneumovirus.
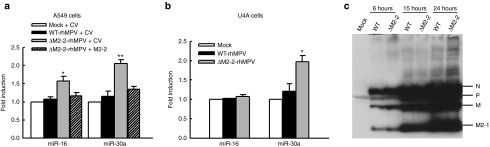
Biogenesis of hMPV-derived sncRNAs
Most studies on the roles of virus-derived sncRNA in controlling viral replication were performed with double-stranded DNA viruses.37 It is commonly believed that negative-sense paramyxoviruses do not produce viral miRNAs as their replication does not occur in the nuclear compartment, where miRNA biogenesis is initialized.23 However, several sncRNAs are known to be generated by alternative pathways and some of them exert miRNA-like functions.8 Indeed, our deep sequencing data demonstrate that there were several hMPV-encoded sncRNAs from hMPV infections. These sncRNAs could be viral miRNAs generated via noncanonical pathways, or other forms of sncRNAs, such as siRNA or recently identified novel microRNA (miRNA)-like molecules generated from virus and host tRNA transcripts.38 To define their functions in the future, it is important to identify the enzymes responsible for their biogenesis. As mentioned, hMPV is a single-negative–stranded RNA virus. Therefore, the transcription and replication of hMPV likely happens in the cytoplasm, and hMPV-derived sncRNAs cannot be generated by nuclear RNases, such as Drosha and RNase P, two enzymes critical for miRNA biogenesis and tRNA cleavage, respectively.39,40 As expected, we did not observe the changes in the expression of hMPV-derived sncRNAs by Drosha silencing (Figure 6), reinforcing our view that the biogenesis of hMPV-derived sncRNAs was distinct from that of siRNA/miRNA. To identify host protein(s) responsible for hMPV-derived sncRNA generation, we paid close attention to the nucleases whose expression was significantly induced by hMPV, using previously collected microarray data.41 One particular enzyme called exoribonuclease 1 (XRN1) was identified as a candidate. Since Dicer is a cytosolic nuclease in the RNase III family that cleaves double-stranded RNA and pre-miRNA into short double-stranded RNA fragments,39 and we observed that its gene transcription is enhanced by hMPV infection,41 it was also identified as a candidate. We found that both XRNI and Dicer are necessary for the expression of hMPV-derived sncRNA upon hMPV infection (Figure 6a). The efficiency of knockdown was confirmed by qRT-PCR measurement of each mRNA (Figure 6b) and western blot (data not shown). Collectively, our data suggest that the production of hMPV-derived sncRNAs upon hMPV infection specifically required XRNI and Dicer.
Figure 6.
Human metapneumovirus (hMPV)-derived sncRNA is generated by Dicer and RXN1. (a) A549 cells were transfected with 120 nmol/l of siRNA against indicated proteins or control siRNA as a negative control. At 48 hours post-transfection, the cells were mock- or hMPV-infected for 24 hours. Total RNAs were then subjected to northern hybridization as described in Figure 3b; 5S rRNA expression is shown for equal loading. (b) The suppression of target mRNAs by each siRNA was confirmed by quantitative real-time polymerase chain reaction measurement. The values are the relative expression levels of each mRNA, with the value of control siRNA set as one. Data are the representative of two independent experiments.
Discussion
The discovery of posttranscriptional control of genes by sncRNAs is a key advance in biology. sncRNAs, such as miRNA and recently identified tRFs, are gene-silencing mediators, which have an important role in the regulation of many important cellular process, including cellular differentiation, innate immunity, apoptosis, and oncogenic transformation.9,10,42 During host–virus interactions, many viruses have been reported to change miRNA expression profile to modulate host responses to viral infection.43,44,45 Whether sncRNAs are changed by hMPV infection is currently unknown. In addition, there is limited knowledge on the relationship between paramyxovirus-induced host responses and sncRNAs, either host or virus encoded. In this study, we found that the expression of miRNAs among sncRNAs in A549 cells was increased post-hMPV infection. In addition, the changes in cellular sncRNA profiling in response to hMPV are different from that of RSV. Following RSV infection, tRFs are the major type of sncRNAs significantly affected, while the change in miRNAs was minimal.10 However, in the context of hMPV infection, hMPV infection did not induce tRF expression but changed miRNA profiling significantly. A total of 375 known miRNAs were cloned in A549 cells, among which, 142 and 32 miRNAs were upregulated and downregulated by hMPV at 15 hours postinfection, respectively.
As mentioned, there are very limited studies on the interaction between miRNAs and paramyxoviruses. Hendra virus, a highly pathogenic zoonotic paramyxovirus, has been reported to make use of miR-146a to negatively regulate NF-κB activity via targeting ring finger protein 11, to favor its replication.46 RSV downregulates miR-221, which leads to enhanced expression of the nerve growth factor in human airway cells, and subsequently favors viral replication by interfering with the apoptotic death of infected cells.47 RSV also uses let-7f induction to promote its replication.14 Since let-7f was the second most abundant miRNAs with cloning frequency >100,000 reads in uninfected cells and was also significantly induced by hMPV infection, we investigated whether let-7f is functionally important. In contrast to the stimulus role of let-7f in RSV replication, we found that hMPV-induced let-7f inhibited hMPV replication (Figure 4). Similar inhibitory effect of let-7f on viral replication has been shown for several other viruses,48,49 suggesting that let-7f is an important and common regulator of viral replication. In addition, in these experiments, the lactate dehydrogenase assay did not reveal the cell toxicity by transfection reagent (data not shown). The opposite regulatory effect of let-7f on hMPV and RSV replication suggests that the role of miRNA in regulating viral replication is pathogen dependent. There are several possible reasons why miRNA functions differently in response to different viral infections. First, miRNA may target different host genes in the context of hMPV and RSV infection. Second, since the sncRNA-viral RNA interaction has been reported to be important in viral RNA synthesis and stability,50 the different role of let-7f in RSV and hMPV replication may result from the difference in the binding of let-7f to RSV and hMPV RNA. To investigate whether miRNAs are able to interact with hMPV genome/antigenome, we mapped miRNAs to the viral genome/antigenome.51 Interestingly, we found that the first 13 bases of let-7f match with hMPV genome 9871–9883, where RNA-dependent RNA polymerase L is located. However, such interaction did not exist between let-7f and RSV genome.14 The mechanism underlying the effect of let-7f on hMPV will be studied in the future. In the future, we will also investigate the functions of other miRNAs whose expression was affected by hMPV infection.
In this study, we also demonstrated that hMPV M2-2 protein inhibited the expression of miR-30a and miR-16, as ▵M2-2 infection resulted in a significant induction of miR-30a and miR-16, while WT infection did not result in induction (Figure 5a). By target prediction algorithms (TargetScan and miRanda), several targets were hit. Given the fact that miR-16 and miR-30a were not sensitive to naive/WT hMPV infection, we selected those targets, whose expression is not changed by naive hMPV in our previous microarray gene transcription data, as targeting candidates.41 For example, we chose cysteinyl-tRNA synthetase (CARS), KIAA1949, CDC37L1, and runt-related transcription factor 1 (RUNX1) as target candidates of miR-30a, because their expression remained unaffected by hMPV infection. RUNX1 actually is required for human immunodeficiency virus infection.52 Whether these targets favor hMPV infection and whether hMPV uses M2-2 to suppress these miRNAs to prevent downregulation of their targets will be studied in the future. It is a common trend that miRNAs with significant change in the expression by viral infection are always paid close attention. This example reminds us that we may need to study those miRNAs that are unchanged by viral infection, as they are probably suppressed by certain viral components to favor viral infection, which, in this case, is hMPV M2-2 protein.
We also identified several hMPV-encoded sncRNAs and corresponding ribonucleases for their biogenesis (Figure 6). Whether these sncRNAs play a role in mediating viral replication and innate immune responses need to be experimentally validated. If so, such virus-encoded small RNAs would be ideal tools for controlling viral replication, as they are nonimmunogenic. They may also regulate multiple host transcripts to varying degrees. Therefore, silencing the action of virus-encoded sncRNAs may enable host cells or the immune system to gain control over the virus and even to eliminate it. Due to the scant homology between viral sncRNA and host cell miRNAs,53 anti-sncRNA oligonucleotides might have minimal off-target effects and great therapeutic potential. A recombinant virus system for hMPV is well established in our laboratory. Any modification to the recombinant genome, such as modifying sequences corresponding to virus-encoded sncRNAs, should help develop live attenuated vaccines against hMPV infections. In addition, hMPV, as a negative sense RNA virus, might be a suitable model to study the biogenesis of virus-encoded sncRNAs for those viruses that do not replicate in the nuclear compartment.
In summary, deep sequencing results provided us an overall big picture without any priori assumptions and guided us which sncRNAs species to be explored further, although data bias may exist. Within reasonable variance, we confirmed the deep sequencing results with qRT-PCR and northern blot experiments which overcame the data bias problems. Overall, our ultrahigh-throughput sequencing study provides novel information on the regulation of sncRNAs by hMPV infection and can serve as a foundation for future investigations on the role of the identified miRNAs and hMPV-derived sncRNAs in initiating antiviral as well as innate and adaptive immune responses to this important pathogen. In addition, sncRNAs identified to be important for hMPV replication have potentials to serve as therapeutic targets, suggesting a potential translational application for this study.
Materials and methods
Cell lines and virus. A549, human alveolar type II-like epithelial cells, and MK-2 cell line (both from ATCC, Manassas, VA) were maintained as described.27,28 U4A cells, a JAK-1-deficient fibrosarcoma cell line (kindly provided by Dr George Stark, Cleveland Clinic Foundation, Cleveland, OH), were cultured as previously described.26,54 hMPV, either naive or recombinant derived from the Canadian isolate hMPV83 (CAN 97–83), was grown in MK-2 cells, purified by sucrose-gradient and titrated by immunostaining in MK-2 cells using homemade rabbit anti-hMPV antibody as described.26,27,28,54–56
Viral infection. A549 cells were infected with hMPV, at multiplicity of infection of 3, in serum-free medium containing antibiotics and 1 µg trypsin/ml for all experiments, unless otherwise stated. Cells treated with infection media plus sucrose were defined as mock infected. Mock- or hMPV-infected cells were scrapped into the medium, followed by centrifugation and RNA extraction using TRIzol Reagents according to manufacturer's instruction (Invitrogen, Carlsbad, CA) as described.10
Deep sequencing and RNA mapping. A549 cells after mock or hMPV (CN 97–83) infection were harvested for total RNA preparation. The RNAs were delivered to Eureka Genomics (Houston, TX) for small RNAs isolation, directional adaptor ligation, cDNA library construction, and sequencing using a Genome Analyzer IIx (Illumina, San Diego, CA). About 2,103 Mb of sequence data with a total of 58,517,786 sequence reads were generated for mock- and hMPV-infected samples, using 36b single-end sequencing reads. The cDNA sequences were aligned to both human (hg19) and hMPV (CAN97-8) reference genomes using NovoalignCS. The alignments were carried out with options to strip 3′ adapter sequences, for a minimum of 15 good quality bases per read, and a tolerance of one mismatch. After initial alignment, further processing was performed using in-house programs and SAMtools.10 First, uniquely aligned reads and sequences aligned to more than one genome location (ambiguously aligned reads) were separated. The ambiguously aligned reads were then randomly assigned to one location and combined with uniquely aligned reads. Sequences with reads ≥10 were used for the downstream analysis shown in Figure 1. We assigned the sequences mapped to both hg19 and hMPV to only one of them with better BLAST score not to duplicate the read number. We performed several normalization methods for comparison including total reads of human genome, total number of each species (such as total miRNA reads or total tRNA reads), and upper quartile normalization. While detailed fold changes differ, the overall trends are similar. Considering the shifting of sncRNAs composition with hMPV infection, we used values normalized by total human reads in the results.
RNA interference. Transfections of siRNA into A549 cells were carried out at a final concentration of 120 nmol/l, targeting Drosha, Dicer, 5′-3′ exoribonuclease 1 (XNR1), or a scrambled negative control (Sigma, Houston, TX), using lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. Fourty-eight hours later, A549 cells were mock infected or infected with hMPV for 18 hours at a multiplicity of infection of 3.
Expression confirmation of sncRNA. Northern hybridization for sncRNAs were performed as described.9,10 Briefly, RNA was separated in 15% denaturing polyacrylamide gel with 7 mol/l urea and then transferred to a positively charged nylon membrane (Amersham Biosciences, Piscataway, NJ). The membrane was hybridized with 32P-labled probes in ULTRAhyb-Oligo solution (Life Technologies, Grand Island, NY), followed by washing according to the manufacturer's instruction.
qRT-PCR was also used to validate miRNA expression data from high-throughput sequencing. Extracted total RNA was reverse transcribed into single-stranded cDNA with a TaqMan microRNA reverse transcription kit (Life Technologies). Real-time PCR was performed using first-strand cDNA with TaqMan Fast Universal PCR Master Mix. The assay numbers for the miRNA endogenous control (U6 snRNA) and miRNAs were as follows: U6 snRNA (001973), miR-16 (000391), let-7f (00382), miR-192 (000491), miR-194 (000493), miR-374a*(002125), miR-30a (000417), and miR-452 (003229). Quantitative PCR was performed on an Applied Biosystems Prism 7500 Fast Sequence Detection System (Applied Biosystems). Quantitative PCR parameters for cycling were as follows: 95 °C for 20 seconds, 40 cycles of PCR at 95 °C for 30 seconds and 60 °C for 30 seconds. All reactions were done in a 10-μl reaction volume in triplicate. The mRNA and miRNA expression levels were determined using the 2–ΔCT method.
Statistical analysis. Statistical significance was analyzed using analysis of variance. P value of less than 0.05 was considered significant. Mean ± standard error is shown.
SUPPLEMENTARY MATERIAL Figure S1. Distribution of hMPV-derived sncRNAs. Figure S2. Let-7f validation. Table S1. miRNA with total sequencing reads >500, and fold induction ≥1.5 or fold reduction ≤0.7 by hMPV at 6 hours postinfection.
Acknowledgments
All authors concur there are no conflicts of interest associated in this published work. This work was supported by grants from the National Institutes of Health-National Institute of Allergy and Infectious Diseases 1K22AI074829-01A2 and 1R56AI07033-01A1, the American Lung Association RG232529N, and American Heart Association 12BGIA12060008 to X.B. Authors thank Animesh Chandra for assistance with manuscript editing.
Supplementary material
Distribution of hMPV-derived sncRNAs.
Let-7f validation.
miRNA with total sequencing reads >500, and fold induction ≥1.5 or fold reduction ≤0.7 by hMPV at 6 hours postinfection.
References
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KM, Zhu Y, Griffin MR, Weinberg GA, Hall CB, Szilagyi PG, et al. New Vaccine Surveillance Network Burden of human metapneumovirus infection in young children. N Engl J Med. 2013;368:633–643. doi: 10.1056/NEJMoa1204630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigal E, Maakaroun-Vermesse Z, Gaudy-Graffin C, Bonnemaison E, Marchand S, Labarthe F, et al. Epidemiological and clinical description of human metapneumovirus infectious diseases in children. Arch Pediatr. 2010;17:26–33. doi: 10.1016/j.arcped.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Falsey AR, Erdman D, Anderson LJ, Walsh EE. Human metapneumovirus infections in young and elderly adults. J Infect Dis. 2003;187:785–790. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Weidmer A, Liu CG, Volinia S, Croce CM, Lieberman PM. Epstein-Barr virus-induced miR-155 attenuates NF-kappaB signaling and stabilizes latent virus persistence. J Virol. 2008;82:10436–10443. doi: 10.1128/JVI.00752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Wang P, Lin L, Liu X, Ma F, An H, et al. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 2009;183:2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Miyoshi T, Siomi H. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol Genet Genomics. 2010;284:95–103. doi: 10.1007/s00438-010-0556-1. [DOI] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther. 2013;21:368–379. doi: 10.1038/mt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölken L, Pfeffer S, Koszinowski UH. Cytomegalovirus microRNAs. Virus Genes. 2009;38:355–364. doi: 10.1007/s11262-009-0347-0. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Benham AL, Zhu H, Khan MF, Reid JG, Nagaraja AK, et al. Discovery of novel microRNAs in female reproductive tract using next generation sequencing. PLoS One. 2010;5:e9637. doi: 10.1371/journal.pone.0009637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakre A, Mitchell P, Coleman JK, Jones LP, Saavedra G, Teng M, et al. Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J Gen Virol. 2012;93 Pt 11:2346–2356. doi: 10.1099/vir.0.044255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL, et al. Differential regulation of the Let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J Immunol. 2012;188:6238–6246. doi: 10.4049/jimmunol.1101196. [DOI] [PubMed] [Google Scholar]
- Edge RE, Falls TJ, Brown CW, Lichty BD, Atkins H, Bell JC. A let-7 MicroRNA-sensitive vesicular stomatitis virus demonstrates tumor-specific replication. Mol Ther. 2008;16:1437–1443. doi: 10.1038/mt.2008.130. [DOI] [PubMed] [Google Scholar]
- Buggele WA, Johnson KE, Horvath CM. Influenza A virus infection of human respiratory cells induces primary microRNA expression. J Biol Chem. 2012;287:31027–31040. doi: 10.1074/jbc.M112.387670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Daucher M, Armistead D, Russell R, Kottilil S. MicroRNA expression profiling in HCV-infected human hepatoma cells identifies potential anti-viral targets induced by interferon-alpha. PLoS One. 2013;8:e55733. doi: 10.1371/journal.pone.0055733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781–4788. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
- Ishida H, Tatsumi T, Hosui A, Nawa T, Kodama T, Shimizu S, et al. Alterations in microRNA expression profile in HCV-infected hepatoma cells: involvement of miR-491 in regulation of HCV replication via the PI3 kinase/Akt pathway. Biochem Biophys Res Commun. 2011;412:92–97. doi: 10.1016/j.bbrc.2011.07.049. [DOI] [PubMed] [Google Scholar]
- Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowton VM, McGivern DR, Fearns R. Unravelling the complexities of respiratory syncytial virus RNA synthesis. J Gen Virol. 2006;87 Pt 7:1805–1821. doi: 10.1099/vir.0.81786-0. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Five questions about viruses and microRNAs. PLoS Pathog. 2010;6:e1000787. doi: 10.1371/journal.ppat.1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JS, Langlois RA, Pham AM, Tenoever BR. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA. 2012;18:1338–1346. doi: 10.1261/rna.032268.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, et al. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 2010;6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Kolli D, Ren J, Liu T, Garofalo RP, Casola A. Human metapneumovirus glycoprotein G disrupts mitochondrial signaling in airway epithelial cells. PLoS One. 2013;8:e62568. doi: 10.1371/journal.pone.0062568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Liu T, Shan Y, Li K, Garofalo RP, Casola A. Human metapneumovirus glycoprotein G inhibits innate immune responses. PLoS Pathog. 2008;4:e1000077. doi: 10.1371/journal.ppat.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Wang Q, Kolli D, Prusak DJ, Tseng CT, Chen ZJ, et al. Human metapneumovirus M2-2 protein inhibits innate cellular signaling by targeting MAVS. J Virol. 2012;86:13049–13061. doi: 10.1128/JVI.01248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa Y, Zhou M, Yamaguchi M, Komatsu T, Takeuchi K, Itoh M, et al. Human metapneumovirus M2-2 protein inhibits viral transcription and replication. Microbes Infect. 2010;12:135–145. doi: 10.1016/j.micinf.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Biacchesi S, Pham QN, Skiadopoulos MH, Murphy BR, Collins PL, Buchholz UJ. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2-2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J Virol. 2005;79:12608–12613. doi: 10.1128/JVI.79.19.12608-12613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ. Antagonism of NF-kappaB-up-regulated micro RNAs (miRNAs) in sporadic Alzheimer's disease (AD)-anti-NF-kappaB vs. anti-miRNA strategies. Front Genet. 2013;4:77. doi: 10.3389/fgene.2013.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassishin L, Loudig O, Bauman A, Shafit-Zagardo B, Suh HS, Lee SC. Interferon regulatory factor 3 inhibits astrocyte inflammatory gene expression through suppression of the proinflammatory miR-155 and miR-155*. Glia. 2011;59:1911–1922. doi: 10.1002/glia.21233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg NJ, Hayward SL, Crowe JE. Respiratory syncytial virus regulates human microRNAs by using mechanisms involving beta interferon and NF-κB. MBio. 2012;3 doi: 10.1128/mBio.00220-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Sen CK. MiRNA in innate immune responses: novel players in wound inflammation. Physiol Genomics. 2011;43:557–565. doi: 10.1152/physiolgenomics.00160.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, et al. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- Moens U. Silencing viral microRNA as a novel antiviral therapy. J Biomed Biotechnol. 2009;2009:419539. doi: 10.1155/2009/419539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TA, Xia J, Johnson LS, Zhou X, Zhang W, Virgin HW. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J Virol. 2010;84:10344–10353. doi: 10.1128/JVI.00707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JC, Brown JW. The RNase P family. RNA Biol. 2009;6:362–369. doi: 10.4161/rna.6.4.9241. [DOI] [PubMed] [Google Scholar]
- Bao X, Sinha M, Liu T, Hong C, Luxon BA, Garofalo RP, et al. Identification of human metapneumovirus-induced gene networks in airway epithelial cells by microarray analysis. Virology. 2008;374:114–127. doi: 10.1016/j.virol.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall-Levin M, Rissland OS, Johnston WK, Perrimon N, Bartel DP, Berger B. Unusually effective microRNA targeting within repeat-rich coding regions of mammalian mRNAs. Genome Res. 2011;21:1395–1403. doi: 10.1101/gr.121210.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J, Berkhout B. RNAi and cellular miRNAs in infections by mammalian viruses. Methods Mol Biol. 2011;721:23–41. doi: 10.1007/978-1-61779-037-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuddenham L, Pfeffer S. Roles and regulation of microRNAs in cytomegalovirus infection. Biochim Biophys Acta. 2011;1809:613–622. doi: 10.1016/j.bbagrm.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Dhuruvasan K, Sivasubramanian G, Pellett PE. Roles of host and viral microRNAs in human cytomegalovirus biology. Virus Res. 2011;157:180–192. doi: 10.1016/j.virusres.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CR, Marsh GA, Jenkins KA, Gantier MP, Tizard ML, Middleton D, et al. Promotion of Hendra virus replication by microRNA 146a. J Virol. 2013;87:3782–3791. doi: 10.1128/JVI.01342-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othumpangat S, Walton C, Piedimonte G. MicroRNA-221 modulates RSV replication in human bronchial epithelium by targeting NGF expression. PLoS One. 2012;7:e30030. doi: 10.1371/journal.pone.0030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow P, et al. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol. 2010;53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- Jin H, Lv S, Yang J, Wang X, Hu H, Su C, et al. Use of microRNA Let-7 to control the replication specificity of oncolytic adenovirus in hepatocellular carcinoma cells. PLoS One. 2011;6:e21307. doi: 10.1371/journal.pone.0021307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman KL, Sarnow P. Modulation of hepatitis C virus RNA abundance and the isoprenoid biosynthesis pathway by microRNA miR-122 involves distinct mechanisms. J Virol. 2010;84:666–670. doi: 10.1128/JVI.01156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz P, Rödelsperger C, Jäger M, Jostins L, Bauer S, Robinson PN. Microindel detection in short-read sequence data. Bioinformatics. 2010;26:722–729. doi: 10.1093/bioinformatics/btq027. [DOI] [PubMed] [Google Scholar]
- Hultquist JF, McDougle RM, Anderson BD, Harris RS. HIV type 1 viral infectivity factor and the RUNX transcription factors interact with core binding factor β on genetically distinct surfaces. AIDS Res Hum Retroviruses. 2012;28:1543–1551. doi: 10.1089/aid.2012.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Yang Z, Skogerbo G, Ren F, Cui H, Zhao H, et al. The properties and functions of virus encoded microRNA, siRNA, and other small noncoding RNAs. Crit Rev Microbiol. 2008;34:175–188. doi: 10.1080/10408410802482008. [DOI] [PubMed] [Google Scholar]
- Ren J, Kolli D, Liu T, Xu R, Garofalo RP, Casola A, et al. Human metapneumovirus inhibits IFN-beta signaling by downregulating Jak1 and Tyk2 cellular levels. PLoS One. 2011;6:e24496. doi: 10.1371/journal.pone.0024496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolli D, Bao X, Liu T, Hong C, Wang T, Garofalo RP, et al. Human metapneumovirus glycoprotein G inhibits TLR4-dependent signaling in monocyte-derived dendritic cells. J Immunol. 2011;187:47–54. doi: 10.4049/jimmunol.1002589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Kolli D, Liu T, Shan Y, Garofalo RP, Casola A. Human metapneumovirus small hydrophobic protein inhibits NF-kappaB transcriptional activity. J Virol. 2008;82:8224–8229. doi: 10.1128/JVI.02584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of hMPV-derived sncRNAs.
Let-7f validation.
miRNA with total sequencing reads >500, and fold induction ≥1.5 or fold reduction ≤0.7 by hMPV at 6 hours postinfection.