Abstract
Connexin43 (Cx43) is the most widely and abundantly expressed gap junction (GJ) protein and it is strongly associated with the regulation of cell cycle progression. Emerging roles for Cx43 in cell adhesion and migration during neural differentiation have also been recently recognized, and this has emphasized the involvement of Cx43 in different physiological process beyond its role as a GJ protein. In this study, we explore the function of Cx43 in the differentiation of human neural progenitor cells (hNPCs) using viral vectors that mediate the overexpression or knockdown of the protein. Results showed that in the absence of this protein fetal cortex-derived hNPCs differentiated toward a neuronal phenotype at expenses of a glial phenotype. Furthermore, the silencing of Cx43 did not affect hNPC proliferation rate or numbers of apoptotic cells. The increase in the number of neurons was not recapitulated when GJ intercellular communications were pharmacologically blocked, and this suggested that Cx43 was influencing hNPCs differentiation with a GJ-independent effect. In addition, Cx43 knockdown significantly increased β-catenin signaling, which has been shown to regulate the transcription of pro-neuronal genes during embryonic neural development. Our results add further support to the hypothesis that Cx43 protein itself regulates key signaling pathways during development and neurogenesis beyond its role as GJ protein.
Keywords: Connexin43, shRNA, neural stem cells, differentiation, β-catenin
Connexin43 (Cx43) protein has been found to be expressed early during mouse embryogenesis, at the preimplantation stages.1 Indeed when organogenesis occurs, the protein has been reported to be preferentially upregulated in the ectoderm rather than mesodermal and endodermal layer.2 Interestingly during neurogenesis, Cx43 is both temporally and spatially regulated with a prominent expression in neural progenitor cells followed by a decrease when neurons start to differentiate.3 The presence of Cx43 at the early stage of development together with its dual regulation during neurogenesis strongly suggest this protein is greatly involved in development events and the precise regulation of the protein expression maybe crucial to ensure a proper maturation. Cx43 is one of the proteins that form Gap junction (GJ) channels between adjacent cells, and this type of cellular communication is critical to several processes ranging from tissue homeostasis and synchronization of cellular activity, to the establishment and maintenance of cell microenvironments or niches.4 Several Cx proteins have been identified that form GJs, intriguingly Cx43 is the most widely and abundantly expressed isoform with a unique long C-terminal (CT) tail. This has been reported to be the binding site of multiple proteins, which dynamically interact, and participate in the regulation of physiological events (for review, see Giepmans5). The importance of Cx43 in regulating cell adhesiveness and directionality of migration has recently been described (for review, see Matsuuchi and Naus6). Cx43 is present on radial glia fibers at the points where they contact with migrating neurons7 and Cx43 conditional knockout mice show delayed neuronal migration8 together with impairment in the lamination of the neocortex.9 A brain transcriptome analysis identified gene transcripts altered in Cx43 null and Cx43 heterozygous mice, highlighting a change in proteins regulating cell growth, neuronal survival and differentiation.10, 11, 12 Therefore, it is clear that together with physically mediating neurogenesis events, Cx43 might also act as a central role in the control of growth.11
Neural stem cells have been identified in both the developing and adult nervous system.13, 14, 15, 16, 17 In this study, we use neural progenitor cells (NPCs) derived from human fetal brain to unravel the role of Cx43 in neuronal differentiation. We found that Cx43 protein knockdown increased the number of neurons differentiated from human NPCs (hNPCs). We further demonstrated that the knockdown of the protein enhanced the transcriptional activity of β-catenin, which is a key mediator of the canonical WNT pathway and it has been shown to regulate transcription of pro-neuronal genes.18, 19, 20
Results
Human Cx43 expression in an in vitro model of neurogenesis
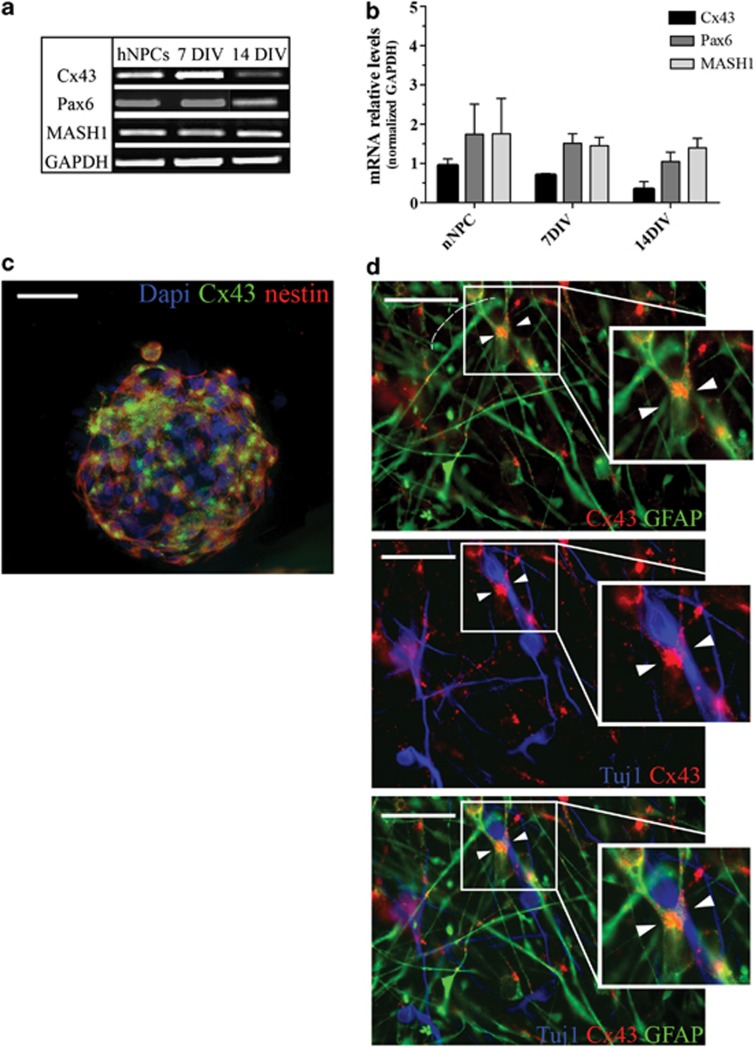
We analyzed the spatiotemporal expression of the human Cx43 (hCx43) protein in hNPCs. Those cells provide a valuable source of neural tissue and an in vitro model for studying neurogenesis.21 RT-PCR was performed in multipotent hNPCs, as well as following 7 and 14 days differentiation. Results indicated a specific pattern of Cx43 mRNA expression during hNPCs differentiation with higher levels of Cx43 mRNA in undifferentiated and 7 days differentiated cells (0.93±0.2; 0.69±0.1), followed by a decrease at 14 days of differentiation (0.3±0.2; Figures 1a and b). There was no change in the mRNA levels of the neural markers Mash1 and Pax6 between undifferentiated and 7 days differentiated hNPCs (undifferentiated Pax6 1.7±0.8, MASH1 1.7±0.9; 7 days differentiated Pax6 1.5±0.2, MASH1 1.4±0.2). At 14 days of differentiation, we observed a small reduction in Pax6 expression (1.0±0.2), whereas Mash1 levels remained consistent with previous expression (1.4±0.3; Figures 1a and b). Those results suggested that as differentiation progressed cell were maintaining a neural phenotype with a decrease in Cx43 expression.
Figure 1.
Cx43 expression in an in vitro model of neurogenesis. (a) Representative RT-PCR bands of undifferentiated, 7 and 14 days differentiated hNPCs for Cx43, Pax6 and MASH1 mRNAs. GAPDH is used as housekeeping gene. (b) Quantification of RT-PCR bands for hCx43, Pax6 and MASH1 genes. Values are the means of three independent experiments and error bars represent S.E.M. Values were expressed as arbitrary units normalized to the GAPDH values. (c) Fluorescence microscopy of undifferentiated hNPCs stained with Cx43 (green), nestin or Tuj1 (red) antibodies. Cell nuclei are indicated by DAPI (blue) staining. Scale bar 50 μm. (d) Higher magnification of the area around 7 days differentiated sphere. Cx43 (red) on GFAP (green)-positive cells expressed at the contact points with migrating Tuj1 (blue) neurons (arrowheads in boxed areas). Scale bar 50 μm
Multipotent hNPCs revealed an intense Cx43 immunolabeling throughout the neurosphere, these cells were all undifferentiated neuroepithelial cells as indicated from the nestin staining (Figure 1c). Following 7 days of differentiation, immunostaining revealed presence of Tuj1 and GFAP-positive cells with the Cx43 protein specifically localized at the contact points of migrating neurons and GFAP-positive process (Figure 1d).
Lentiviral vector mediated specific knockdown of hCx43
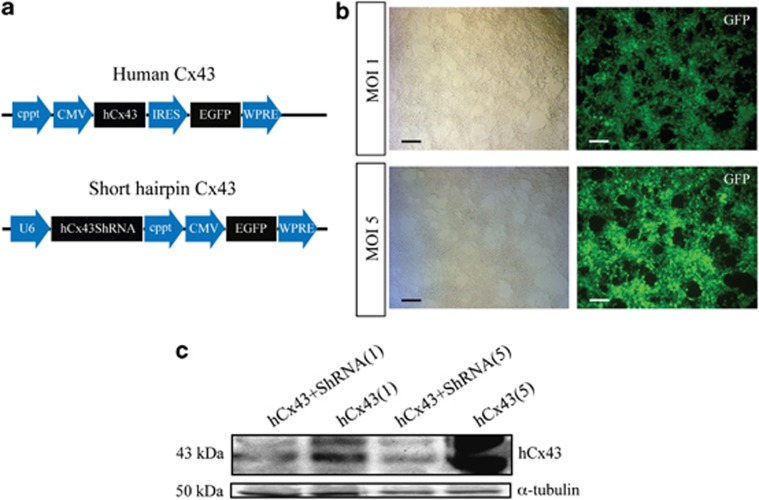
To induce a specific hCx43 protein knockdown, we designed four short hairpin RNA (shRNA) constructs (Supplementary Table 1) and cloned them into pCR 2.1. TOPO. The efficiency of knockdown mediated by each construct was tested in HEK293 cells by co-transfection of shRNA plasmid together with an hCx43-overexpressing vector. This co-transfection was required as HEK293 cells do not express detectable levels of endogenous Cx43 protein. Immunoblot analysis was performed to detect the shRNA sequence allowing the maximum hCx43 protein knockdown (Supplementary Figures 2A and B). Subsequently, the shRNA sequence mediating the most effective knockdown was cloned into a lentiviral vector backbone to generate viral particles (Figure 2a). Viral efficacy was tested using multiplicity of infections (MOIs) of 1 and 5 in HEK293 cells. Again an hCx43shRNA virus was used together with an hCx43-overexpressing virus (Figures 2b and c). High transduction efficiency was obtained at both MOIs tested (Figure 2b), however, western blot results revealed a higher hCx43 protein knockdown at MOI 5 (Figure 2c).
Figure 2.
Validation of lentiviral expression cassette. (a) Schematic of lentiviral vector expression cassette for hCx43 and hCx43ShRNA. (b) Bright field (left panels) and EGFP expression (right panels) of viral MOI 1 and 5 transduced HEK293 cells. Scale bar 50 μm. (c) Western blot bands of hCx43 protein in viral MOI 1 and 5 transduced HEK293 cells
Knockdown of hCx43 regulates hNPC neuronal differentiation
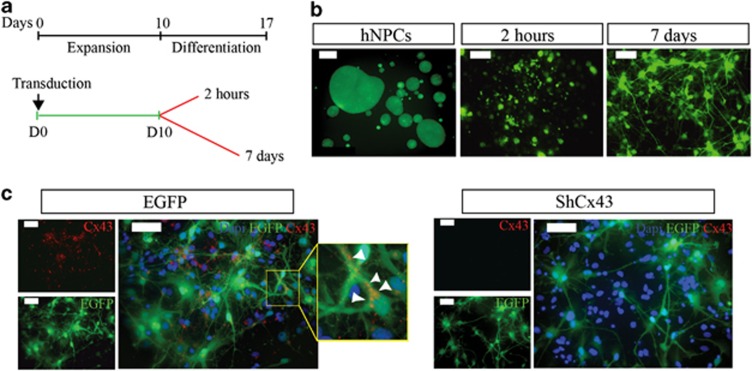
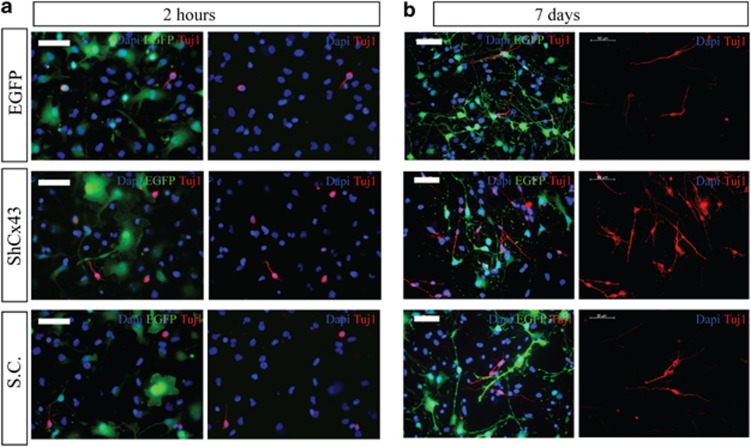
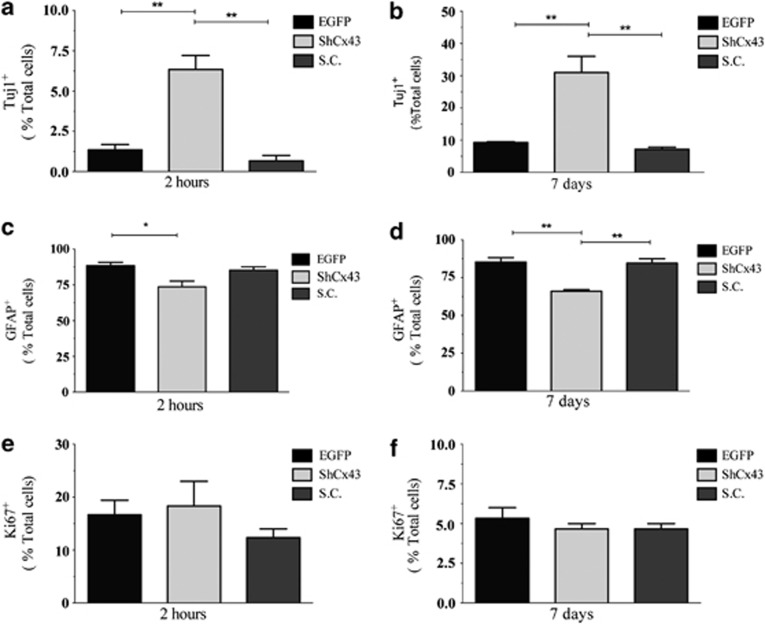
We sought to investigate the contribution of hCx43 to neurogenesis by knocking down this protein in hNPCs and then inducing their differentiation. Figure 3a illustrates the experimental model used in which viral transduced hNPCs were expanded for 10 days from the day of lentiviral vector transduction followed by differentiation, which was carried out for 2 h and for 7 days, respectively. Strong enhanced green fluorescent protein (EGFP) fluorescence was detectable at all-time points tested (Figure 3b) indicating continuous shRNA expression. Immunocytochemistry performed in 7 days differentiated cells revealed a robust knockdown of hCx43 protein compared with EGFP control (Figure 3c). Quantification of nestin-positive cells after 2 h of differentiation revealed that the majority of cells still retained an immature neural phenotype (Supplementary Figure 3A). The shRNA-mediated knockdown of hCx43 resulted in a significant increase in the number of Tuj1-positive cells compared with cells transduced with either EGFP or a scrambled control (SC; Figures 4a and b and Figures 5a and b) for both 2 h (shCx43 6±0.9% EGFP 2±0.3% SC 1±0.2% P≤0.01) and 7 days (shCx43 31.4±5.0% EGFP 9.1±0.4% SC 7.0±0.7% P≤0.01) differentiation. The number of GFAP-positive cells was significantly reduced in cells differentiated for 2 h compared with EGFP control (shCx43 74±4.0% EGFP 88±2.1% SC 86.0±2.2% P≤0.05) and in 7 days differentiated cells compared with EGFP and to SC transduced cells (shCx43 66±1.0% EGFP 85.6±2.8% SC 84.4±3.0% P≤0.01), (Figures 5c and d). Characterization of neurons derived from 7 days differentiated hNPCs revealed a great majority of GABA-positive neurons (Supplementary Figure 4). However, hCx43 knockdown did not affect the number of actively dividing cells as indicated by Ki67 expression (Figures 5e and f). As we detected an alteration in cell fate commitment already at 2 h of hNPCs differentiation, we decided to investigate whether Cx43 knockdown could affect the viability and proliferation at this earlier stage. To assess cell proliferation, 5′-bromo-2-deoxyuridine (BrdU) was added to hNPC medium at the moment of viral transduction and left in culture for the expansion time (10 days). Cells were differentiated for 2 h and analysis indicated that hCx43 absence did not affect the proliferation (Supplementary Figure 3B). For cell viability analysis, TUNEL assay was performed on cells differentiated for 2 h, and this also revealed no difference between conditions (Supplementary Figure 3C). Finally, morphometric analysis of neurons derived from hCx43 and control transduced hNPCs did not reveal any change in neuronal complexity between groups (Supplementary Figure 5), indicating that the absence of Cx43 increased the number of differentiated neurons, however, it did not advance neuronal maturation.
Figure 3.
Experimental model and validation of ShRNAhCx43 virus in neural stem cell culture. (a) Schematic representation of experimental method used to transduce hNPCs. hNPCs were transduced at day 0 (D0) and expanded until D10 (green line). Subsequently, they were differentiated for 2 h or 7 days, respectively, (red lines). (b) EGFP expression in undifferentiated, 2 h and 7 days differentiated hNPCs. Scale bar 50 μm. (c) Fluoresce microscopy for Cx43 (red) and EGFP (green) of 7 days differentiated hNPCs in EGFP and shRNAhCx43 transduced cells. Cell nuclei are indicated by DAPI (blue) staining. Scale bar 50 μm
Figure 4.
Tuj1 expression in viral transduced hNPCs and differentiated for 2 h or 7 days. Fluorescence microscopy of viral transduced hNPCs 2 h (a) and 7 days (b) differentiated cultures. Left panels represent EGFP protein (green) expressing cells stained for Tuj1 (red). Cell nuclei are indicated by DAPI (blue) staining. Right panels indicate Tuj1 (red) and DAPI (blue) staining only. Scale bar 50 μm
Figure 5.
Quantification of Tuj1, GFAP and Ki67 expression in viral transduced hNPCs. (a and b) Number of Tuj1-expressing cells in 2 h (a) and 7 days (b) differentiated cultures. (c and d) Number of GFAP-expressing cells in 2 h (c) and 7 days (d) differentiated cultures. (e and f) Number of Ki67-expressing cells in 2 h (e) and 7 days (f) differentiated cultures. Values are the means of three independent experiments and error bars represent S.E.M. Values were expressed as % on total cell number. *P≤0.05, **P≤0.01
Increase in neuronal differentiation is caused by the absence of hCx43 protein
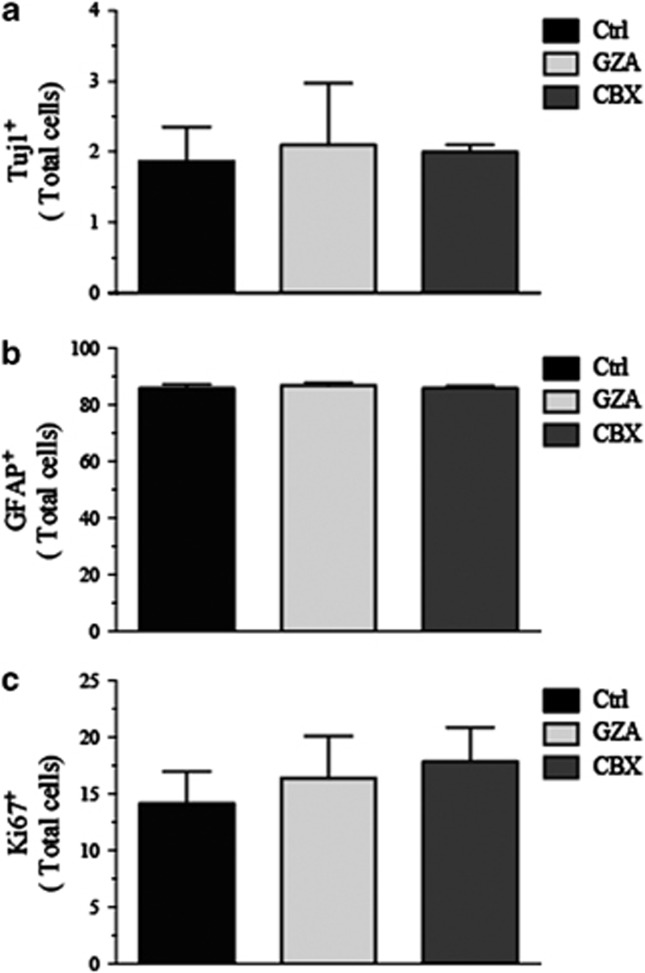
In order to evaluate whether the increase in neuronal number was due to the specific absence of hCx43 protein or to a general reduction of hCx43-mediated GJ intercellular communication, we used a pharmacological blockade of GJ using a well-established GJ blocker carbenoxolone (CBX).22 CBX used at 50 μM largely impaired the spread of GJ permeable dye Cascade blue (Supplementary Figure 6), thus we incubate hNPCs for 10 days with CBX and we used its inactive analog glycyrrhizic acid (GZA) as control. Cells were differentiated for 2 h and results showed in Figure 6 indicated that CBX did not affect the number of Tuj1 or GFAP-positive cells (Figure 6a and b), nor cell proliferation (Figure 6c). Thus, these results suggested that the effect on hNPCs neuronal fate was independent of GJ intercellular communications, but dependent on the expression of the hCx43 protein itself.
Figure 6.
Quantification of Tuj1, GFAP and Ki67 expression in hNPCs expanded in the presence of CBX. Number of Tuj1 (a), GFAP (b) and Ki67 (c) expressing cells in 2 h differentiated cells. GJs were blocked by addition of CBX 50μM. GZA was used as inactive analog of CBX. Values are the means of three independent experiments and error bars represent S.E.M. Values were expressed as % on total cell number
hCx43 contributes to the regulation of β-catenin transcription
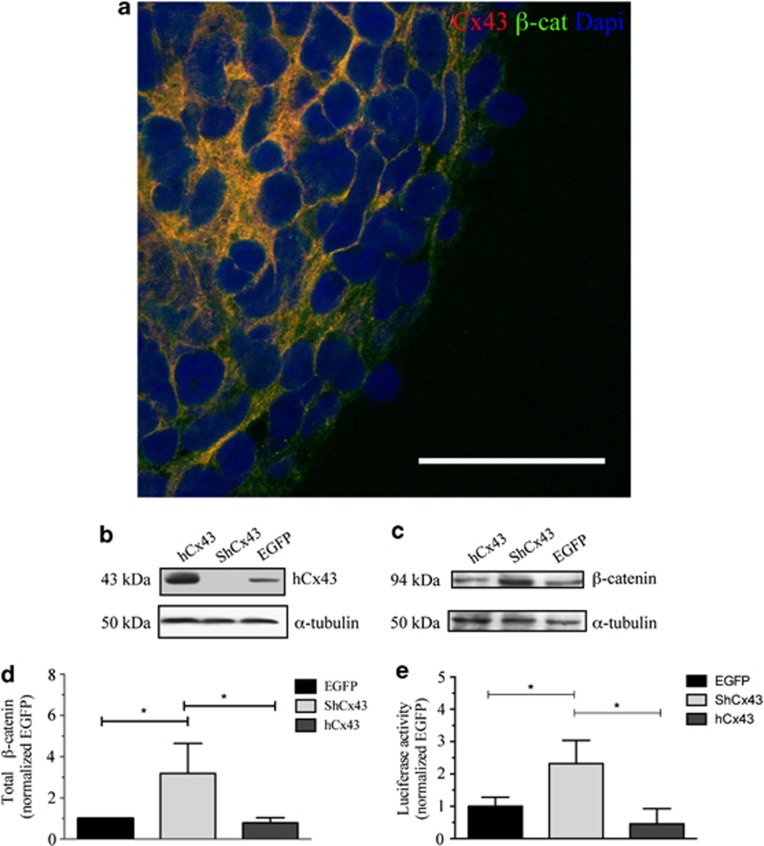
We confirmed a role for hCx43 in hNPCs differentiation and we also demonstrated that the protein itself was responsible for the increase in the number of neurons and a concomitant decrease in glial cells. We next wanted to investigate a possible contribution of hCx43 to the canonical Wnt/β-catenin pathway, which has been shown to be involved in many aspects of embryonic development and neurogenesis.18, 19, 20 In addition, β-catenin has also been reported to regulate hCx43 transcription.23 We first performed immunocytochemistry to determine the levels of hCx43 and β-catenin proteins in hNPCs derived neurospheres, which had been differentiated for 2 h. Results revealed intense immunolabeling of both proteins, which was mainly localized in the cell membrane (Figure 7a). We then assessed if the knockdown of endogenous hCx43 or its forced expression was affecting β-catenin protein expression. We transduced hNPCs with our shCx43 or our viral vector containing the hCx43 full-length sequence or our EGFP control and we collected proteins for immunoblot analysis. Results indicated a significant increase in β-catenin protein expression when hCx43 was silenced (Figures 7b–d; shCx43 3.2±0.83; Cx43 0.8±0.14; EGFP 1±0.0; P≤0.01). Then, we tested the hypothesis that in the absence of hCx43 the increase in β-catenin protein expression observed was due to an increase in the uncomplexed form of the protein. Free cytoplasmic β-catenin accumulates in the cytoplasm and then translocates to the nucleus where it mediates gene transcription binding to the TCF/LEF transcription factor motifs. Therefore, to measure β-catenin transcriptional activity we used a well-established gene reporter assay.24 In the assay, a plasmid containing the TCF/LEF-binding site upstream of a Firefly luciferase ORF (Addgene, Cambridge, MA, USA) is used (TOPFLASH), together with a mock control plasmid (FOPFLASH). In the absence of hCx43, we found a significant increase in TCF/LEF luciferase activity when stimulated with WNT1 agonist CHIR (6-[[2-[[4-(2,4-dichlorophenyl)-5-(5-methyl-1H-imidazol-2-yl)-2-pyrimidinyl]amino]ethyl]amino]-3-pyridinecarbonitrile) at 1.5 μM25 (Figure 7e), compared with control EGFP transduced cells (shCx43 2.2±0.7; EGFP 1.1±0.3) and compared with hCx43-overexpressing cells (shCx43 2.2±0.7; hCx43 0.6±0.4; P≤0.05). Interestingly, we also found a significant decrease in TCF/LEF luciferase activity when hCx43 was overexpressed compared with EGFP control (hCx43 0.6±0.4; EGFP 1.1±0.3; P≤0.05). Thus, we can conclude that the knockdown of Cx43 protein increased β-catenin protein transcriptional activity, which enhanced neuronal differentiation.
Figure 7.
hCx43 protein knockdown increases β-catenin transactivation. (a) Confocal microscopy of 2 h differentiated hNPCs. Cells were stained for β-catenin (red) and Cx43 (green) proteins. Cell nuclei are indicated by DAPI (blue) staining. Scale bar 50 μm. (b and c) Representative western blot bands of hCx43 (b) and β-catenin proteins (c) in viral transduced hNPCs. α-Tubulin was used as internal control. (d) Quantification of β-catenin protein expression by densitometry. Values were normalized to values of internal control. *P≤0.05 (e) Quantification of active β-catenin by luciferase gene reporter assay. Values of transcriptional active β-catenin were determined by the level of luciferase activity and normalized to the internal control (Renilla luciferase plasmid). Values are the means of three independent experiments and error bars represent S.E.M. Values expressed as relative activation compared with cells transfected with a β-catenin-LEF/TCF insensitive (FOP) reporter vector. *P≤0.05
Discussion
Cx proteins are differently regulated during neurogenesis.26, 27, 28, 29, 30 Cx43 is one of the major isoforms in the developing brain,31 being highly expressed in (E10.5) multipotent neural stem cells32 and in proliferating cells during development.27 Furthermore, its levels decrease as NPCs differentiate into neurons,33 thus suggesting Cx43 has an important role in neurogenesis. This is a very dynamic process orchestrated by proliferation and differentiation of NPCs, which contribute to the development and maturation of a highly complex neural network. We used human NPCs isolated from the developing cortex as an in vitro model of human neurogenesis.34 hNPCs are EGF and FGF-2 responsive and can be expanded for >200 days in culture while retaining the ability to differentiate into neurons and glial cells.35 In this study, we confirmed that the expression of Cx43 was found in undifferentiated hNPCs but decreased as differentiation progressed out to 14 days. This is in keeping with what has been reported for the rodent brain.33 Following 7 days of differentiation, Cx43 was only present in GFAP-positive cells, indicating that when cells start to differentiate Cx43 expression is restricted to the glial phenotype. Interestingly, we found Cx43 punctate immunostaining at contact points between GFAP fibers and Tuj1-positive cells. This was in line with other rodent studies in which Cx43 was found to mediate tangential and radial migration of newborn neurons from the ventricular zone to the cortical plate.7, 30, 36 We designed and validated an shRNA construct to specifically knockdown hCx43 protein expression and then we generated lentiviral particles to deliver this construct into hNPCs. Cx43 knockdown resulted in a significant increase in the number of neurons derived from hNPCs both after 2 h and 7 days of differentiation. Interestingly, we also found a significant decrease in the number of GFAP-positive cells at the two time points examined. In vivo neurogenesis involves differentiation of neurons first, followed by glial cells,37 which suggests that in our system the increase in the number of neurons might reduce the pool of progenitor cells available to acquire an astrocytic phenotype at later time points. Alternatively as Cx43 in the mature brain is only expressed in astrocytes, it could be possible that the absence of the protein impaired or delayed astrocytic differentiation. Morphometric analysis revealed that the increase in neuronal phenotype when Cx43 was silenced did not correspond to an increase of neuronal complexity including cell body size, number and length of neurites. This suggests that this protein is not involved in the process of neuronal maturation, findings similar to what we recently reported for Cx36 in neurons derived from rat NPCs.38 In addition, we show that knockdown of Cx43 does not affect cell proliferation or cell death. Taken together, these results suggest that Cx43 knockdown increases neuronal commitment rather than influencing neuronal complexity, cell death or NPC proliferation.
Similarly, in vivo models of neurogenesis have shown that hCx43 is highly expressed at early stages of neurogenesis30, 39 and Cx43 has been reported to mediate GJ coupling between neuroblasts in the hippocampus in mice.26 However, as progenitors start to differentiate, the degree of cellular communication together with Cx43 protein expression is decreased.29 The reduction in Cx43 expression and consequent disruption in neural progenitor coupling as differentiation occurred, suggests that Cx43 and GJ intracellular communication are important in the early stages of neurogenesis to maintain progenitors in an undifferentiated.29 Therefore, we investigated if the increased neuronal fate commitment we observed following hCx43 knockdown was due to a decrease in cellular communications or to the absence of the protein itself. Pharmacological blockade of GJICs with a generic GJ blocker (CBX) did not recapitulate the effects of Cx43 knockdown, suggesting a channel-independent role of Cx43 in the regulation of neuronal differentiation. Previous studies have established that the Cx43 adhesive, scaffolding and cell signaling-mediating properties are active factors during neurogenesis.7, 8, 9, 36, 40, 41 Interestingly, Cx43-CT region contains multiple phosphorylation sites and binding domains, which not only modulate Cx43 protein functions but also allow Cx43 interaction with cytoskeletal elements and other cell membrane proteins including ZO1-2, adherens junction (AJ) proteins, cadherins and β-catenin (reviewed in Matsuuchi and Naus6). β-Catenin signaling has a key role in neuronal differentiation,18 indeed, β-catenin through the TCF/LEF complex regulates the transcription of genes involved in cell adhesion, proliferation and differentiation such as E-cadherin, neuronal cell adhesion molecule, c-MYC and cyclin D1.42, 43 Furthermore, Cx43 promoter contains three TCF/LEF binding sites.23 We stained Cx43 and β-catenin in hNPCs differentiated for 2 h and found that both proteins were abundantly expressed. We also detected areas in which the two proteins were colocalized, indicating a possible functional cross-regulation. Further experiments showed an increase in β-catenin protein expression following hCx43 knockdown and more importantly no changes in β-catenin were observed using different shRNA constructs, confirming direct link between hCx43 knockdown and β-catenin protein expression. We also confirmed that the transcriptional active form of the β-catenin protein was enhanced in the absence of hCx43. In the cell, β-catenin is found at the cell membrane in complex with E-cadherin to form AJs. In the absence of WNT ligands, cytoplasmic β-catenin levels are kept low as the protein is phosphorylated by glycogen synthase kinase 3b (GSK-3b) and then targeted for ubiquitination and degradation. The activation of WNT pathway induces β-catenin dephosphorylation, nuclear translocation and the formation of a transcriptionally active complex with LEF and TCF sites. In cardiac cells, a complex of Cx43/β-catenin proteins have been described at the cell membrane following stimulation of WNT pathway.44 This evidence together with the presence of β-catenin binding sites in the Cx43 promoter region raised the possibility of a dual regulation between the two proteins. Cx43 may serve to negatively regulate its own transcription through β-catenin, as well as participate in the regulation of other β-catenin targets such as pro-neuronal genes18, 19, 20 by sequestering β-catenin to the cell membrane. Thus, in hNPCs, hCx43 knockdown disrupted the protein junctional complex with β-catenin leading to an increase in β-catenin nuclear translocation, transcription of pro-neuronal genes and an increase in the number of neurons. A dual regulation between gene transcription and cell adhesion molecules has already been described in a model proposed by Orsulic et al.,45 in which E-cadherin antagonized β-catenin signaling activity. In conclusion, in our investigation we have provided evidence for an interaction occurring between Cx43 and β-catenin proteins during neurogenesis. Furthermore, our results suggest a novel approach to increase the number of neurons from hNPCs, opening novel frontiers for research applications.
Materials and Methods
Design and construction of hCx43 shRNA lentiviral vector
The hCx43 sequence (accession no. NM.000165.3) was used to generate four hCx43 shRNAs (Supplementary Table 1). Sh expression cassette comprised a sense and an antisense (antisense specific to the target mRNA) strand spaced by a loop sequence, a RNA polymerase III transcription termination signal and XhoI restriction site. A sequence coding for the mouse U6 promoter was inserted upstream of the antisense strand (Supplementary Figure S1A). A PCR-based method was used to generate double-stranded shRNA expression cassettes using the pSilencer 1.0-U6 expression vector (Life Technologies, Carlsbad, CA, USA) as a transcription template and reverse primer encoding the shRNA oligonucleotides. shRNA constructs were cloned into pCR2.1-TOPO vector as previously described46 and validated for specific hCx43 protein knockdown (Supplementary Figures S2A and B). Successful shRNA cassette was then excised by digestion with EcoRI and inserted into the lentiviral backbone plasmid via MfeI site generating the pRRLsincppt.U6.hCx43shRNA.CMV.EGFP.wpre construct (Figure 2a).
Construction of lentiviral vectors expressing hCx43, EGFP or SC
The hCx43 coding sequence was excised from pDest40.hCx43 plasmid (GlaxoSmithKline, Harlow, UK) using NdeI and AgeI restriction endonucleases and cloned into pRRLsincppt.CMV.polyAB.IRES2.EGFP.wpre construct via NdeI and AgeI sites generating pRRLsincppt.CMV.hCx43.IRES2.EGFP.wpre (Figure 2a).
Lentiviral vector coding for EGFP under CMV promoter pRRLsincppt.CMV.EGFP.wpre and a sh SC sequence under a U6 promoter pRRLsincppt.U6.SC.CMV.EGFP.wpre were constructed as previously described.21
Human neurosphere cell cultures and lentiviral vector transduction
hNPCs were derived from 9 to 10 weeks old (post-menstrual) human fetus in line with the UK Department of Health guidelines and according to the Local Research Ethic Committee approval. hNPCs were expanded as neurospheres in presence of growth factors EGF and FGF-2 (20 ng/ml) and heparin (5 μg/ml) as previously described.34 Neurospheres were expanded for a maximum of four passages (2 months) before being transduced with lentiviral vectors. This was performed in order to eliminate any primary cells from the culture. To induce differentiation, neurospheres were dissociated with Accutase (Sigma-Aldrich, St. Louis, MO, USA) and plated onto poly-D-lysine/laminin-coated glass coverslips. Medium consisted in DMEM/F12 (7 : 3; GIBCO, Life Technologies), 2% B27 and 1% penicillin/streptomycin as previously described.35 Cell differentiation was carried out for 2 h and 7 days, respectively. For lentiviral vector transduction, neurospheres were dissociated into single cells using Accutase (37 °C 15 min; Sigma) and seeded at 105 cells per well in poly-HEMA (120 g/l) coated 96-well plate. Viruses were added while cells were in single-cell suspension using a viral MOI of 5. Medium was doubled the following day and cells were transferred to a 12-well plate at 4 days after transduction and expanded for further 6 days before differentiation.
Immunocytochemistry, BrdU and TUNEL assays
Immunocytochemistry was performed on cells fixed with 4% PFA (15 min). Cells were blocked for 2 h in 10% goat serum, 2% BSA in 0.1%Triton-X100 PBS solution and then processed for primary and secondary antibodies immunoreaction. Primary and secondary antibodies are detailed in Supplementary Table 2. BrdU incorporation analysis was performed in cells differentiated for 24 h. Briefly, 0.2 μM BrdU (Sigma) was added in the expansion medium at the time of viral transduction. Following cell fixation, cells were permeabilized with ice-cold methanol for 20 min at −20 °C and then treated with 2 N HCl for 20 min at 37 °C HCl (1 M, 30 min). Subsequently, cells were washed in borate buffer (0.1 M, 12 min) and blocked for 2 h in 10% goat serum and 2% BSA. Incubation with anti-BrdU antibody (Supplementary Table 2) was performed overnight at 4 °C. TUNEL cell death assay was carried out following the manufacturer's guidelines (Roche, Mannheim, Germany).
Western blot and densitometry analysis
For western blot, cells were lysed in RIPA buffer (50 mM Tris-HCL, pH7.4; 150 mM NaCl; 1% Triton-X100; 1% sodium deoxycholate; 0.1% sodium dodecyl sulfate supplemented with protease and phosphatase inhibitors (Roche). Protein concentration was determined by a BCA protein assay (Thermo Scientific, Waltham, MA, USA) and 30 μg of protein were resolved on 10% SDS-Tris glycine gel. Proteins were blotted onto PVDF membranes (Roche) and membranes blocked in 10% non-fat dry milk in 0.1% Tween-20 (Sigma) TBS overnight. Primary and secondary (HRP conjugated; Amersham, GE Healthcare, Little Chalfont, UK) antibodies were prepared in 5% non-fat dry milk in 0.1% Tween-20 (Sigma) TBS buffer (Supplementary Table 2). Protein bands were visualized with ECL plus Western blotting detection reagent (Amersham). Autoradiographic film (Amersham) was exposed to the membrane and developed using a Kodak (Rochester, NY, USA) film processor. Densitometry analysis was carried out using ImageJ free image processing software (http://rsb.info.nih.gov/ij/) and protein values were quantified and normalized to the alpha-tubulin internal control signals. The mean of three individual experiments was calculated.
Reverse transcription and PCR
RNA was isolated from hNPCs or differentiated cells using the RNeasy mini system (Qiagen, Hilden, Germany). Isolated RNA was treated with RNAse-free DNAseI (Roche) to remove genomic DNA. Samples were cleaned up using the RNeasy minelute kit (Qiagen) and RNA quantity and quality was determined by spectrophotometry. RNA (1 μg) was retro-transcribed using the Superscript III system (Life Technologies) according to manufacturer's instructions. PCR was carried out using the GoTaq system (Promega, Madison, WI, USA) according to manufacturer's instructions. Primer sequences are indicated in Supplementary Table 3. PCR products were separated on 1.5% agarose gel containing 200 μg/ml ethidium bromide (Sigma), and visualized using a GelDoc-IT UV imaging system (UVP, Upland, CA, USA). PCR band intensity was analyzed by densitometry using ImageJ software and mRNA levels were normalized to the GAPDH mRNA. The mean of three different experiments was plotted and transcript level expressed as arbitrary units.
Pharmacological blockade of GJ intercellular channels
hNPCs were treated with 50 μM of CBX (Sigma) and 50 μM of the CBX inactive analog GZA (Sigma) for 10 days. CBX or GZA containing media was replaced every 3 days to avoid drug depletion. hNPCs were differentiated for 24 h.
Luciferase reporter assay
β-Catenin transcription was assessed using the TOP FLASH/FOP FLASH reporter luciferase assay.24 For TOP FLASH/FOP FLASH reporter luciferase assay, hNPCs were dissociated with Accutase (37 °C 15 min; Sigma) and plated onto poly-D-lysine-coated 24-well plates at 105 cells per well in differentiation medium. WNT pathway was stimulated adding 1.5 μM CHIR (WNT1 agonist, TOCRIS Biosciences, Minneapolis, MN, USA) at the time of plate down and kept in the medium until the cells were harvested. The following day cells were co-transfected using Fugene 6 (Roche) with 0.99 μg of β-catenin-LEF/TCF-sensitive (TOP) or β-catenin-LEF/TCF insensitive (FOP) reporter vector (Addgene) together with 0.1 μg of a constitutively active Renilla luciferase construct (Promega) for normalization of transfection efficiency. Cells were processed 48 h after co-transfection for Luciferase reporter activity using the Dual Luciferase Reporter System (Promega) according to the manufacturer's instructions.
Validation of hCx43 shRNA constructs
Knockdown activity of hCx43 shRNA constructs was tested in HEK293 cells by transient transfection. Briefly, cells were plated at 7 × 104 cells per well and co-transfected with 0.8 μg of hCx43 shRNA constructs together with 0.2 μg of pDest40.hCx43 using Fugene 6 (Roche). Cells were harvested 48 h following transfection and processed for western blot analysis.
Image acquisition and data analysis
Fluorescence microscopy was performed on 4% PFA fixed cells using Leica DM IRB inverted or LEICA DM RB upright microscopes (Leice, Wetzlar, Germany). Images were taken using × 20, × 40 and × 60 lenses. Neuronal cell body area and neurite branching were analyzed using the ImageJ plugin Simple neurite tracer (http://rsb.info.nih.gov/ij/). All experiments were performed at least three times on samples derived from three different fetuses. Values were expressed as mean±S.E.M. P-values were calculated using Graphpad Prism 4 software (GraphPad, San Diego, CA, USA) by one-way ANOVA followed by Newman–Keuls post-hoc test. *P≤0.05, **P≤0.01.
Acknowledgments
This work was supported by an MRC studentship to Dr Federica Rinaldi. Work in the Caldwell laboratory is supported by Parkinson UK, BBSRC, BRACE and James Tudor foundation.
Glossary
- Cx43
Connexin43
- hNPC
human neural progenitor cell
- GJ
Gap junction
- Cx
connexin
- CT
C-terminal
- shRNA
short hairpin RNA
- MOI
multiplicity of infection
- BrdU
5′-bromo-2-deoxyuridine
- CBX
carbenoxolone
- GZA
glycyrrhizic acid
- AJ
adherens junction
- GSK-3b
glycogen synthase kinase 3b
- CHIR
6-[[2-[[4-(2,4-dichlorophenyl)-5-(5-methyl-1H-imidazol-2-yl)-2-pyrimidinyl]amino]ethyl]amino]-3-pyridinecarbonitrile
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Author Contributions
Federica Rinaldi: conception and design of the study, performed the experiments and wrote the paper. Elizabeth M Hartfield: collection and assembly of data, final approval of the manuscript. Lucy A Crompton: data analysis and interpretation, final approval of the manuscript. Jennifer L Badger: provision of study material, final approval of the manuscript. Colin P Glover: provision of study material, final approval of the manuscript. Claire M Kelly: provision of study material, final approval of the manuscript. Anne E Rosser: provision of study material, final approval of the manuscript. James B Uney: conception and design, financial support, data analysis and interpretation, final approval of the manuscript. Maeve A Caldwell: conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of the manuscript.
Edited by A Verkhratsky
Supplementary Material
References
- Nishi M, Kumar NM, Gilula NB. Developmental regulation of gap junction gene expression during mouse embryonic development. Dev Biol. 1991;146:117–130. doi: 10.1016/0012-1606(91)90452-9. [DOI] [PubMed] [Google Scholar]
- Yancey SB, Biswal S, Revel JP. Spatial and temporal patterns of distribution of the gap junction protein connexin43 during mouse gastrulation and organogenesis. Development. 1992;114:203–212. doi: 10.1242/dev.114.1.203. [DOI] [PubMed] [Google Scholar]
- Trosko JE, Chang CC, Wilson MR, Upham BL, Hayashi T, Wade M. Gap junctions and the regulation of cellular function of stem cells during development and differentiation. Methods. 2000;20:245–264. doi: 10.1006/meth.1999.0941. [DOI] [PubMed] [Google Scholar]
- Simon AM, Goodenough DA. Diverse functions of vertebrate gap junctions. Trends Cell Biol. 1998;8:477–483. doi: 10.1016/s0962-8924(98)01372-5. [DOI] [PubMed] [Google Scholar]
- Giepmans BN. Gap junctions and connexin-interacting proteins. Cardiovasc Res. 2004;62:233–245. doi: 10.1016/j.cardiores.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Matsuuchi L, Naus CC. Gap junction proteins on the move: connexins the cytoskeleton and migration. Biophys Acta. 2013;1828:94–108. doi: 10.1016/j.bbamem.2012.05.014. [DOI] [PubMed] [Google Scholar]
- Elias LA, Wang DD, Kriegstein AR. Gap junction adhesion is necessary for radial migration in the neocortex. Nature. 2007;7156:901–907. doi: 10.1038/nature06063. [DOI] [PubMed] [Google Scholar]
- Fushiki S, Perez Velazquez JL, Zhang L, Bechberger JF, Carlen PL, Naus CC. Changes in neuronal migration in neocortex of connexin43 null mutant mice. J Neuropathol Exp Neurol. 2003;62:304–314. doi: 10.1093/jnen/62.3.304. [DOI] [PubMed] [Google Scholar]
- Cina C, Maass K, Theis M, Willecke K, Bechberger JF, Naus CC. Involvement of the cytoplasmic C-terminal domain of connexin43 in neuronal migration. J Neurosci. 2009;29:2009–2021. doi: 10.1523/JNEUROSCI.5025-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobas DA, Scemes E, Spray DC. Gene expression alteration in connexin null mice extended beyond the gap junction. Neurochem Int. 2004;45:243–250. doi: 10.1016/j.neuint.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Iacobas DA, Iacobas S, Urban-Maldonado M, Spray DC. Sensitivity of the brain transcriptome to connexin ablation. Biochimica et Biofisica Acta. 2005;1711:183–196. doi: 10.1016/j.bbamem.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Kardami E, Dang X, Iacobas DA, Nickel BE, Jeyaraman M, Srisakuldee W, et al. The role of connexins in controlling cell growth and gene expression. Prog Biophys Mol Biol. 2007;94:245–264. doi: 10.1016/j.pbiomolbio.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Wolfram T, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cells produces neurons and astrocytes. J Neurosci. 1992;12:4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons, astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- Carpenter MK, Cui X, Hu Z, Jackson J, Sherman S, Seiger A, et al. In vitro expansion of a multipotent population of human neural progenitor cells. Exp Neurol. 1999;158:265–278. doi: 10.1006/exnr.1999.7098. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Seri B, Doetsch F. Identification of neural stem cells in the adult vertebrate brain. Brain Res Bull. 2000;57:751–758. doi: 10.1016/s0361-9230(01)00770-5. [DOI] [PubMed] [Google Scholar]
- Gao Z, Ure K, Ables JL, Lagace DC, Nave KA, Goebbles S, et al. Neurod1 is necessary for the survival and maturation of adult-born neurons. Nat Neurosci. 2009;12:1090–1092. doi: 10.1038/nn.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluske KK, Vue TY, Kawakami Y, Taketo MM, Yoshikawa K, Johnson JE, et al. β-Catenin signaling specifies progenitor cell identity in parallel with Shh signaling in the developing mammalian thalamus. Dev Stem Cells. 2012;15:2692–2702. doi: 10.1242/dev.072314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Muotr A, Yeo G, Warashina M, Lie DC, et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci. 2009;12:1097–1105. doi: 10.1038/nn.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth JL, Kelly S, Keasey MP, Glover CP, Lee YB, Mitropanous K, et al. Hsp40 molecules that target to the ubiquitin-proteasome system decrease inclusion formation in models of polyglutamine disease. Mol Therapy. 2007;15:1100–1105. doi: 10.1038/sj.mt.6300163. [DOI] [PubMed] [Google Scholar]
- Tovar KR, Maher BJ, Westbrook GL. Direct action of carbenoxolone on synaptic transmission and neuronal membrane proprieties. J Neurophysiol. 2009;102:974–978. doi: 10.1152/jn.00060.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heyden MA, Rook MB, Hermans MM, Rijksen G, Boonstra J, Defize LH, et al. Identification of connexin43 as a functional target for Wnt signalling. J Cell Sci. 1998;111:1741–1749. doi: 10.1242/jcs.111.12.1741. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, Winchen WD, de Weger R, Kinzler KV, et al. Constitutive transcriptional activation by a β-Catenin–TCF complex in APC2/2 colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Liu J, Wu X, Mitchell B, Kintner C, Ding S, Shulz PG. A small-molecule agonist of the WNT1 pathway. Angewandte Chimie Int Edn. 2005;44:1987–1990. doi: 10.1002/anie.200462552. [DOI] [PubMed] [Google Scholar]
- Rozental R, Morales M, Mehler MF, Urban M, Kremer M, Dermietzel R, et al. Changes in the properties of gap junctions during neuronal differentiation of hippocampal progenitor cells. J Neurosci. 1998;18:1753–1762. doi: 10.1523/JNEUROSCI.18-05-01753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittman KS, LoTurco JJ. Differential regulation of connexin 26 and 43 in murine neocortical precursors. Cerebral Cortex. 1999;9:188–195. doi: 10.1093/cercor/9.2.188. [DOI] [PubMed] [Google Scholar]
- Rozental R, Srinivas M, Gokhan S, Urban M, Dermietzel R, Kessler JA, et al. Temporal expression of neuronal connexins during hippocampal ontogeny. Brain Res. 2000;32:57–71. doi: 10.1016/s0165-0173(99)00096-x. [DOI] [PubMed] [Google Scholar]
- Rozental R, Giaume C, Spray DC. Gap junctions in the nervous system. Brain Res. 2000;32:11–15. doi: 10.1016/s0165-0173(99)00095-8. [DOI] [PubMed] [Google Scholar]
- Cina C, Bechberger JF, Ozog MA, Naus CC. Expression of connexins in embryonic mouse neocortical development. Jf Comp Neurol. 2007;504:298–313. doi: 10.1002/cne.21426. [DOI] [PubMed] [Google Scholar]
- Dermietzel R, Spray DC. Gap junctions in the brain: where, what type, how many and why. Trends Neurosci. 1993;16:189–192. doi: 10.1016/0166-2236(93)90151-b. [DOI] [PubMed] [Google Scholar]
- Cai J, Cheng A, Luo Y, Lu C, Mattson MP, Rao MS, et al. Membrane proprieties of rat embryonic multipotent neural stem cells. J Neurochem. 2004;88:212–226. doi: 10.1046/j.1471-4159.2003.02184.x. [DOI] [PubMed] [Google Scholar]
- Leung DS, Unsicker K, Reuss B. Expression and developmental regulation of gap junction connexins cx26, cx32, cx43 and cx45 in the rat midbrain-floor. Int J Dev Neurosci. 2002;20:63–75. doi: 10.1016/s0736-5748(01)00056-9. [DOI] [PubMed] [Google Scholar]
- Svendsen CN, ter Borg MG, Armstrong RJ, Rosser AE, Chandran S, Ostenfeld T, et al. A new method for the rapid and long term growth of human neural precursor cells. J Neurosci Methods. 1998;85:141–152. doi: 10.1016/s0165-0270(98)00126-5. [DOI] [PubMed] [Google Scholar]
- Caldwell MA, He X, Wilkie N, Pollack S, Marshall G, Wafford KA, et al. Growth factors regulate the survival and fate of cells derived from human neurospheres. Nat Biotechnol. 2001;19:475–479. doi: 10.1038/88158. [DOI] [PubMed] [Google Scholar]
- Liu X, Sun L, Torii M, Rakic P. Connexin43 controls the multipolar phase of neuronal migration to the cerebral cortex. Proc Natl Acad Sci USA. 2012;109:8280–8285. doi: 10.1073/pnas.1205880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller FD, Gauthier AS. Timing is everything: making neurons versus glia in the developing cortex. Neuron. 2007;54:357–369. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Hartfield EM, Rinaldi F, Glover CP, Wong LF, Caldwell MA, Uney JB. Connexin 36 expression regulates neuronal differentiation from neural progenitor cells. PLoS One. 2001;6:e14746. doi: 10.1371/journal.pone.0014746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah B, Jones AM, Evans WH, Parnavelas JG. Differential expression of connexins during neocortical development and neuronal circuit formation. J Neurosci. 1997;17:3096–3111. doi: 10.1523/JNEUROSCI.17-09-03096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekkadan B, Berdichevsky Y, Irimia D, Leeder A, Yarmush G, Toner M, et al. Cell-cell interaction modulates neuroectodermal specification of embryonic stem cells. Neurosci Lett. 2008;438:190–195. doi: 10.1016/j.neulet.2008.03.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago MF, Alcami P, Striedinger K, Spray DC, Scemes E. The carboxyl-terminal domain of connexin43 is a negative modulator of neuronal differentiation. J Biol Chem. 2010;285:11836–11845. doi: 10.1074/jbc.M109.058750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- Ai Z, Fischer A, Spray DC, Brown AM, Fishman GI. Wnt-1 regulation of connexin43 in cardiac myocytes. J Clin Invest. 2000;105:161–171. doi: 10.1172/JCI7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/ LEF-1-mediated transactivation. J Cell Sci. 1999;112:1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Davidson BL. Plasmid-based RNA interference: construction of small-hairpin RNA expression vectors. Methods Mol Biol. 2005;309:219–235. doi: 10.1385/1-59259-935-4:219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.