Abstract
Overexpression of the human epidermal growth factor receptor-2 (HER2) in breast cancer strongly correlates with aggressive tumors and poor prognosis. Recently, a positive correlation between HER2 and MIF (macrophage migration inhibitory factor, a tumor-promoting protein and heat-shock protein 90 (HSP90) client) protein levels was shown in cancer cells. However, the underlying mechanistic link remained unknown. Here we show that overexpressed HER2 constitutively activates heat-shock factor 1 (HSF1), the master transcriptional regulator of the inducible proteotoxic stress response of heat-shock chaperones, including HSP90, and a crucial factor in initiation and maintenance of the malignant state. Inhibiting HER2 pharmacologically by Lapatinib (a dual HER2/epidermal growth factor receptor inhibitor) or CP724.714 (a specific HER2 inhibitor), or by knockdown via siRNA leads to inhibition of phosphoactivated Ser326 HSF1, and subsequently blocks the activity of the HSP90 chaperone machinery in HER2-overexpressing breast cancer lines. Consequently, HSP90 clients, including MIF, AKT, mutant p53 and HSF1 itself, become destabilized, which in turn inhibits tumor proliferation. Mechanistically, HER2 signals via the phosphoinositide-3-kinase (PI3K)–AKT– mammalian target of rapamycin (mTOR) axis to induce activated pSer326 HSF1. Heat-shock stress experiments confirm this functional link between HER2 and HSF1, as HER2 (and PI3K) inhibition attenuate the HSF1-mediated heat-shock response. Importantly, we confirmed this axis in vivo. In the mouse model of HER2-driven breast cancer, ErbB2 inhibition by Lapatinib strongly suppresses tumor progression, and this is associated with inactivation of the HSF1 pathway. Moreover, ErbB2-overexpressing cancer cells derived from a primary mouse ErbB2 tumor also show HSF1 inactivation and HSP90 client destabilization in response to ErbB2 inhibition. Furthermore, in HER2-positive human breast cancers HER2 levels strongly correlate with pSer326 HSF1 activity. Our results show for the first time that HER2/ErbB2 overexpression controls HSF1 activity, with subsequent stabilization of numerous tumor-promoting HSP90 clients such as MIF, AKT and HSF1 itself, thereby causing a robust promotion in tumor growth in HER2-positive breast cancer.
Keywords: MIF, HSP90, HSF1, mTOR, HER2, ErbB2
The human epidermal growth factor receptor-2 (HER2; ErbB2/Neu) is overexpressed in 20–30% of invasive breast cancers, which correlates with poor prognosis.1 Compared with normal HER2 expression levels, HER2-overexpressing cells can exhibit up to 100-fold higher concentrations. The high receptor density triggers ligand-independent spontaneous and constitutive dimerization, leading to subsequent kinase activation of the cytoplasmic tail.2, 3
Activated kinase signaling stimulates autophosphorylation, which in turn promotes downstream signaling through the phosphoinositide-3-kinase (PI3K)–AKT and Ras–Raf–MEK–ERK1/2 (rat sarcoma–rat fibrosarcoma–mitogen-activated protein kinase kinase– extracellular signal-regulated kinase 1/2) pathways.4, 5 Activation of these pathways triggers proliferation, blocks apoptosis and promotes invasion and angiogenesis to boost tumor growth.6 Treatment of HER2-positive tumors with Trastuzumab has revolutionized clinical outcome for these patients, but despite all its remarkable success, long-term response rates are limited due to primary or acquired resistances.6 Thus, a detailed understanding of HER2 signaling is essential to enable further advances in treating these cancers.
In our recent study with the transgenic mouse model of HER2 human breast cancer, we showed that the tumor enhancer MIF (macrophage migration inhibitory factor) promoted breast cancer progression and reduced overall survival of ErbB2 mice.7 In general, MIF is strongly implicated as a human cancer promoter with a central role in the inflammation–tumorigenesis axis.8, 9, 10 Human cancers of the breast, colon, ovary, prostate, liver, lung, pituitary and brain frequently express elevated MIF levels.7, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27 Importantly, elevated MIF levels correlate with clinical aggressiveness of the breast, lung, liver, brain, ovary and prostate, implicating MIF in poor prognosis and resistance to chemotherapeutic drugs.16, 18, 22, 23, 25, 26, 28 In clear genetic support of MIF as an important pathophysiologic tumor driver, MIF deletion delays tumor progression and lowers tumors stages, and decreases angiogenesis in several mouse cancer models, including Myc-induced lymphomagenesis,29 UVB-exposed skin cancer progression,30 adenomatous polyposis coli protein (Apc)MIN/+-mediated intestinal adenomas31 and nitrosamine-induced bladder tumorigenesis.32
The link between MIF and HER2 we found earlier7 led us to the question whether elevated MIF levels might be specifically linked to and perhaps causally dependent on the molecular subtype of HER2-postive breast cancers. In line with this notion, correlation studies found elevated MIF levels specifically in HER2-positive human breast cancers.33 To test whether HER2 signaling directly regulates MIF levels, we chose in the current study a set of HER2-overexpressing human breast cancer cell lines and treated them pharmacologically with HER2 inhibitors. Indeed, MIF was downregulated in a PI3K– mammalian target of rapamycin (mTOR)-dependent manner exclusively at the level of MIF protein. As we recently identified MIF as a new heat-shock protein 90 (HSP90) chaperone client,7 we next asked whether HER2-mediated MIF regulation is controlled by the HSP90 chaperone machinery.
Cancer cells are in a perennial state of proteotoxic stress due to cell-extrinsic (hypoxia, acidosis) and cell-intrinsic (aberrant/misfolded proteins, massive oxidative stress, high levels of DNA damage and genomic instability, stoichiometric imbalances among components of multiprotein complexes) aberrant conditions. Their proteins, especially their mutated and deregulated oncoproteins, require constant massive heat-shock protein (HSP) chaperone support, especially from the HSP90 system, to prevent protein aggregation and illicit interactions.34, 35, 36 Thus, cancer cells become addicted to the HSP machinery, which is highly upregulated and activated specifically in cancer but not in normal cells, turns it in a powerful prosurvival and anti-apoptotic system.34 Mechanistically, cancer cells respond to their proteotoxic stress by constitutive activation of heat-shock factor 1 (HSF1), the master transcription factor for the synthesis of classic inducible HSPs such as Hsp90α, Hsp70, numerous co-chaperones and adaptor proteins. Inactive monomeric HSF1 is sequestered by cytoplasmic HSP90 chaperones, whereas active HSF1 is a phosphoactivated trimer in the nucleus. Thus, the HSF1-HSP90 axis has a key role in the stabilization of oncogenic proteins, including receptor tyrosine kinases (ErbB1 and ErbB2/HER2),37 signaling kinases (Bcr-Abl and AKT),38 c-Raf,39 mutated p53,40 MIF7 and monomeric HSF1 itself.41 Moreover, HSF1 was shown to be one of the main determinants of oncogenesis in mouse cancer models (HSF1 knockout mice are markedly resistant to a number of oncogenes42, 43, 44, 45, 46), not only by inducing the adaptive proteotoxic stress response but also by modulating the expression of a broad set of genes involved in cell-cycle regulation, signaling, metabolism, adhesion and protein translation.47
Of note, a correlation between human breast cancer subtypes and HSF1 level was observed. In detail, HER2-positive tumors with additional high levels of nuclear HSF1 correlates with poorer prognosis.42, 48 Another study showed an increase of total HSF1 and HSF1 trimer formation after HER2 overexpression in MCF7 cells.49 Furthermore, HSF1 is necessary in HER2-induced cell transformation in breast epithelial MCF-10A cells in vitro.44 Another hint for cooperation between HER2/ErbB2 and HSF1 came from the ErbB2-transgenic mouse model, where depletion of HSF1 significantly reduces tumorigenesis.45 However, whether a linear signaling pathway exists between HER2 and HSF1 in HER-positive breast cancers remains unknown. Mechanistically, our study identifies a direct axis between HER2 and HSF1 activation, where HER2 signaling controls HSF1 activity and thereby stabilizes HSP90 clients.
Results
Specific inhibition of HER2 signaling in HER2-overexpressing breast cancer cells reduces MIF protein levels
Recently, we demonstrated that genetic MIF deletion markedly protects ErbB2-overexpressing transgenic mice from developing breast cancer,7 suggesting a causal link between ErbB2/HER2 status and MIF levels. In support, another group recently reported a correlation between elevated MIF levels and HER2-positive human breast cancers.33
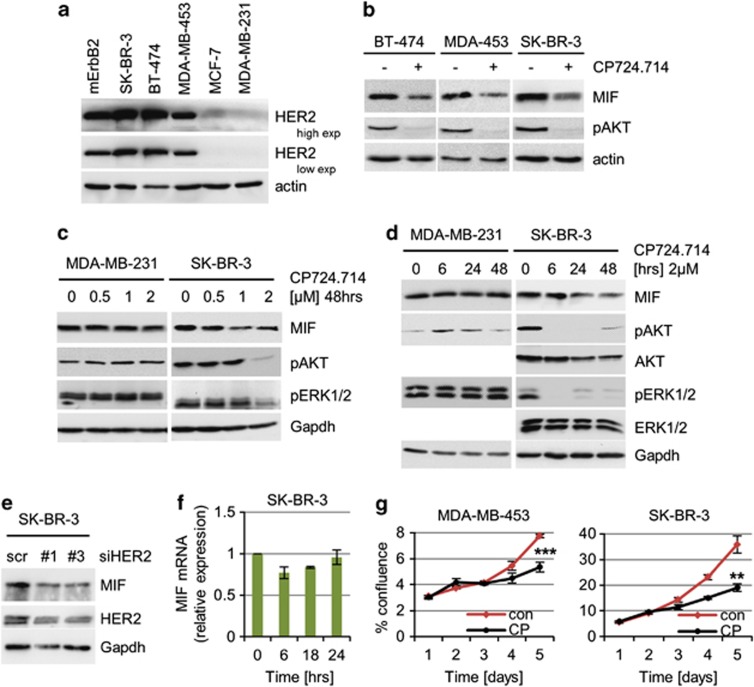
To test for a putative causal connection, we used a random panel of human breast cancer cell lines showing either overexpression (SK-BR-3, BT-474 and MDA-MB-453) or normal expression of HER2 protein (MCF7 and MDA-MB-231; Figure 1a). Indeed, in HER2-overexpressing lines we observed a reduction of MIF protein in response to specific HER2 inhibition by the small molecule inhibitor CP724.714 (Figures 1b–d) in a dose- and time-dependent manner (Figures 1c and d). Both main branches of HER2 signaling, that is, HER2–PI3K/AKT and HER2–MEK–ERK1/2, were inhibited, confirming the functionality of the pharmacological inhibitor (Figures 1b–d). Moreover, specific HER2 depletion by siRNAs also abolished MIF stabilization (Figure 1e), confirming that HER2 regulates MIF stabilization. On the other hand, MIF transcript levels were not impaired (Figure 1f), indicating destabilization of MIF protein on HER2 inhibition. In contrast, the basal-like cell line MDA-MB-231 with normal HER2 expression failed to show a response to HER2 inhibition (Figures 1c and d). HER2 inhibition by CP724.714 in HER2-overexpressing cells caused growth inhibition, confirming the importance of enhanced HER2 signaling in such cells (Figure 1g). Thus, MIF protein stabilization is controlled by HER2 signaling.
Figure 1.
Specific inhibition of HER2 signaling in HER2-overexpressing breast cancer cells reduces MIF protein levels. (a) Endogenous HER2 protein levels in human breast cancer cells. Representative immunoblot of cell lysates from the indicated cell lines. mErbB2, murine ErbB2 cell line. Actin, loading control. (b) HER2 inhibition destabilizes endogenous MIF protein. The indicated cells were treated with 2 μM CP724.714 or DMSO for 48 h. Immunoblot. Actin, loading control. (c and d) The specific HER2 inhibitor CP724.714 reduces endogenous MIF levels in a dose- (c) and time- (d) dependent manner. MDA-MB-231 serves as negative control. pAKT and pERK1/2 are functional controls for HER2 inhibition. Immunoblot analyses, Gapdh as loading control. (e) Depletion of HER2 in HER2-overexpressing cells leads to reduced MIF levels. SK-BR-3 cells were transfected with two different siRNA against HER2 (1 and 3) or control siRNA (scr). After 3 days, protein levels were assessed by immunoblots. Gapdh, loading control. (f) In contrast to reduced MIF protein, corresponding MIF mRNA levels remain unchanged after CP724.714 treatment (2 μM). SK-BR-3 cells, qRT-PCR normalized to 36B4. Relative values in (ratio (2−ddCT)). Error bars indicate S.E.M. of two independent experiments in triplicates each. (g) HER2 inhibition causes growth inhibition in HER2-overexpressing cells. Cells were seeded (day 0) and cultured for 24 h (day 1), then treated with 2 μM of CP724.714 (CP) for 24 h or left untreated, and followed up to day 5. Cell confluence measured daily by CELIGO Cytometer for MDA-MB-453 (***P=0.0006) and SK-BR-3 (**P=0.0037). Error bars indicate the ±S.E.M of two independent experiments in duplicates each. Student's t-test of day 5, one-tailed, P-value: *P<0.05, **P<0.01 and ***P<0.001
PI3K and mTOR activity are required for MIF stabilization in HER2-overexpressing cancer cells
Normal (non-overexpressing) HER2 signaling is dependent on ligand binding and subsequent receptor dimerization, and promotes downstream activation of the PI3K-AKT and/or the Ras–Raf–MEK–ERK1/2 axes.4, 5 Importantly, strong HER2 overexpression in cancer triggers spontaneous and constitutive ligand-independent dimerization that can lead to activation of both axes.2, 3
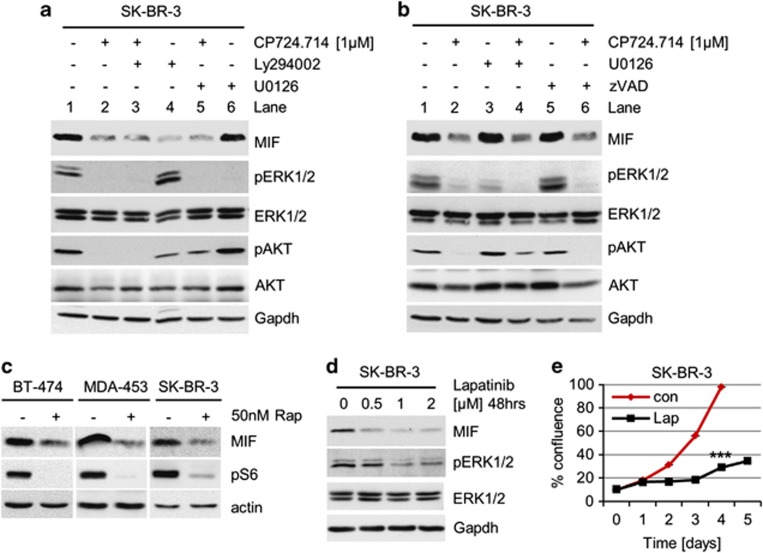
To elucidate which downstream axis of HER2 signaling is specifically involved in MIF stabilization, we used inhibitors of PI3K (Ly294002) and MEK (U0126), alone or in combination with HER2 inhibitor CP274.714 (Figures 2a and b, and overview Figure 6c). Compared with Figure 1, we lowered the CP724.714 concentration to see possible additive or synergistic effects by co-treatments. PI3K inhibition (Figure 2a, lane 4), but not MEK inhibition (lane 6), decreased MIF levels. Proof of effective HER2/MEK inhibition was provided by a sharp drop in the pERK1/2 levels (lanes 2, 3, 5 and 6). Further, effective HER2/PI3K inhibition was confirmed by pAKT staining (lane 2, 3, 4 and 5). pERK1/2 activation was not blocked by inhibition of PI3K via Ly294002 (Figure 2a, lane 4) and pAKT activation was not blocked by U0126 (lane 6), indicating the independence of these signaling systems. We excluded additive effects of HER2 and PI3K signaling, as the combination of CP724.714 and Ly294002 (Figure 2a, lane 3) did not decrease MIF levels further compared with CP724.714 alone (lane 2), confirming that HER2 and PI3K are part of the same signaling axis. To analyze whether apoptotic signaling could influence MIF levels, we used the pan-Caspase inhibitor zVAD (Figure 2b). As zVad alone did not reduce MIF levels (lane 5), and a combination of CP724.714 plus zVad (lane 6) did not rescue MIF protein destabilization, we excluded an involvement of apoptotic effects in our system.
Figure 2.
Blocking the HER2–PI3K–mTOR pathway reduces MIF protein levels. (a and b) Inhibition of the HER2–PI3K axis, but not the HER2–MEK–ERK1/2 axis, destabilizes MIF protein. SK-BR-3 cells were treated with 1 μM CP724.714 (specific HER2 inhibitor), 25 μM Ly294002 (PI3K inhibitor) or 10 μM U0126 (MEK1/2 inhibitor) (a), or with 1 μM CP724.714, 10 μM U0126 or 50 μM zVAD (panCaspase inhibitor) (b) alone or in combination as indicated. Immunoblot analyses. pERK1/2 and pAKT staining serve as positive control for respective inhibition. Gapdh, loading control. (c) Inhibition of mTOR destabilizes MIF protein. HER2-overexpressing BT-474, MDA-MB-453 and SK-BR-3 cells were treated with 50 nM of Rapamycin for 48 h or left untreated. Immunoblot analyses. pS6 serves as functional control of mTOR inhibition. Actin, loading control. (d) The dual HER2/EGFR inhibitor Lapatinib markedly reduces MIF levels. SK-BR-3 cells were treated or not with Lapatinib for 48 h. Immunoblot analysis. pERK1/2 is a functional control for HER2 inhibition. Gapdh, loading control. (e) Lapatinib blocks survival. SK-BR-3 cells were seeded (day 0) and cultured for 24 h (day 1). Cells were then treated or left untreated with 2 μM Lapatinib for 48 h and followed up to day 5. Confluence measured daily by CELIGO Cytometer. Error bars indicate the S.E.M. of a triplicate experiment. Student's t-test of day 4, two-tailed, P-value: ***P<0.001
The major downstream target of PI3K/AKT activation is mTOR. Indeed, Rapamycin, the classic inhibitor of mTOR, strongly destabilized MIF protein in HER2-overexpressing breast cancer cells (Figure 2c). Ribosomal protein S6 as a main target of mTOR phosphorylation served as control for mTOR inhibition. Likewise, Lapatinib, a dual HER2/epidermal growth factor receptor (EGFR) inhibitor also induced strong downregulation of MIF (and pERK1/2) levels (Figure 2d). Concomitantly, Lapatinib induced growth defects in HER2-overexpressing SK-BR-3 cells (Figure 2e). We conclude that MIF is a novel downstream target of HER2–PI3K–AKT–mTOR signaling in HER2-overexpressing breast cancer cells.
The activity of HSF1 and its transcriptional program is controlled by HER2 signaling
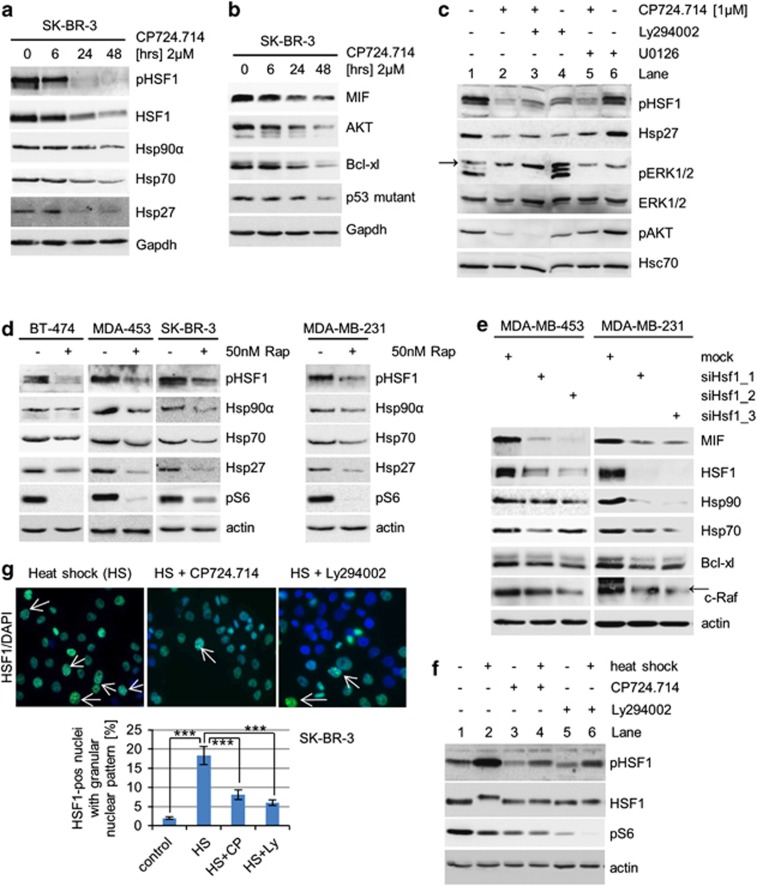
As MIF mRNA is not reduced after HER2 inhibition (Figure 1f), we concluded that MIF protein levels are regulated. In agreement, we recently identified MIF as a novel stabilized client of the HSP90 chaperone.7 HSF1, the master transcriptional regulator of the inducible heat-shock response, controls many stress-inducible chaperones, including Hsp90α, Hsp70 and Hsp27. HSF1 is frequently activated in human tumors and the HSF1-mediated adaptive proteotoxic stress response has a key role in mammalian tumorigenesis.42, 43 In cancers, HSF1 is constitutively phosphorylated, leading to its trimerization and nuclear translocation to activate chaperone expression, among other programs. The critical phosphorylation site for HSF1 activation is the Ser326 residue, seen after, for example, heat shock and chemical stress.50 This prompted us to ask whether HER2 signaling also induces phosphoSer326 activation of HSF1. In support, it was recently reported that in HeLa cells mTOR phosphorylates HSF1 at Ser326, leading to induction of HSP90 chaperones.51 Indeed, in response to HER2 inhibition by CP724.714, HSF1 underwent dephosphorylation at Ser326, already underway at 6 hrs, and indicated by a phosphoSer326-specific HSF1 antibody (Figure 3a). Of note, downregulation of total HSF1 protein occurred later, clearly indicating that the reduction of pHSF1 is not simply a consequence of reduced total HSF1 levels. In agreement with HSF1 inactivation (i.e., dephosphorylation at Ser326), HSF1 target gene expression, including Hsp90α, Hsp70 and Hsp27, was downregulated (Figure 3a). Moreover, in response to CP724.714, HSP90 clients, including MIF, AKT, Bcl-xl and mutant p53, were destabilized, confirming the inactivation of the HSF1-HSP90 stress response on HER2 inhibition (Figure 3b). Please note that AKT itself is a prominent Hsp90 target protein and, subsequently, total AKT level are always reduced after 24–48 h of HER2 inhibition, as expected (Figure 1d for SkBr-3 and Figure 3b). Importantly, the dephosphorylation of AKT occurs much early than the reduction of total AKT (Figure 1d for SkBr-3), which confirms that the inactivation of the PI3K-AKT-HSF1 signaling pathway is a prerequisite for Hsp90 client destabilization. Further and because of AKT as an Hsp90 client, we see a reduction of total AKT in experiments where we used CP724.714 for 24 h or longer (e.g., Figures 2a and b).
Figure 3.
HER2 inhibition leads to HSF1 inactivation and subsequent inactivation of the HSP90 chaperone. (a and b) Inhibition of HER2 inactivates HSF1 (a) and thereby destabilizes HSP90 clients (b). SK-BR-3 cells were treated with 2 μM CP724.714 for 48 h. Protein levels of pSer326 HSF1, total HSF1, Hsp90α, Hsp70 and Hsp27 (a) and MIF, AKT, Bcl-xl and mutant p53 R175H (b) were assessed by immunoblots. Gapdh, loading control. (c) Inhibition of the HER2–PI3K axis, but not the HER2–ERK1/2 axis, inactivates HSF1 and downstream chaperones. SK-BR-3 cells were treated with 1 μM CP724.714 (specific HER2 inhibitor), 25 μM Ly294002 (PI3K inhibitor) or 10 μM U0126 (MEK inhibitor) alone or in combination as indicated. Immunoblot analysis. Arrow indicates unrelated band. Hsc70, loading control. (d) Inhibition of mTOR prevents HSF1 activation. Cells were treated or left untreated with 50 nM Rapamycin for 48 h. Immunoblot analyses. pS6 serves as functional control of mTOR inhibition. Actin, loading control. (e) HSF1 silencing destabilizes MIF protein. MDA-MB-453 and MDA-MB-231 cells were transfected with different siRNAs against HSF1 for 3 days. Cell lysates were immunoblotted for MIF, panHsp90, Hsp70, Bcl-xl and c-Raf. Representative blots are shown. Actin, loading control. Arrow indicates related band. (f) The heat-shock response is impaired in response to inhibition of the HER2 pathway. SK-BR-3 cells were left untreated or treated with 2 μM CP724.714 or 25 μM Ly294002 for 48 h, followed by heat shock at 42 °C for 1 h where indicated. Protein lysates were prepared after a recovery of 3 h. Immunoblot analysis for pSer326 HSF1 and total HSF1. Note that the HS-induced hyperphosphorylated species of total HSF1 (slower migrating band) is blocked by HER2 and PI3K inhibition. pS6 serves as control for HER2 and PI3K inhibition. Actin, loading control. (g) The heat-shock response is markedly attenuated in response to HER2 inhibition. SK-BR-3 were treated with 2 μM CP724.714 or 25 μM Ly294002 for 48 h. After heat shock at 42 °C for 1 h followed by a 3-h recovery, cells were immunostained for total HSF1 with DAPI counterstain. Top, representative immunofluorescence of heat-shocked control cells and heat-shocked/CP274.714-treated cells. Arrows indicate HSF1-positive nuclei with a granular nuclear pattern. Only those were counted. Bottom, quantification of HSF1-positive nuclei. For every sample, eight random fields ( × 20 magnification) in duplicates were counted. The number of HSF1-positive cells as percent of total nuclei (DAPI stained) is shown. Error bars indicates ±S.E.M. Student's t-test, two-tailed, P-value: ***P<0.001
Mechanistically, HSF1 phosphorylation at Ser326 was again dependent on PI3K (Figure 3c, lane 4), but not on MEK–ERK1/2 (lane 6). Rapamycin treatment confirmed the involvement of mTOR in regulating HSF1 activity in HER2-overexpressing cells (Figure 3d, left). Interestingly, inhibition of mTOR also induced HSF1 dephosphorylation in HER2 non-overexpressing MDA-MB-231 cells (Figure 3d, right), confirming that downstream targets of HER2 can also act independently in HSF1 regulation. On the other hand, siRNA-mediated knockdown of HSF1, which in turn downregulates Hsp90 and Hsp70, abolished MIF stabilization and other clients such as Bcl-xl and c-Raf in both HER2-overexpressing and non-overexpressing breast cancer cells (Figure 3e). Taken together, cancer cells develop several different strategies to increase HSF1 activity. HER2 signaling is one of them.
To obtain further support that HSF1 is controlled by HER2, we heat shocked SK-BR-3 cells to increase HSF1 activity, indicated by induction of pSer326 HSF1. Indeed, HER2 (and PI3K) inhibition markedly attenuated this heat-shock response (Figure 3f, compare lanes 2 and 4), once again confirming the functional link between HER2 signaling and HSF1 activation. Similarly, immunofluorescence staining of activated HSF1, indicated by granular nuclear staining, confirmed the impairment of the heat-shock response after HER2 inhibition (Figure 3g).
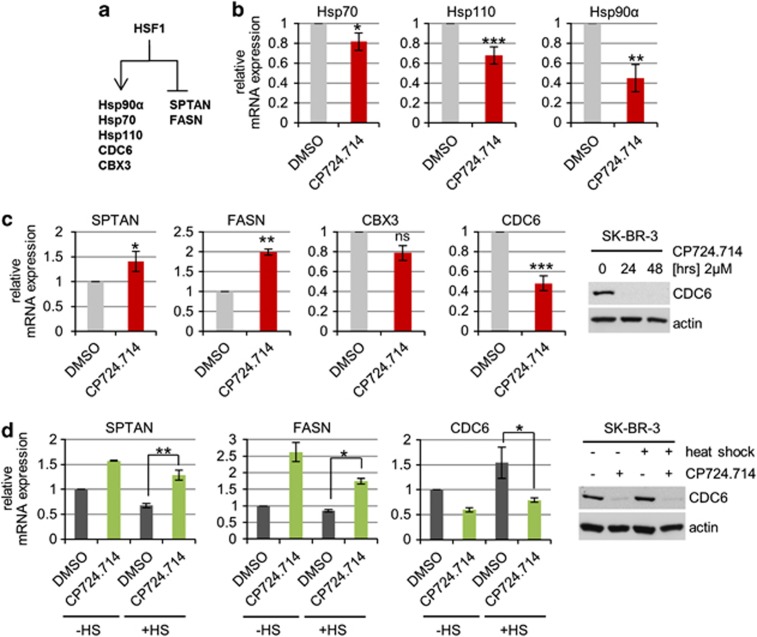
HSF1 not only orchestrates the cellular stress response. In cancer cells, HSF1 also broadly modulates (via transactivation and transrepression) tumor-promoting genes involved in cell cycle regulation, DNA repair, signaling, metabolism, adhesion and translation.47 Thus, we analysed mRNA levels of randomly selected representative target genes of these different HSF1 programs (Figure 4a). Notably, in response to HER2 inhibition we observed repression of classic activated HSF1 targets, including Hsp90α, Hsp70 and Hsp110 (Figure 4b). Moreover, expression of the HSF1 targets CDC6 (cell division cycle 6) and CBX3 (chromobox protein homolog 3; chromatin remodeling) was also repressed (Figure 4c). CDC6 repression was confirmed on the protein level (Figures 4c and d). Conversely, expression of the repressed targets SPTAN (spectrin alpha (non-erythrocytic; apoptosis)) and FASN (fatty acid synthase; metabolism) was upregulated on HER2 inhibition (Figure 4c). Most importantly, HER2 inhibition also specifically influenced the transcriptional HSF1 program after heat shock, as shown by SPTAN, FASN and CDC6 transcript levels and CDC6 protein levels (Figure 4d panels, compare bars 3 and 4). Taken together, our results clearly indicate that HER2 signaling mediates activation of the HSF1 transcriptional program in HER2-overexpressing cancer cells.
Figure 4.
The HSF1 transcriptional program is impaired by HER2 inhibition. (a) Schematic overview of HSF1-regulated gene expression. Transcriptionally activated and -repressed target genes analyzed here are indicated. (b and c) HER2 inhibition impairs the HSF1-dependent inducible heat-shock response (b) and the tumor-promoting (c) transcriptional program of HSF1. SK-BR-3 cells were treated with 2 μM CP724.714 or DMSO for 48 h and mRNA was isolated. qRT-PCR of Hsp70 (*P=0.0396), Hsp110 (***P=0.001), Hsp90α (**P=0.0049), SPTAN (*P=0.0362), CDC6 (***P=0.001), FASN (**P=0.0069) and CBX3 (P=0.067), each normalized to 36B4 mRNA. Relative values are given in (ratio (2−ddCT)). Error bars indicate ±S.E.M. of three independent experiments, repeated twice each with all in triplicates. Student's t-test, two-tailed, P-value: *P<0.05, **P<0.01 and ***P<0.001. (d) HER2 inhibition also attenuates the HSF1-mediated oncogenic program after heat shock. SK-BR-3 cells were treated with 2 μM CP724.714 or DMSO for 48 h. Heat shock (HS) at 42 °C for 1 h with 3 h recovery before mRNA isolation. qRT-PCRs of SPTAN (**P=0.0048), CDC6 (*P=0.0373) and FASN (*P=0.0161), normalized to 36B4. Relative values are given in (ratio (2−ddCT)). Error bars indicate ±S.E.M. of two or three independent experiment in triplicates. Student's t-test, two-tailed, P-value: *P<0.05, **P<0.01 and ***P<0.001
In the HER2 mouse model, ErbB2 inhibition delays cancer progression, which is associated with HSF1 inactivation and destabilization of HSP90 clients
To date, a causal tumor-promoting role of MIF in vivo has been established in murine cancer models. Specifically, MIF gene ablation specifically impairs B-cell lymphomagenesis in EμMyc mice,29 ApcMIN/+-induced intestinal tumor growth,31 nitrosamine-induced bladder cancer,32 UVB-induced skin cancer30 and, notably, breast tumor development in mouse mammary tumor virus (MMTV)-ErbB2 mice.7
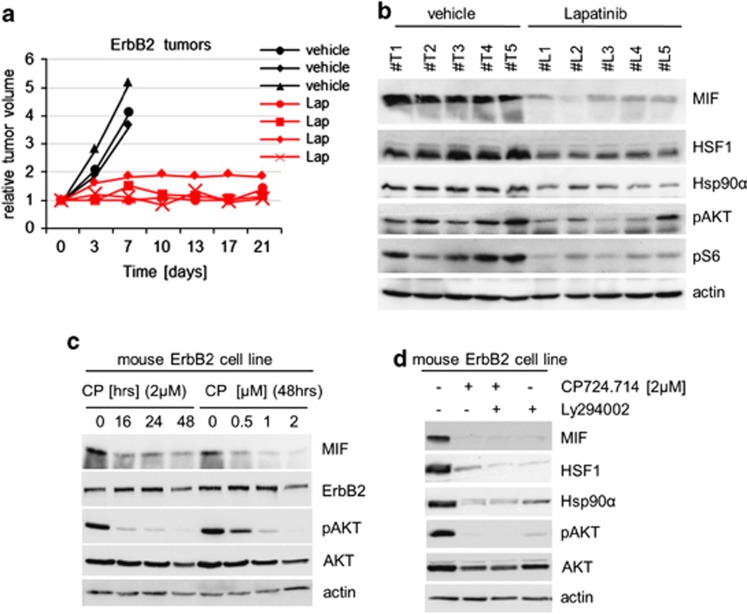
To test whether ErbB2 inhibition also downregulates the HSF1-HSP90-MIF axis and consequently impairs tumor progression in spontaneous HER2/ErbB2-driven breast cancers in vivo, we treated MMTV-ErbB2 mice systemically with Lapatinib, a dual HER2/EGFR inhibitor that is clinically used in HER2-overexpressing breast cancers. All ErbB2-derived tumors treated with Lapatinib showed significant reduction or even cessation of tumor progression compared with vehicle-treated controls (Figure 5a). Importantly, all Lapatinib-treated tumors also showed significant depletion of HSF1 and Hsp90α protein levels and, consequently, of MIF levels in response to ErbB2 inhibition, confirming the mechanistic link in vivo (Figure 5b). Next, we established a stable mouse cell line derived from a primary MMTV-ErbB2 tumor. In agreement with our data in human breast cancer cells, specific HER2/ErbB2 inhibitor CP724.714 again strongly reduced MIF levels in these mouse tumor cells in a dose- and time-dependent manner, and was associated with abolishing PI3K/AKT signaling (Figure 5c). Moreover, as shown by Ly294002 treatment, HSF1 inactivation is dependent on blocking PI3K activity (Figure 5d, lane 4).
Figure 5.
In ErbB2-driven breast cancers in mice, ErbB2 inhibition by Lapatinib inhibits the HSF1-Hsp90-MIF axis and strongly impairs tumor growth. (a) Response to systemic Lapatinib treatment in breast cancer-bearing ErbB2-overexpressing transgenic mice. Time course (in days) of median tumor volumes. Mice with comparably sized spontaneous breast tumors were treated by oral gavage with 100 mg/kg Lapatinib (red lines) or vehicle (black lines) for 5 days per week for 3 contiguous weeks. Tumors were analyzed side-by-side. Response rates of tumors were normalized to their respective starting volume. (b) Lapatinib treatment inactivates the HSF1-HSP90 axis. Immunoblot analysis of lysates of tumors of Lapatinib- and vehicle-treated mice. Actin, loading control. ErbB2/vehicle, five tumors in three mice. ErbB2/Lapatinib, five tumors in four mice. (c) Stable ErbB2-overexpressing tumor cells (derived from a primary mouse MMTV-ErbB2 tumor) confirms HSF1 inactivation and destabilization of HSP90 clients in response to ErbB2 inhibition. Cancer cells were treated in a time- and dose-dependent manner as indicated. Immunoblot analyses for MIF and ErbB2. pAKT is a functional control for ErbB2 inhibition. Actin, loading control. (d) Inhibition of ErbB2 and PI3K leads to inactivation of HSF1 and HSP90. Mouse mammary cancer cells from c were treated with 1 μM CP724.714 or 25 μM Ly294002, alone or in combination as indicated. Immunoblot analysis. pAKT serves as positive control for ErbB2 and PI3K inhibition. Actin, loading control
In human HER2-positive breast cancers, HER2 levels correlate with pSer326 HSF1 activity
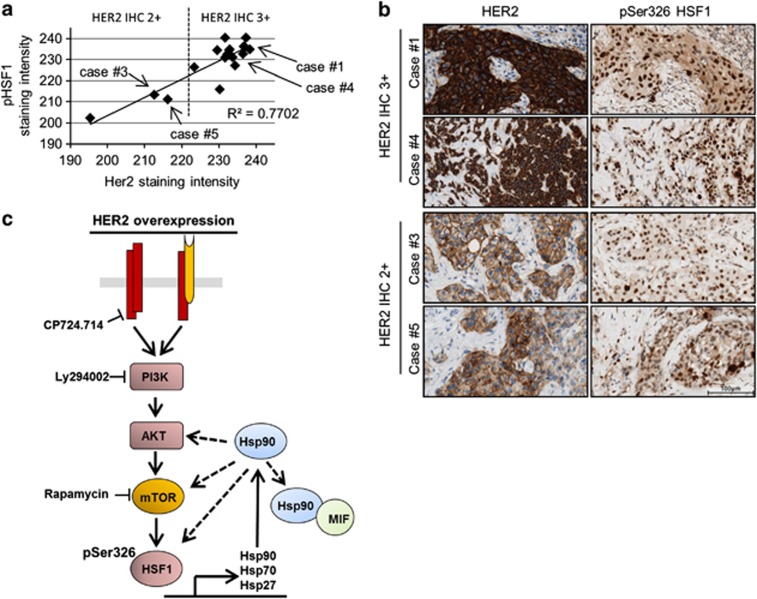
Finally, in agreement with the above data, we also detected a strong correlation in primary human HER2-positive breast cancer samples between their immunohistochemical staining intensities of HER2 (2+ versus 3+) and, specifically, nuclear pSer326 HSF1 as a bona fide tissue marker of HSF1 activity42, 46, 47 (Pearson's correlation coefficient R2=0.8817; n=19 patients; Figures 6a and b).
Figure 6.
In human HER2-positive breast cancers, HER2 levels correlate with pSer326 HSF1 activity. (a and b) Correlation in staining intensity between HER2 and activated HSF1 (pSer326) in human breast cancer. (a) Quantitative immunohistochemistry of HER2 and pSer326 HSF1 protein in HER2-amplified invasive ductal carcinoma with 2+ and 3+ HER2 staining intensity, respectively. Two regions encompassing >60 cells were quantified per case; DAB color intensity displayed as values on an inverted 8 bit gray scale, 0=white, 255=black. (b) Representative photomicrographs of cases of invasive ductal carcinoma quantified in a. Immunohistochemical staining for HER2 and pSer326 HSF1. DAB with hematoxylin counterstain, × 400 magnification. Bar, 100 μm. (c) Proposed model summarizing the findings of this study. In HER2-overexpressing cells, the PI3K–AKT–mTOR pathway is the main signaling axis that leads to phosphorylation of HSF1 at Ser326, which activates HSF1 transcriptional activity and induces, among other target genes, expression of heat-shock proteins. The activated HSP90 machinery stabilizes a broad panel of oncogenic and tumor-promoting proteins. Dashed lines mean feedback loop
Taken together, our data from cultured human cell lines, and murine and human breast cancer tissues, support the model that HSF1 is regulated by HER2 receptor signaling and thus acts as a major downstream tumor promoter in HER2-driven breast cancers in vivo.
Discussion
The transcription factor HSF1 is a major determinant of oncogenesis, as established by genetic cancer models in mice.42, 43, 44, 45, 46 This is based on two mechanisms. HSF1 induces the adaptive proteotoxic stress response of heat-shock chaperones essential for cancer cell survival. Moreover, HSF1 also broadly modulates expression of genes regulating cell cycle signaling, metabolism, adhesion and protein translation.47 Here we provide evidence that overexpression of the receptor tyrosine kinase HER2 in breast cancer leads to concomitant constitutive activation of the HSF1-HSP90 axis, with subsequent stabilization of numerous tumor-promoting HSP90 clients such as MIF, AKT, mTOR and HSF1 itself, thereby causing a robust acceleration in tumor growth. In human and mouse HER2-overexpressing breast cancer systems in vitro and in vivo, we show that inhibiting HER2/ErbB2 leads to inhibition of phosphoactivated Ser326 HSF1, and subsequently blocks the activity of the HSP90 (and likely the HSP27) chaperone machinery. This leads to destabilization of Hsp90 clients, including MIF, AKT mutant p53 and HSF1, which in turn inhibits cancer cell growth. Mechanistically, HER2 signals via the PI3K–AKT–mTOR axis to activate HSF1. Attenuation of the heat-shock/HSF1 response on HER2 inhibition confirms the functional link between HER2 and HSF1.
Our findings provide an explanation for older hints in the literature towards a possible connection between HER2 and HSF1. First, in HER2-overexpressing mice, genetic HSF1 deletion reduces mammary tumorigenesis.45 Second, HER2 promotes glycolysis at least in part through the HSF1-mediated upregulation of LDH-A.49 Third, Her2-induced p21 inactivation and survivin activation is necessary for mammary cell transformation by HSF1.44 And finally, in HER2 non-overexpressing breast cancer cells, Heregulin β1, a ligand for ErbB3/ErbB4, induces increased HSF1 levels, which depends on ErbB2 and the PI3K–AKT axis.52 However, a definitive link and specific mechanism between HER2 overexpression signaling and HSF1 activation did not exist. Our study now provides some answers to both questions. This study demonstrates for the first time the existence of a contiguous signaling axis of a multicomponent HER2–HSF1 pathway, which is constitutively active in HER2-overexpressing cancer cells. Previously, only individual segments of this axis were known. It was reported that in HER2 non-overexpressing cancer cells, PI3K and MAPK/ERK1/2 can activate or repress, respectively, HSF1 transcriptional activity via posttranslational modifications of HSF1, dependent on cell type and cell signal.53, 54, 55 An interesting study last year in HeLa cells showed that mTOR, one of the main targets of AKT, can phosphorylate HSF1 at Ser326.51 We now extend these observations and showed that HER2-overexpressing breast cancer cells require mTOR for HSF1 Ser326 phosphorylation, expression of heat-shock proteins and stabilization of HSP90 clients (Figures 2c and 3d).
In HER2 non-overexpressing cells, the PI3K–AKT–mTOR, as well as the Ras–Raf–MEK–ERK pathway is activated after ligand-dependent homo- or heterodimerization of HER2-receptor family members.5, 6 HER2 homodimers are known to activate the Ras–Raf–MEK–ERK1/2 axis, whereas heterodimers activate both the PI3K–AKT–mTOR and Ras–Raf–MEK–ERK1/2 pathways.5 In contrast, in HER2-overexpressing cells the HER2–PI3K–AKT axis appears to be the major route of HER2 downstream signaling (Figures 2a and b, and 3c and 5d). This finding is further supported by the fact that PI3K inhibition suppresses tumor growth in multiple HER2-amplified and PI3K-mutant breast cancer cell lines.56 Importantly though, overexpression and/or amplification of HER2 renders breast cancer cells independent of ligand binding,2, 3, 6, 48 and thus could be predicted to lead to constitutive HSF1 activation (causing further strong promotion of tumor growth). Our study confirmed this hypothesis. HER2-overexpressing breast cancer cells signal predominantly through the PI3K–AKT axis to activate HSF1, enhancing the co-oncogenic HSP90 chaperone machinery, which endows these cells with increased proteotoxic stress tolerance and stabilizes downstream effectors of HER2-driven mitogenic and survival pathways (Figure 6c).
Over the last decade, aberrantly stabilized MIF in tumor cells has been established as an important tumor promoter with pleiotropic actions in several pathways. Elevated MIF levels occur in multiple human malignancies.11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 57 Recently, we identified MIF as a highly stabilized novel HSP90 client in cancer cells.7 Degradation of MIF protein by HSP90 inhibition via 17AAG strongly inhibits growth of spontaneous breast cancers in MMTV-ErbB2 mice, identifying MIF as a potential drug target for breast cancer therapy.7, 58 Our study now suggests a second, clinically immediately actionable way to inhibit MIF function, based on the individual molecular profile of a given tumor. Indirect MIF destabilization via HER2 inhibition is a ‘clean' targeted treatment embedded in a signaling pathway and appears most attractive in HER2-overexpressing cancers. On the other hand, direct MIF destabilization via HSP90 inhibition is a pleiotropic ‘dirty' treatment7, 58 and could be used in HER2 non-overexpressing cancers. Overall, this renders MIF an even more attractive drug target.
In sum, we showed that HER2/ErbB2 overexpression controls HSF1 activity via a PI3K–AKT–mTOR signaling cascade and, consequently, the aberrant stabilization of tumor-promoting HSP90 clients in cancer cells. This novel pathway opens up actionable therapeutic possibilities, given that HER2 can be selectively targeted.
Materials and Methods
Mouse model
The ErbB2 transgenic mouse FVBN-Tg(MMTV-ErbB2)NK1Mul/J (Jackson Lab, Bar Harbor, ME, USA) was used as spontaneous breast cancer models because of its clear phenotype that mimicks the human disease.59, 60 The expression of the activated ErbB2 (c-neu) oncogene (Val664 to Glu664 mutation) is controlled by the MMTV promoter and, therefore, randomly expressed in mammary gland epithelium from hemizygous mice. Tumor formation is multifocal, stochastic and matches the transgene expression. Homozygous MMTV-ErbB2 mice were crossed for two generations with wild-type 129SV mice. For analysis, heterozygous mice with a mixed FVBN/129SV (25% FVBN:75% 129SV) was used. Palpation for tumors was done twice a week and, as expected, mice developed breast tumors starting from 25 weeks of age. Mice with small, comparably sized tumors were treated with 100 mg/kg Lapatinib (Tyverb, GlaxoSmithKline GmbH & Co. KG, Munich, Germany) by oral gavage (6 days per week with 1 day rest, for 3 weeks) via a gastric cannula by dissolving a 250-mg tablet of Tyverb in HPMC buffer (0.5% hydroxypropyl-methyl-cellulose/0.1%Tween-80 in water). For vehicle treatment, HPMC buffer were given alone. During treatment, tumor sizes were monitored twice a week using a caliper and tumor volumes calculated as ellipsoid (V=abc × 4/3π). At endpoint (tumor size of 15 × 15 × 15 mm for vehicle group), mice were euthanized and tumors were dissected for analysis. All experiments were carried out in full agreement with the Göttingen University Animal Care Committee and the Institutional Guidelines for Humane Use of Animals in Research.
Cell culture, reagents and siRNAs
Human breast cancer cells SK-BR-3 and MDA-MB-231 were cultured in RPMI/10% FBS. BT-474 cells were cultured in RPMI/20% FBS/insulin. Human breast cancer cells MDA-MB-453 were cultured in Leibovitz/10% FBS under CO2-free conditions. Mouse cell line mErbB2, generated from a primary MMTV-HER2 mouse tumor, were cultured on gelatin-coated plates in RPMI media with 15% FCS and PS, non-essential aminoacids, 1% pyruvate and EGF (0.02 μg/ml). Lapatinib for cell culture and CP724.714 was ordered by SelleckBiochem (Munich, Germany) and used as indicated. 17AAG (Calbiochem, Darmstadt, Germany), Ly294002 and U0126 (both Cell Signaling, Frankfurt, Germany) was used as indicated. siRNAs against HER2 (validated, IDs: s611 and s613), MIF (validated, IDs: s8780 and s194615) and HSF1 (pre-designed, IDs: s6950 and s6952) were purchased from Ambion (Hamburg, Germany) (Silencer select siRNAs). siRNAs were transfected with Lipofectamine2000 (Invitrogen, Darmstadt, Germany).
Quantitative RT-PCR
Total RNA from cells was isolated using Trizol reagent (Invitrogen) according to manufacturer's guidelines. Equal amounts of RNA were reverse-transcribed (M-MuLV Reverse Transcriptase from NEB) and real-time PCR analysis was performed using qPCR Master-Mix (75 mM Tris-HCl, pH 8.8, 20 mM (NH4)2SO4, 0.01% Tween-20, 3 mM MgCl2, SYBR Green 1:80,000, 0.2 mM dNTPs, 20 U/ml Taq-polymerase, 0.25% TritonX-100, 0.3 M Trehalose and 0.3 μM primers). Primers: MIF, 5′-AGCAGCTGGCGCAGGCCAC-3′ and 5′-CTCGCTGGAGCCGCCGAAGG-3′ Hsp90AA1, 5′-GCCCAGAGTGCTGAATACCC-3′ and 5′-GTGGAAGGGCTGTTTCCAGA-3′ Hsp70, 5′-TCAAGGGCAAGATCAGCGAG-3′ and 5′-TGATGGGGTTACACACCTGC-3′ Hsp110, 5′-ACTGCTTGTTCAAGAGGGCTGTGA-3′ and 5′-AACATCCACACCCACACACATGCT-3′ SPTAN, 5′-GGATGAAGTGGCAGCTCGTA-3′ and 5′ -CTGGAGGTTCTGCACATTGG-3′ CBX3, 5′-GTTGAAGAGGCAGAGCCTGA-3′ and 5′-TCTTTGCCAGCTTTCTGAGAGT-3′ CDC6, 5′-TAAAAGCCCTGCCTCTCAGC-3′ and 5′-TGAGTGAGGGGGACCATTCT-3′ FASN, 5′-GTGCCCATCCTGGAGAAGTT-3′ and 5′-GGTCACCTTGGTCTGCAGTG-3′ and 36B4, 5′-GCAGATCGGGTACCCAACTGTTG-3′ and 5′-CAGCAGCCGCAAATGCAGATG-3′. Primers were used in a two-step protocol (2 min at 95 °C pre-heating; 40 cycles at 95 °C for 15 s followed by 60 °C for 1 min).
Immunoblotting
Whole-cell lysates were made with RIPA buffer (1% TritonX-100, 1% Desoxycholat, 0.1% SDS, 150 mM NaCl, 10 mM EDTA, 20 mM Tris-HCl, pH 7.5, and complete protease inhibitor mix). Tumor tissues were minced and lysed with RIPA buffer followed by sonication. After a centrifugation step and measuring of protein amounts with a BCA protein assay (Pierce, Bonn, Germany), equal amounts of total protein were separated by SDS gel electrophoresis, transferred onto nitrocellulose membrane (Millipore, Darmstadt, Germany), blocked and probed with the following antibodies: MIF, Sigma, Munich, Germany, human and mouse), Hsp70 (W-27), Hsp90 (H-114), phospho-ERK (E4), total ERK (K23) and p53 (FL393; all for human and mouse, Santa Cruz, Heidelberg, Germany), AKT (9271), phospho-AKT (D9E XP), phospho-S6 (Ser235/236; 2211), total HSF1 (4356), CDC6 (3387), c-Raf (9422), Bcl-xl (2764) and HER2 (29D8; all for human and mouse, Cell Signaling), phospho-Ser326 HSF1 (human, ADI-SPA-902, ENZO, Lörrach, Germany), Hsp27 (G31, Cell Signaling), Hsp90α (human and mouse, Millipore), and Gapdh (8245) and actin (8227; both Abcam, Cambridge, UK).
Immunofluorescence
Cells were seeded on gelatine-coated glass slides and treated or not treated. After fixation with 3.7% paraformaldehyde/PBS for 1 h, samples were permeabilized with 0.05% Tween-20/PBS for 3–5 min at RT and blocked with 10% FBS/1% BSA/ PBS for 1 h. For staining, samples were incubated with total anti-HSF1 antibody (4356, Cell Signaling) overnight at 4 °C. Primary antibodies were detected by AlexaFluor488-conjugated secondary antibody (Molecular Probes, Darmstadt, Germany). Samples were mounted in Fluoromount-G (SouthernBiotech, Birmingham, AL, USA) with DAPI staining solution.
Images were made using a standard fluorescence microscope (Carl Zeiss AG, Jena, Germany). Fluorochromes were visualized with adequate filtersets for DAPI (364/454 nm) and AlexaFluor488 (488/546 nm). Figures were prepared using Adobe Photoshop software.
CELIGO survival assay
For cell survival, equal numbers of treated or untreated cells were plated into 12-well plates. With the Celigo Cytometer (Cyntellect, San Diego, CA, USA), cell confluence was measured over the indicated time periods und calculated with the Celigo software program.
Quantitative immunohistochemistry and human sample analysis
Clinical samples of invasive ductal carcinoma were classified by a board-certified pathologist. HER2 status was determined according to current ASCO/CAP guidelines. Clinical samples of hyperplastic breast tissue were used as reference. Immunohistochemistry was performed on a BenchMark XT autostainer (Ventana Medical Systems Inc., Tucson, AZ, USA). Primary antibodies against HER2 (4B5, Ventana Medical Systems Inc.) and phospho-Ser326 HSF1 (EP1713Y, Abcam) and OptiView DAB IHC detection kits (Ventana Medical Systems Inc.) were used. The glass slides were digitized (Aperio ScanScope XT, Oxford, UK) and quantified with ImageJ image analysis software (http://rsb.info.nih.gov/ij). Areas covered by 3,3′-diaminobezidine precipitate around the cell membranes (HER2) and in the nuclei (pHSF1) of 60 cells in two regions per sample were manually selected. Intensity values (8 bit) were obtained using color deconvolution.1
Acknowledgments
This work was supported in parts by a stipend from the Dorothea Schlözer Program (to RS) and grants from the Deutsche Forschungsgemeinschaft (MO1998 2-1), the National Cancer Institute (R01CA176647) and the Carol Baldwin Breast Cancer Research Fund (to UMM). We thank Marina Pesic (Helmholtz Centre for Infection Research, Braunschweig, Germany) for establishing the stable mouse HER2-overexpressing cell line.
Glossary
- HSF1
heat-shock factor 1
- HER-2
human epidermal growth factor receptor 2
- HSP90
heat-shock protein 90
- HSP
heat-shock protein
- MIF
macrophage migration inhibitory factor
- PI3K
phosphoinositide-3-kinase
- AKT/PKB
proteinkinase B
- mTOR
mammalian target of rapamycin
- EGFR
epidermal growth factor receptor
- Ras
rat sarcoma
- Raf
rat fibrosarcoma
- MEK
mitogen-activated protein kinase kinase
- ERK
extracellular signal-regulated kinase
- Apc
adenomatous polyposis coli protein
- SPTAN (protein-coding gene)
spectrin alpha (non-erythrocytic)
- FASN (protein-coding gene)
fatty acid synthase
- CBX3 (protein-coding gene)
chromobox protein homolog 3
- CDC6 (protein-coding gene)
cell division cycle 6
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Hudziak RM, Schlessinger J, Ullrich A. Increased expression of the putative growth factor receptor p185HER2 causes transformation and tumorigenesis of NIH 3T3 cells. Proc Natl Acad Sci USA. 1987;84:7159–7163. doi: 10.1073/pnas.84.20.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore PP, Pierce JH, Kraus MH, Segatto O, King CR, Aaronson SA. erbB-2 is a potent oncogene when overexpressed in NIH/3T3 cells. Science. 1987;237:178–182. doi: 10.1126/science.2885917. [DOI] [PubMed] [Google Scholar]
- Ursini-Siegel J, Schade B, Cardiff RD, Muller WJ. Insights from transgenic mouse models of ERBB2-induced breast cancer. Nat Rev Cancer. 2007;7:389–397. doi: 10.1038/nrc2127. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer. 2012;12:553–563. doi: 10.1038/nrc3309. [DOI] [PubMed] [Google Scholar]
- Nahta R. Molecular mechanisms of trastuzumab-based treatment in HER2-overexpressing breast cancer. ISRN Oncol. 2012;2012:428062. doi: 10.5402/2012/428062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz R, Marchenko N, Holembowski L, Fingerle-Rowson G, Pesic M, Zender L, et al. Inhibiting the HSP90 chaperone destabilizes macrophage migration inhibitory factor and thereby inhibits breast tumor progression. J Exp Med. 2012;209:275–289. doi: 10.1084/jem.20111117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucala R, Donnelly SC. Macrophage migration inhibitory factor: a probable link between inflammation and cancer. Immunity. 2007;26:281–285. doi: 10.1016/j.immuni.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Conroy H, Mawhinney L, Donnelly SC. Inflammation and cancer: macrophage migration inhibitory factor (MIF) - the potential missing link. Q J Med. 2010;103:831–836. doi: 10.1093/qjmed/hcq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemajerova A, Moll UM, Petrenko O, Fingerle-Rowson G. Macrophage migration inhibitory factor coordinates DNA damage response with the proteasomal control of the cell cycle. Cell Cycle. 2007;6:1030–1034. doi: 10.4161/cc.6.9.4163. [DOI] [PubMed] [Google Scholar]
- Bando H, Matsumoto G, Bando M, Muta M, Ogawa T, Funata N, et al. Expression of macrophage migration inhibitory factor in human breast cancer: association with nodal spread. Jpn J Cancer Res. 2002;93:389–396. doi: 10.1111/j.1349-7006.2002.tb01269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bini L, Magi B, Marzocchi B, Arcuri F, Tripodi S, Cintorino M, et al. Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis. 1997;18:2832–2841. doi: 10.1002/elps.1150181519. [DOI] [PubMed] [Google Scholar]
- Cheng RJ, Deng WG, Niu CB, Li YY, Fu Y. Expression of macrophage migration inhibitory factor and CD74 in cervical squamous cell carcinoma. Int J Gynecol Cancer. 2011;21:1004–1012. doi: 10.1097/IGC.0b013e31821c45b7. [DOI] [PubMed] [Google Scholar]
- Cludts S, Decaestecker C, Johnson B, Lechien J, Leroy X, Kindt N, et al. Increased expression of macrophage migration inhibitory factor during progression to hypopharyngeal squamous cell carcinoma. Anticancer Res. 2010;30:3313–3319. [PubMed] [Google Scholar]
- Hagemann T, Robinson SC, Thompson RG, Charles K, Kulbe H, Balkwill FR. Ovarian cancer cell-derived migration inhibitory factor enhances tumor growth, progression and angiogenesis. Mol Cancer Ther. 2007;6:1993–2002. doi: 10.1158/1535-7163.MCT-07-0118. [DOI] [PubMed] [Google Scholar]
- Hagemann T, Wilson J, Kulbe H, Li NF, Leinster DA, Charles K, et al. Macrophages induce invasiveness of epithelial cancer cells via NF-kappa B and JNK. J Immunol. 2005;175:1197–1205. doi: 10.4049/jimmunol.175.2.1197. [DOI] [PubMed] [Google Scholar]
- He XX, Yang J, Ding YW, Liu W, Shen QY. Xia HHX. Increased epithelial and serum expression of macrophage migartion inhibitory factor (MIF) in gastric cancer carcinogenesis. Gut. 2006;55:797–802. doi: 10.1136/gut.2005.078113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira E, Ono T, Dhar DK, El-Assal ON, Hishikawa Y, Yamanoi A, et al. Overexpression of macrophage migration inhibitory factor induces angiogenesis and deteriorates prognosis after radical resection for hepatocellular carcinoma. Cancer. 2005;103:588–598. doi: 10.1002/cncr.20818. [DOI] [PubMed] [Google Scholar]
- Meyer-Siegler KL. Increased stability of macrophage migration inhibitory factor (MIF) in DU-145 prostate cancer cells. J Inter Cytokine Res. 2000;20:769–778. doi: 10.1089/10799900050151030. [DOI] [PubMed] [Google Scholar]
- Meyer-Siegler KL, Iczkowski KA, Leng L, Bucala R, Vera PL. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J Immunol. 2006;177:8730–8739. doi: 10.4049/jimmunol.177.12.8730. [DOI] [PubMed] [Google Scholar]
- Pyle ME, Korbonits M, Gueorguiev M, Jordan S, Kola B, Morris DG, et al. Macrophage migration inhibitory factor expression is increased in pituitary adenoma cell nuclei. J Endocrinol. 2003;176:103–110. doi: 10.1677/joe.0.1760103. [DOI] [PubMed] [Google Scholar]
- Reome JB, Hylind JC, Dutton RW, Dobrzanski MJ. Type 1 and type 2 tumor infiltrating effector cell subpopulations in progressive breast cancer. Clin Immunol. 2004;111:69–81. doi: 10.1016/j.clim.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Tomiyasu M, Yoshino I, Suemitsu R, Okamoto T, Sugimachi K. Quantification of Macrophage migration inhibitory factor mRNA expression in non-small cell lung cancer tissues and its clinical signficance. Clin Cancer Res. 2002;8:3755–3760. [PubMed] [Google Scholar]
- Verjans E, Noetzel E, Bektas N, Schütz AK, Lue H, Lennartz B, et al. Dual role of macrophage migration inhibitory factor (MIF) in human breast cancer. BMC Cancer. 2009;9:1–18. doi: 10.1186/1471-2407-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Tian XY, Li Y, Li B, Li Z. Elevated expression of macrophage migration inhibitory factor correlates with tumor recurrence and poor prognosis of patients with gliomas. J Neurooncol. 2012;106:43–51. doi: 10.1007/s11060-011-0640-3. [DOI] [PubMed] [Google Scholar]
- Xu X, Wang B, Ye C, Yao C, Lin Y, Huang X, et al. Overexpression of macrophage migration inhibitory factor induces angiogenesis in human breast cancer. Cancer Lett. 2008;261:147–157. doi: 10.1016/j.canlet.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Zhou S, Xu S, Zhang H, Liu Z, Liang Z, Song X, et al. [Prognostic significance of angiogenesis and blood vessel invasion in stage I non-small cell lung cancer after complete surgical resection] Zhongguo Fei Ai Za Zhi. 2007;10:29–33. doi: 10.3779/j.issn.1009-3419.2007.01.07. [DOI] [PubMed] [Google Scholar]
- Petrenko O, Moll UM. Macrophage migration inhibitory factor MIF interferes with the Rb-E2F pathway. Mol Cell. 2005;17:225–236. doi: 10.1016/j.molcel.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Talos F, Mena P, Fingerle-Rowson G, Moll UM, Petrenko O. MIF loss impairs Myc-induced lymphomagenesis. Cell Death Differ. 2005;12:1319–1328. doi: 10.1038/sj.cdd.4401653. [DOI] [PubMed] [Google Scholar]
- Martin J, Duncan FJ, Keiser T, Shin S, Kusewitt DF, Oberyszyn T, et al. Macrophage migration inhibitory factor (MIF) plays a critical role in pathogenesis of ultraviolet-B (UVB)- induced nonmelanoma skin cancer (NMSC) FASEB J. 2009;23:720–730. doi: 10.1096/fj.08-119628. [DOI] [PubMed] [Google Scholar]
- Wilson JM, Coletta PL, Cuthbert RJ, Scott N, MacLennan K, Hawcroft G, et al. Macrophage migration inhibitory factor promotes intestinal tumorigenesis. Gastroenterology. 2005;129:1485–1503. doi: 10.1053/j.gastro.2005.07.061. [DOI] [PubMed] [Google Scholar]
- Taylor JA, Kuchel GA, Hedge P, Voznesensky OS, Claffey K, Tsimikas J, et al. Null mutation for macrophage migration inhibitory factor (MIF) is associated with less agressive bladder cancer in mice. BMC Cancer. 2007;7:1–8. doi: 10.1186/1471-2407-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Jung WH, Koo JS. Metabolism-related proteins are differentially expressed according to the molecular subtype of invasive breast cancer defined by surrogate immunohistochemistry. PathobiolJ Immunopathol Mol Cell Biol. 2013;80:41–52. doi: 10.1159/000339513. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nature reviews. Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Trepel J, Mollarpour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimnaugh EG, Chavany C, Neckers L. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldamycin. J. Biol Chem. 1996;271:22796–22801. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, Rosen N. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. JBC. 2002;227:39858–39866. doi: 10.1074/jbc.M206322200. [DOI] [PubMed] [Google Scholar]
- Schulte TW, Blagosklonny MV, Ingui C, Neckers L. Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J Biol Chem. 1995;270:24585–24588. doi: 10.1074/jbc.270.41.24585. [DOI] [PubMed] [Google Scholar]
- Li D, Marchenko ND, Schulz R, Fisher V, Velasco-Hernandez T, Talos F, et al. Functional inactivation of endogenous MDM2 and CHIP by Hsp90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res. 2011;9:577–588. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Bharadwaj S, O'Carroll R, Ovsenek N. HSP90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes. Mol Cell Biol. 1998;18:4949–4960. doi: 10.1128/mcb.18.9.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci USA. 2011;108:18378–18383. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Gabai VL, Sherman MY. Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene. 2010;29:5204–5213. doi: 10.1038/onc.2010.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi C, Hu Y, Buckhaults P, Moskophidis D, Mivechi NF. Heat shock factor Hsf1 cooperates with ErbB2 (Her2/Neu) protein to promote mammary tumorigenesis and metastasis. J Biol Chem. 2012;287:35646–35657. doi: 10.1074/jbc.M112.377481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Santagata S, Tang Z, Shi J, Cao J, Kwon H, et al. Loss of tumor suppressor NF1 activates HSF1 to promote carcinogenesis. J Clin Invest. 2012;122:3742–3754. doi: 10.1172/JCI62727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK, Gong J. Molecular chaperones in mammary cancer growth and breast tumor therapy. J Cell Biochem. 2012;113:1096–1103. doi: 10.1002/jcb.23461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YH, Zhou M, Liu H, Ding Y, Khong HT, Yu D, et al. Upregulation of lactate dehydrogenase A by ErbB2 through heat shock factor 1 promotes breast cancer cell glycolysis and growth. Oncogene. 2009;28:3689–3701. doi: 10.1038/onc.2009.229. [DOI] [PubMed] [Google Scholar]
- Guettouche T, Boellmann F, Lane WS, Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SD, Prince T, Gong J, Calderwood SK. mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PLoS One. 2012;7:e39679. doi: 10.1371/journal.pone.0039679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleque MA, Bharti A, Sawyer D, Gong J, Benjamin IJ, Stevenson MA, et al. Induction of heat shock proteins by heregulin beta1 leads to protection from apoptosis and anchorage-independent growth. Oncogene. 2005;24:6564–6573. doi: 10.1038/sj.onc.1208798. [DOI] [PubMed] [Google Scholar]
- Bijur GN, Jope RS. Opposing actions of phosphatidylinositol 3-kinase and glycogen synthase kinase-3beta in the regulation of HSF-1 activity. J Neorochem. 2000;75:2401–2408. doi: 10.1046/j.1471-4159.2000.0752401.x. [DOI] [PubMed] [Google Scholar]
- Chu B, Soncin F, Price BD, Stevenson MA, Calderwood SK. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J Biol Chem. 1996;271:30847–30857. doi: 10.1074/jbc.271.48.30847. [DOI] [PubMed] [Google Scholar]
- He B, Meng YH, Mivechi NF. Glycogen synthase kinase 3beta and extracellular signal-regulated kinase inactivate heat shock transcription factor 1 by facilitating the disappearance of transcriptionally active granules after heat shock. Mol Cell Biol. 1998;18:6624–6633. doi: 10.1128/mcb.18.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She QB, Chandarlapaty S, Ye Q, Lobo J, Haskell KM, Leander KR, et al. Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS One. 2008;3:e3065. doi: 10.1371/journal.pone.0003065. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang C, Liang T, Song J, Jiang S, Qu L, Hou G. Evaluation of macrophage migration inhibitory factor as an imaging marker for hepatocellular carcinoma in murine models. Scand J Gastroenterol. 2011;46:720–726. doi: 10.3109/00365521.2011.568517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz R, Dobbelstein M, Moll UM. HSP90 inhibitor antagonizing MIF: The specifics of pleiotropic cancer drug candidates. Oncoimmunology. 2012;1:1425–1426. doi: 10.4161/onci.21173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrington RE, Subler MA, Rands E, Omer CA, Miller PJ, Hundley JE, et al. A farnesyltransferase inhibitor induces tumor regression in transgenic mice harboring multiple oncogenic mutations by mediating alterations in both cell cycle control and apoptosis. Mol Cell Biol. 1998;18:85–92. doi: 10.1128/mcb.18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiff RD, Anver MR, Gusterson BA, Hennighausen L, Jensen RA, Merino MJ, et al. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene. 2000;19:968–988. doi: 10.1038/sj.onc.1203277. [DOI] [PubMed] [Google Scholar]