Abstract
We provide the first biochemical evidence of a direct interaction between the glutathione transferase P1-1 (GSTP1-1) and the TRAF domain of TNF receptor-associated factor 2 (TRAF2), and describe how ligand binding modulates such an equilibrium. The dissociation constant of the heterocomplex is Kd=0.3 μM; however the binding affinity strongly decreases when the active site of GSTP1-1 is occupied by the substrate GSH (Kd≥2.6 μM) or is inactivated by oxidation (Kd=1.7 μM). This indicates that GSTP1-1's TRAF2-binding region involves the GSH-binding site. The GSTP1-1 inhibitor NBDHEX further decreases the complex's binding affinity, as compared with when GSH is the only ligand; this suggests that the hydrophobic portion of the GSTP1-1 active site also contributes to the interaction. We therefore hypothesize that TRAF2 binding inactivates GSTP1-1; however, analysis of the data, using a model taking into account the dimeric nature of GSTP1-1, suggests that GSTP1-1 engages only one subunit in the complex, whereas the second subunit maintains the catalytic activity or binds to other proteins. We also analyzed GSTP1-1's association with TRAF2 at the cellular level. The TRAF2–GSTP1-1 complex was constitutively present in U-2OS cells, but strongly decreased in S, G2 and M phases. Thus the interaction appears regulated in a cell cycle-dependent manner. The variations in the levels of individual proteins seem too limited to explain the complex's drastic decline observed in cells progressing from the G0/G1 to the S–G2–M phases. Moreover, GSH's intracellular content was so high that it always saturated GSTP1-1. Interestingly, the addition of NBDHEX maintains the TRAF2–GSTP1-1 complex at low levels, thus causing a prolonged cell cycle arrest in the G2/M phase. Overall, these findings suggest that a reversible sequestration of TRAF2 into the complex may be crucial for cell cycle progression and that multiple factors are involved in the fine-tuning of this interaction.
Keywords: TRAF, GST, MAPK, NBDHEX, osteosarcoma
The TNF receptor-associated factors, also known as TRAFs, are a family of proteins that directly or indirectly interact with and modulate the function of many receptors involved in the regulation of various cellular responses, including death, survival and differentiation.1, 2, 3 Human TRAFs share a trimeric structure and mushroom-shape with a conserved carboxy-terminal region (TRAF-C domain) as the cap, and a coiled-coil region (TRAF-N domain) as the stalk. Both domains form the TRAF domain responsible for self-association and for interaction with upstream receptors. The TRAFs' amino-terminal region contains a RING finger and several zinc-finger motifs, and is responsible for interaction with downstream molecules.4 Among the TRAF family, TRAF2 is the most ubiquitously expressed and widely studied member. TNF receptor activation induces TRAF2 recruitment, which in turn leads to the activation of downstream signaling cascades including those of the c-Jun N-terminal kinase (JNK)5 and nuclear factor (NF)-κB.6, 7 These cascades represent two different responses to TNF receptor activation; the bifurcation of their pathways occurs at the level of TRAF2.8 TRAF2 mediates JNK activation via the apoptosis signal-regulating kinase 1 (ASK1), a mitogen-activated protein kinase kinase kinase (MAP3K).9, 10 In resting cells, ASK1 forms an inactive complex together with the redox-sensitive protein thioredoxin, which blocks its interaction with TRAF2.11 On the other hand, TNF receptor activation and the subsequent TRAF2 recruitment to the plasma membrane stimulates the production of reactive oxygen species (ROS),12 which, by disrupting the ASK1–thioredoxin complex, allow the binding of ASK1 to the TRAF domain of TRAF2 and the activation of the ASK1–JNK signaling cascade. The formation of the TRAF2–ASK1 complex is essential for the apoptotic cell death response triggered by TNF receptor stimulation and oxidative stress.13, 14
More recently, GSTP1-1 has emerged as a new inhibitor of the TRAF2 signaling pathway, having an antiapoptotic role. GSTP1-1 is overexpressed in many cancers, this having been related to the development of multidrug resistance. In fact, several anticancer drugs can be neutralized by the GST-catalyzed reaction, and extruded from the cell. Moreover, GSTP1-1 is able to bind directly to the mitogen-activated protein kinase (MAPK) JNK, thus inhibiting its pro-apoptotic action.15, 16 Wu et al.17 demonstrated that GSTP1-1 is also associated with TRAF2 in cervical carcinoma (HeLa) and human embryonic kidney (HEK293) cells. Moreover, we have recently demonstrated the interaction of GSTP1-1 with TRAF2 in human osteosarcoma cell lines by immunoprecipitation.18 Of note, GSTP1-1 seems to interact specifically with the ASK1–JNK pathway, as it does not inhibit the activation of NF-κB or the formation of the complex between TRAF2 and MEKK1, another MAP3K mediating JNK activation.17, 19 Furthermore, evidence has been provided by Wu et al.17 that the ASK1–JNK pathway activation and cell death induced by TNFα are both inhibited by GSTP1-1 overexpression, whereas apoptosis of TNFα-treated cells is enhanced by GSTP1-1 knockdown. Given these premises, the present study has been designed to characterize the TRAF2–GSTP1-1 complex at a molecular level and to investigate the complex's formation in situ.
Results
TRAF2 purification and structure analysis
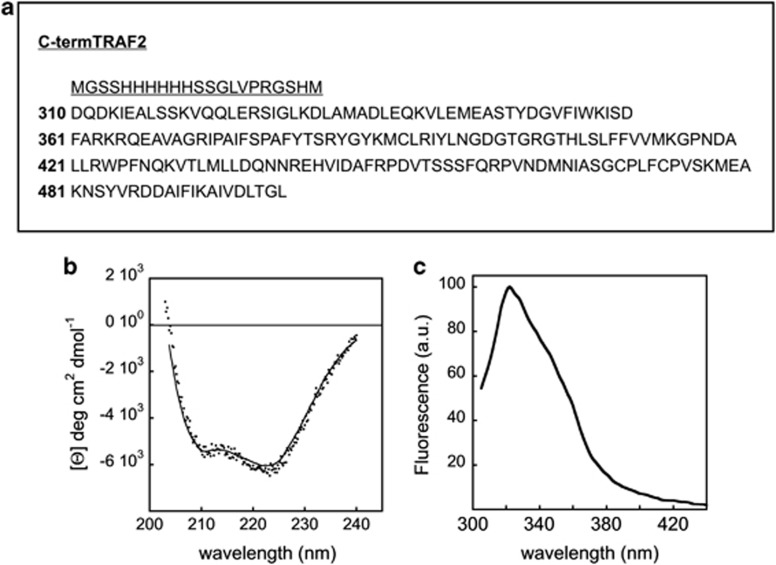
We expressed and purified a His-tagged C-terminal domain of TRAF2 (residues 310–501) (Figure 1a) comprising a portion of the coiled-coil α-helix domain (TRAF-N domain) and the conserved TRAF-C domain that is capable of binding to the cytoplasmic region of receptors as well as to cytoplasmic signaling proteins.4 The TRAF-C domain is known to form an eight-stranded antiparallel β-sandwich structure containing two twisted, four-stranded, antiparallel β-sheets and a long helical segment linking strands β1 and β2.4 In order to check whether the truncated TRAF2 maintains the folded conformation of the full-length protein, we investigated its secondary and tertiary structure content through spectroscopic measurements. Figure 1b shows the circular dichroism (CD) in the peptidic region of truncated TRAF2. The protein spectrum has the typical features (shape and minima) exhibited by α/β proteins.20 A quantitative analysis of the spectrum yields the values reported in Table 1. It is worth mentioning that the estimated secondary structure content is in line with previously published crystallographic data.4
Figure 1.
Structure and spectroscopic analysis of purified TRAF2. (a) Sequence coding for the His-tagged C-terminal domain of TRAF2 (residues 310–501); underlined is reported as the His-tagged sequence. The sequence was cloned into the pET28a(+) (Novagen) expression vector. The resulting plasmid was used to transform E. coli BL21 (DE3) cells. (b) CD spectrum in the peptidic region of the truncated TRAF2 (2 μM). The protein spectrum has the typical features (shape and minima) exhibited by α/β proteins. The spectrum was recorded on a JASCO J-710 spectropolarimeter at 20 °C. (c) Steady-state fluorescence spectrum of truncated TRAF2 (2 μM); excitation was set at 292 nm. The spectrum exhibits a peak around 320 nm, diagnostic of buried tryptophan residues. The spectrum was recorded at 20 °C on the PC1-ISS photon counting fluorometer
Table 1. Spectroscopic versus structural properties of TRAF2.
The fluorescence properties of the two tryptophan residues located in the TRAF domain have also been studied. In particular, the steady-state spectrum shown in Figure 1c exhibits a pronounced peak around 320 nm, indicating that the two tryptophan residues present in each TRAF2 subunit are fully buried.21 The resulting center of mass and the measured steady-state anisotropy are reported in Table 1. Both CD and fluorescence measurements provide evidence that the TRAF2 construct used in this study retained the structural features expected for the complete and folded protein.
Evidence of the direct binding between GSTP1-1 and TRAF2
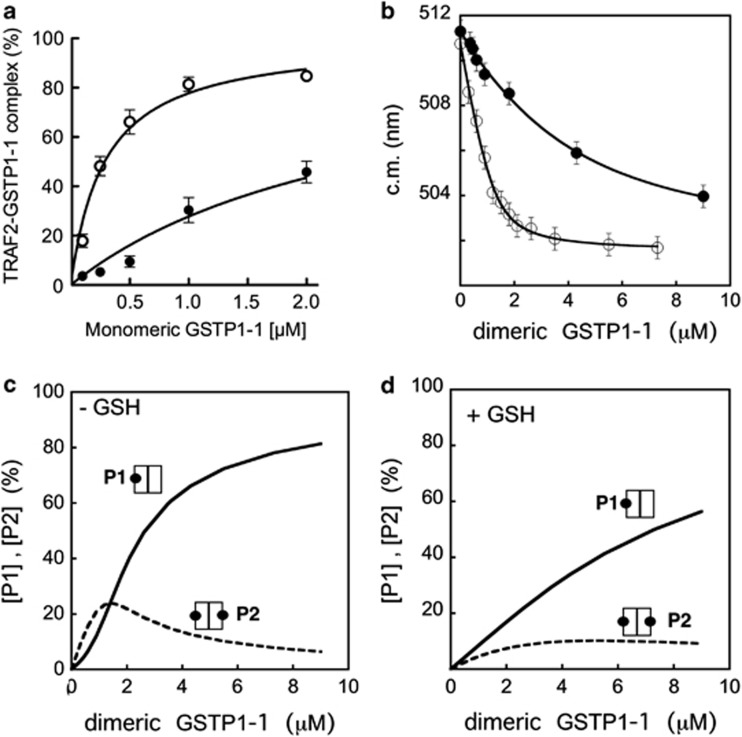
To obtain evidence for the formation of a complex between GSTP1-1 and TRAF2, we used an enzyme-linked immuno sorbent assay (ELISA), where increasing amounts of GSTP1-1 were added to His-tagged TRAF2 immobilized on a Ni-NTA-coated plate. The amount of bound GSTP1-1 was then revealed by an anti-GSTP1-1 specific antibody. A dose-dependent increase in the antibody signal was observed on addition of GSTP1-1, proving the formation of a complex (Figure 2a). To analyze the binding data, we used equation (1), describing a 1 : 1 interaction between one TRAF2 monomer and one GSTP1-1 subunit, the binding sites being equal and independent (see Materials and Methods section). The estimated equilibrium dissociation constant for the TRAF2–GSTP1-1 complex was Kd=0.29±0.02 μM (Table 2). The same model was previously used to analyze the interaction between GSTP1-1 and JNK1α2 obtaining a Kd value of 0.39±0.14 μM.15
Figure 2.
Evidence of the direct binding between TRAF2 and GSTP1-1. (a) ELISA detection of the TRAF2–GSTP1-1 interaction. His-tagged TRAF2 (0.005 μM) was immobilized on Ni-NTA-coated plates and incubated with increasing amounts of GSTP1-1 (from 0.1 to 2 μM (-○-) refers to the concentration monomeric protein). The same experiment was repeated in presence of saturating GSH concentrations (1 mM) (-●-). The curves represent the best fit of data to equation (1) that fulfills the Kd values for the TRAF2–GSTP1-1 complex (Table 2). (b) Fluorometric detection of the TRAF2–GSTP1-1 interaction. Dansylated TRAF2 (2.5 μM) was incubated with increasing amounts of GSTP1-1 (from 0.2 to 9 μM (-○-) refers to the concentration of dimeric protein). The interaction of dansylated TRAF2 (5.9 μM) with GSTP1-1 was also analyzed in the presence of 1 mM GSH (-●-). The curves represent the best fit of data to equation (2) that fulfills the Kd values for the TRAF2–GSTP1-1 complex (Table 2). Each point represents the mean±S.E.M. of at least three different experimental sets. (c) Theoretical dependence of the P1 (one TRAF2 monomer bound to dimeric GSTP1-1) and P2 species (two TRAF2 monomers bound to dimeric GSTP1-1) from the total dimeric GSTP1-1 concentration, assuming a Kd value of 0.3 μM for the TRAF2–GSTP1-1 complex (i.e. in the absence of GSH). (d) The same as (c) assuming a Kd value of 3 μM for the TRAF2–GSTP1-1 complex (i.e. in the presence of GSH)
Table 2. Dissociation constants (μM) of the TRAF2–GSTP1-1 complex.
| GSTP1-1 | GSTP1-1 ligand | Kd by ELISA | Kd by fluorometry |
|---|---|---|---|
| Native |
None |
0.29±0.02 |
0.30±0.02 |
| |
GSH 0.03 mM |
0.49±0.06 |
nd |
| |
GSH 0.3 mM |
0.93±0.10 |
nd |
| |
GSH 1 mM |
2.60±0.30 |
4.60±0.70 |
| |
GSH 0.03 mM, NBDHEX 8 μM |
1.20±0.10 |
nd |
| |
GSH 0.3 mM, NBDHEX 8 μM |
2.70±0.20 |
nd |
| |
GSH 1 mM, NBDHEX 8 μM |
≥ 5 |
nd |
| Oxidized | None | 1.70±0.20 | nd |
Abbreviation: Nd, not determined
Fluorescence analysis confirms the binding between GSTP1-1 and TRAF2
The interaction of TRAF2 with GSTP1-1 was also monitored by steady-state fluorescence, measuring the perturbation of the emission spectrum of dansylated TRAF2 occurring on addition of GSTP1-1. A GSTP1-1 concentration-dependent blue shift of the dansylated TRAF2 spectrum was observed, diagnostic of the interaction between the two proteins (Figure 2b). The fluorescence data were fitted to both the general equations (1) and (2) (see Materials and Methods section); the latter describes the equilibrium between the species P1 (one TRAF2 monomer bound to one GSTP1-1 dimer) and P2 (two TRAF2 monomers bound to one GSTP1-1 dimer), the GSTP1-1-binding sites being equal and independent. Both fits gave an apparent dissociation constant of 0.3 μM (Table 2), a value that overlaps the Kd obtained with the ELISA assay by using equation (1). In addition, the second model allows the determination of the relative amounts of the P1 and P2 species at different GSTP1-1 concentrations. Figure 2 shows that, in the absence of GSH, P2 is the prevalent form at low-GSTP1-1 concentrations (panel c), while in the presence of saturating GSH (see below), P1 is always the most abundant species (panel d).
The GST substrate GSH decreases the interaction between GSTP1-1 and TRAF2
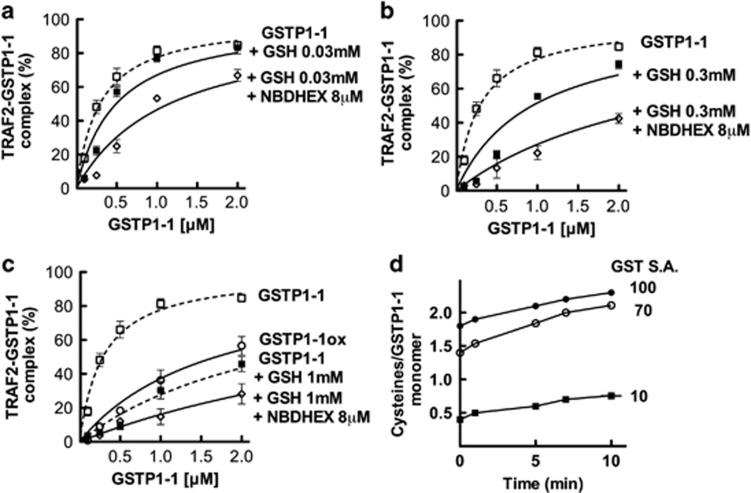
Under physiological conditions, protein–protein interactions may be influenced by ligands such as enzyme substrates or inhibitors. We have previously shown that the specific GST substrate, GSH, strongly reduces the affinity of GSTP1-1 for JNK1.15 In order to examine the role of GSH in the TRAF2–GSTP1-1 interaction, we monitored the formation of the TRAF2–GSTP1-1 complex, by using ELISA, in the presence of GSH concentrations ranging between 0.03 and 1 mM, that is, from 1/3 to 10 times the Km of GSTP1-1 for GSH.22 In the presence of GSH the amount of the protein–protein complex decreased in a dose-dependent manner. In particular, the apparent dissociation constant rose from a value ∼0.3 μM, in the absence of GSH, to a value ∼3 μM, under saturating GSH concentrations (Figure 2a, Figures 3a–c and Table 2). The effect of GSH was confirmed by fluorometric experiments: in the presence of saturating GSH (1 mM), the addition of GSTP1-1 to dansyl-TRAF2 caused a lower perturbation of the fluorescence emission spectrum (Figure 2b), the dissociation constant, calculated using both equations (1) and (2), being 4.6±0.7 μM (Table 2).
Figure 3.
Effect of ligands and of oxidative state of GSTP1-1 on the TRAF2–GSTP1-1 interaction. (a) His-Tagged TRAF2 (0.005 μM) was immobilized on Ni-NTA-coated plates and incubated with increasing amounts of GSTP1-1 (from 0.1 to 2 μM, the concentration refers to the monomeric protein) in the absence (-□-) or in the presence of subsaturating (0.03 mM) GSH, with (-◊-) or without (-▪-) 8 μM NBDHEX. The same experiment was repeated in the presence of increasing amounts of GSH: 0.3 mM (b) and 1 mM (c). In addition, c shows the binding of TRAF2 to the oxidized form of GSTP1-1 (-○-). The curves represent the best fit of data to equation (1) that fulfills the Kd values for the TRAF2–GSTP1-1 complex (Table 2). Each point represents the mean±S.E.M. of at least three different experimental sets. (d) GST activity and cysteine reactivity of GSTP1-1 before and after oxidative inactivation. The fully active enzyme (-●-) shows two fast reacting sulfhydryls/subunit and a third slow reactive thiol, whereas the inactivated GSTP1-1 (-▪-) has only one slow reactive -SH group/subunit. The inactivation was reverted by incubation with 100 mM DTT and 1 mM GSH (-○-). GSTP1-1 SA expressed as μmol/min/mg protein
‘Oxidized' GSTP1-1 has low affinity for TRAF2
Cellular oxidative stress is known to cause the inactivation and release of GSTP1-1 from the complex with JNK.23 We thus evaluated the effect of GSTP1-1's oxidative inactivation on its interaction with TRAF2. To this end, GSTP1-1 was incubated under conditions that promote the formation of one intra-chain disulfide bond between cysteine 47 and 101 residues.24 Figure 3d shows the specific activity (SA) and the sulfhydryl group reactivity of GSTP1-1 before and after the oxidative reaction. After incubation with DTT and GSH, both the reactivity of Cys-47 and Cys-101 sulfhydryls and the enzymatic activity were restored, thus confirming the thiol-reversible nature of the inactivation. The ‘oxidized' form of GSTP1-1 exhibited low affinity for TRAF2 in the ELISA assay (Kd=1.7±0.2 μM); the binding curve was highly similar to that obtained with the fully active GSTP1-1 saturated with GSH (Figure 3c and Table 2).
The GST inhibitor 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) decreases the interaction between GSTP1-1 and TRAF2
We also analyzed the effect of the strong GST inhibitor NBDHEX25, 26 on the amount of the complex between GSTP1-1 and TRAF2. We had previously shown by immunoprecipitation that NBDHEX treatment triggers the dissociation of the TRAF2–GSTP1-1 complex in U-2OS cells.18 Here, we show that addition of NBDHEX to a mixture of GSTP1-1, TRAF2 and GSH of different concentrations (between 0.03 and 1 mM) further decreases the amount of protein complex, compared with the case when GSH is the only ligand (Figures 3a–c and Table 2). Specifically, in the presence of subsaturating GSH, NBDHEX leads to a value of Kd∼2 μM, whereas in the presence of saturating GSH, NBDHEX leads to a value of Kd≥5 μM. Therefore the effect of NBDHEX is strictly correlated to the concentration of GSH.
The TRAF2–GSTP1-1 complex is affected by cell cycle progression
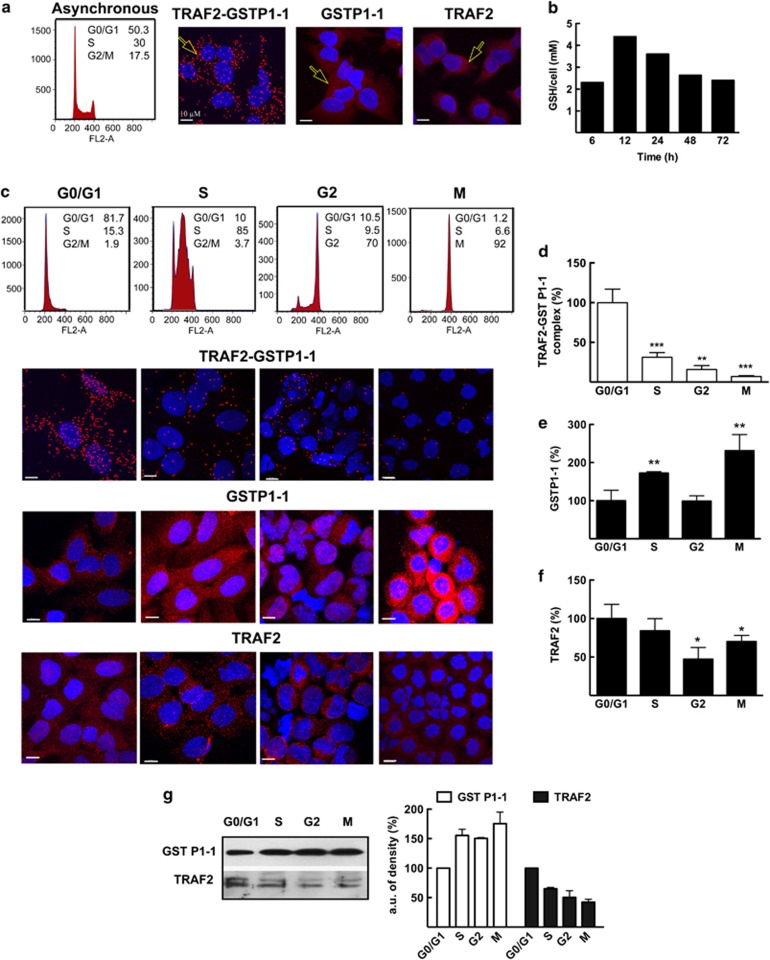
The in situ formation and localization of the TRAF2–GSTP1-1 complex was investigated by the proximity ligation assay (PLA) in U-2OS osteosarcoma cells. The PLA procedure makes it possible to visualize individual protein–protein interaction events, generating a fluorescent spot only when two proteins are in close proximity (<40 nm).27, 28 We found that the complex between GSTP1-1 and TRAF2 is constitutively present in U-2OS cells, being localized in both the cytoplasm and nucleus (Figure 4a). Next, given GSH's role in modulating the TRAF2–GSTP1-1 interaction, we analyzed the intracellular GSH content in proliferating U-2OS cells. We noted that the thiol fluctuations, over a time period of 72 h, were limited and never attained values below 2 mM (Figure 4b); in other words, the intracellular GSH always saturated GSTP1-1.22 On the other hand, experiments performed on synchronized U-2OS-cell cultures revealed that the interaction between TRAF2 and GSTP1-1 was markedly affected by cell cycle progression. Cell cycle synchronization was achieved by treating the cultures with thymidine to arrest cells in the G0/G1 phase. This was followed by a treatment with deoxycytidine in order to promote entry into the S phase then, subsequently, by treatment with the microtubule inhibitor nocodazole to arrest cells in the G2–M phases. The association between TRAF2 and GSTP1-1 was significantly higher in the G0/G1 phase, whereas it decreased to a minimum in the G2 and M phases (Figures 4c and d); therefore, the amount of TRAF2 associated with GSTP1-1 appears to be regulated in a cell cycle-dependent fashion. None of the molecules utilized for cell synchronization inhibited GSTP1-1's catalytic activity (data not shown); therefore we can rule out these molecules' direct interaction with the TRAF2–GSTP1-1 complex. The findings reported above prompted us to assess whether the observed cell cycle-dependent variations in the amount of the TRAF2–GSTP1-1 complex could be related to the intracellular levels of either GSTP1-1 or TRAF2. Confocal microscopy revealed an increase of GSTP1-1 in the S and G2–M phases compared with the G0/G1 phase (Figures 4c and e). This result was further confirmed by western blot analysis (Figure 4g). Conversely, both methods showed a decrease in TRAF2 during the progression from the G0/G1 to the S–G2–M phases (Figures 4c, f and g).
Figure 4.
In situ detection of the TRAF2–GSTP1-1 complex and evaluation of intracellular GSH, GSTP1-1 and TRAF2 levels. (a) Cell cycle analysis and confocal fluorescence imaging of U-2OS cells 24 h after plating. The TRAF2–GSTP1-1 complex was visualized by PLA, which generates red dots when the two proteins are in close proximity (see arrow). GSTP1-1 and TRAF2 were detected with specific primary antibodies and Alexa 647-conjugated secondary antibodies, which generate a diffuse red fluorescence (see arrows). DAPI (blue fluorescence) was used to counterstain cell nuclei. (b) The GSH content of proliferating cells was determined by HPLC analysis. The molar concentration of GSH was obtained using the U-2OS-cell volume of 4000 μm3. (c) Cell cycle analysis and confocal fluorescence imaging of synchronized U-2OS cells. The amount per cell of (d)TRAF2–GSTP1-1 complex, (e) GSTP1-1 and (f) TRAF2 were normalized to the content obtained at the G0/G1 phase. (g) Synchronized cells were also subjected to immunoblot analysis with anti-GSTP1-1 and anti-TRAF2 antibodies. Each point represents the mean±S.E.M. of at least three different experimental sets. *P<0.05, **P<0.005 and ***P<0.0005
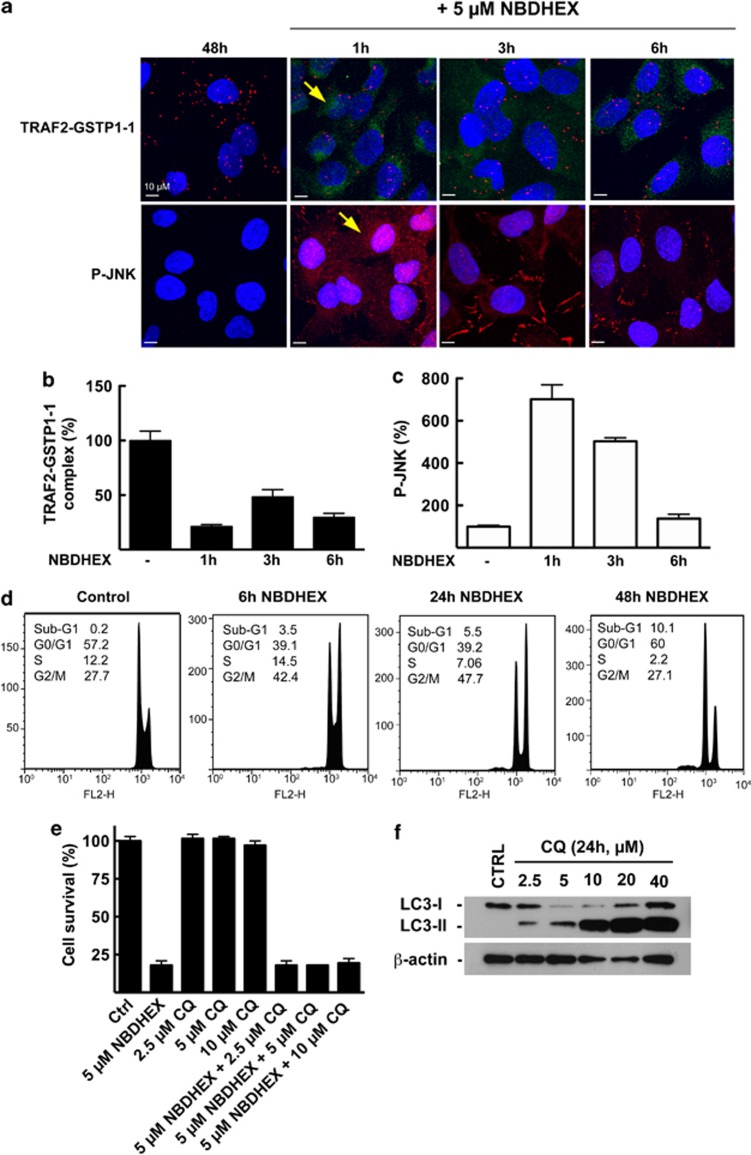
In situ evidence of the TRAF2–GSTP1-1 complex's NBDHEX-induced dissociation and of cellular outcomes following NBDHEX treatment
NBDHEX's ability to induce the dissociation of the complex between GSTP1-1 and JNK1, leading to JNK1 activation, is well documented.25 In a previous study, we demonstrated through immunoprecipitation that NBDHEX induced the dissociation of the complex between GSTP1-1 and TRAF2 as well.18 Here, following the TRAF2–GSTP1-1 interaction in situ through PLA, we confirm NBDHEX's ability to induce the dissociation of this complex; we also show that such an event is paralleled by JNK activation. Indeed, in U-2OS cells treated with 5 μM NBDHEX, a massive decrease in the association of TRAF2 with GSTP1-1 was evident even after 1 h of treatment (Figures 5a and b). The intrinsic fluorescence (green) of NBDHEX was localized in the cell cytoplasm; the presence of the drug persisted up to 6 h, maintaining the TRAF2–GSTP1-1 complex at low levels (Figures 5a and b). At the same time, the level of phospho-active JNK was assessed by confocal imaging, a strong activation of the kinase being evident within 1 h of treatment (Figures 5a and c).
Figure 5.
Effects of NBDHEX on the interaction between GSTP1-1 and TRAF2, JNK phospho-activation and cell cycle progression/cell death. (a) U-2OS cells were treated with 5 μM NBDHEX 48 h after plating and stained with specific antibodies at 1, 3 or 6 h of treatment, to detect the TRAF2–GSTP1-1 complex (PLA: red dots) and the phospho-activated form of JNK (diffuse red fluorescence, see arrow). The intrinsic fluorescence (green) of NBDHEX was localized in the cell cytoplasm (see arrow). (b) Analysis of the amount of TRAF2–GSTP1-1 complex and (c) of phospho-JNK. NBDHEX induces a rapid dissociation of the TRAF2–GSTP1-1 complex (a and b) paralleled by the phosphorylation of JNK (a and c). (d) NBDHEX causes a sustained cell cycle arrest in the G2/M phase followed by apoptosis. (e) U-2OS cells were treated with NBDHEX (5 μM), the autophagy inhibitor CQ (2.5–10.0 μM) or NBDHEX-CQ combinations. Cell survival was assessed after 48 h by the SRB assay; the effect of NBDHEX on cell survival was not modified by simultaneous treatment with autophagy-inhibiting, non-cytotoxic doses of CQ. (f) Cell lysates from U-2OS cultures, untreated or treated with CQ (2.5–40.0 μM dose range) for 24 h, were subjected to immunoblot analysis with an anti-LC3 antibody recognizing both the cytosolic LC3-I and the autophagosome-associated LC3-II. β-actin was used to ensure equal loading and transfer of samples
Regarding the cellular outcomes following NBDHEX treatment, cytofluorimetric analysis revealed a cell cycle arrest in the G2/M phase at 24 h, followed by apoptosis at 48 h (Figure 5d). Our group had previously demonstrated NBDHEX's ability to induce cell death by apoptosis in U-2OS cells as well as in other tumor cell types.16, 18, 25, 29 Therefore, to investigate whether NBDHEX could also trigger autophagic cell death, we performed cell survival studies on U-2OS-cell cultures treated with NBDHEX in the presence of the autophagy inhibitor chloroquine diphosphate (CQ). As illustrated in Figure 5e, the effect of NBDHEX on cell survival was not modified by the simultaneous treatment with autophagy-inhibiting, non-cytotoxic concentrations of CQ. Of note, CQ's ability to inhibit autophagy, in the concentration range used in cell survival studies, was verified by assessing the levels of the autophagosome-associated LC3-II protein in lysates from CQ-treated cells (Figure 5f), as previously described.30 Indeed, CQ is a late-phase autophagy inhibitor that blocks the autophagic flux, thus causing the accumulation of autophagosomes and the LC3-II protein.31 In conclusion, based on the results of cell survival studies, autophagic cell death does not seem to contribute to the cytotoxic effect of NBDHEX.
Discussion
Evidence of the interaction between GSTP1-1 and TRAF2 had been originally obtained in human HEK293 and HeLa cells.17 We have recently demonstrated by western blot that the GST inhibitor NBDHEX is able to dissociate the TRAF2–GSTP1-1 complex in U-2OS cells, leading to a rapid and sustained activation of MKK4 and the downstream kinases JNK and p38.18 Although p38 causes an increase in p21 levels and a concomitant cell cycle arrest, JNK is crucial for the induction of apoptosis.18 However, up to now, a biochemical proof of the direct interaction between TRAF2 and GSTP1-1 has been lacking.
In the present study, we expressed and purified both GSTP1-1 and the TRAF domain of TRAF2 (region 310–501) and provided evidence that this truncated TRAF2 retains the structural features expected for the complete and folded protein. Indeed, the values estimated on the basis of the experimental CD spectrum are very close to those available from the X-ray crystallographic data of TRAF2 (pdb file 1ca4).4 The fluorescence measurements also suggest that the protein is in a folded state, as (i) the spectrum is blue-shifted, indicating that the two tryptophan residues present in each TRAF2 subunit are located in a compact, hydrophobic core; (ii) the anisotropy is much larger than the value expected for fully exposed and freely rotating tryptophilic residues (≈0.05) in fully unfolded polypeptidic chains.21
Next, we used ELISA and fluorometric assay procedures to investigate the binding of GSTP1-1 to TRAF2. Both the experimental approaches demonstrated the formation of a complex characterized by an apparent dissociation constant of 0.3 μM. The evidence for an interaction between GSTP1-1 and our truncated form of TRAF2 (residues 310–501) narrows down to the binding region of TRAF2 suggested by Wu et al. (aa 271–501).17
We also analyzed the effects of different redox conditions on the interactions involving GSTP1-1. The mechanism traditionally reported for the dissociation of the JNK–GSTP1-1 complex is based on the ROS-mediated oxidation of GSTP1-1.23 We here report that the oxidative inactivation of GSTP1-1 reduces the enzyme's affinity for TRAF2 as well. Interestingly, GSH has a similar effect on in vitro assays, inducing a concentration-dependent decrease of the complex involving the two proteins. In particular, the addition of 1 mM GSH to either ELISA or the fluorometric assay leads to a Kd value at least nine times higher as compared with the Kd value obtained in the absence of GSH. From a mechanistic point of view, these results may be explained by taking the following into account: Wu et al. reported that the protein region spanning residues 38–41 of GSTP1-1 contains a TRAF2-binding motif (TWQE) and is involved in the interaction with TRAF2.17 Of note, this region is also part of the α-helix 2 (residues 35–46), which forms the external wall of the GSH-binding site (G-site). We previously analyzed the flexibility of α-helix 2 in the apo-structure of GSTP1-1 and in the binary complex with GSH.32 In the apo-form, the irregular α-helix 2 displayed high flexibility and fluctuated rapidly between two families of conformations. However, under saturating GSH concentrations the equilibrium was completely (83%) shifted toward a highly rigid GST–GSH complex. The same protein region undergoes structural changes during GSTP1-1's oxidative inactivation, in which Cys-47 (located on α-helix 2) and Cys-101 (located on α-helix 4) approach each other to form a disulfide bond.24 Overall, these data indicate that this enzyme's binding ability depends on the conformation and flexibility of a protein region involved in the GSH binding process. We also show that addition of the GST inhibitor NBDHEX to a mixture of GSTP1-1, TRAF2 and GSH further decreases the affinity between the two proteins compared with when GSH is the only ligand, the effect being more evident at high levels of GSH. Crystallographic data of GSTP1-1 in a complex with NBDHEX and GSH show a number of hydrophobic interactions involving NBDHEX and protein residues shaping the hydrophobic portion of the GSTP1-1 active site (H-site).33 Thus, the TRAF2-binding region involves the enzyme active site at both the G- and H-sites; as a consequence the interaction with TRAF2 likely inactivates GSTP1-1. On the other hand, the analysis of fluorometric data using a model describing an equilibrium between a monoligated GSTP1-1 dimer and a GSTP1-1 with both binding sites occupied by TRAF2 (Figures 2b–d), leads us to conclude that the monoligated GSTP1-1 is the predominant species at high-GSTP1-1 concentrations, as in the case of tumor cells.34 Therefore, we might expect that GSTP1-1 engages only one subunit in the complex, while the other subunit may maintain the catalytic activity or interact with other proteins such as JNK. Indeed, we have previously shown that the binding of GSTP1-1 to JNK1 also involves the GSTP1-1 active site.15 Intriguingly, by binding and bringing into proximity different members of the same signaling pathway such as TRAF2 and JNK, GSTP1-1 may act as a scaffold protein, thus contributing to the coordinated regulation of their activity.
Wu et al.17 reported that in HeLa cells the complex between GSTP1-1 and TRAF2 was not affected by the addition of either the GSH precursor n-acetyl-cysteine or BSO, the specific inhibitor of GSH synthesis. Therefore, they hypothesized that the intracellular GSH levels, and more generally the redox state of the cell, may not be critical for this interaction. These findings only apparently contradict our own evidence, which shows that GSH affects the enzyme's affinity for TRAF2. Indeed, the lack of complex sensitivity to the intracellular redox state, reported by Wu et al.,17 can be explained by our proof that opposite redox conditions (i.e. the oxidative inactivation of GSTP1-1 and high concentrations of GSH), have converging effects on the TRAF2–GSTP1-1 interaction; both conditions cause a similar increase in the complex's Kd value.
To demonstrate our findings' physiological relevance, we complemented our analysis with cell-based experiments. The TRAF2–GSTP1-1 complex was constitutively present in U-2OS cells. However, its amount did not appear to be modulated by the intracellular GSH content, as under non-stressed conditions the intracellular concentration of GSH was so high that it always saturated GSTP1-1.22 Interestingly, the association between TRAF2 and GSTP1-1 was regulated in a cell cycle-dependent manner. The amount of the complex was high in the G0/G1 phase, whereas it strongly decreased in cells synchronized in the S, G2 and M phases. The individual proteins moved in opposite directions during the progression from the G0/G1 to S–G2–M phases; GSTP1-1 increased, whereas TRAF2 showed a descending profile. The reduction of TRAF2 levels may contribute to cause the observed cell cycle-dependent decrease of the TRAF2–GSTP1-1 complex. This is also supported by the low intracellular concentration of TRAF2, which we found to be at least five times lower than that of GSTP1-1 (data not shown), indicating that TRAF2 is the limiting factor in the TRAF2–GSTP1-1 interaction. On the other hand, variations in the levels of the individual proteins seem too limited to explain the drastic decline of the complex observed in cells progressing from the G0/G1 to S–G2–M phases. These findings suggest that additional factors are involved in the fine-tuning of the TRAF2–GSTP1-1 interaction during cell proliferation. It is worth noting that the lowest amount of the TRAF2–GSTP1-1 complex was found in the G2/M phase and that NBDHEX treatment, which hampers the TRAF2–GSTP1-1 interaction, leads to a prolonged cell cycle arrest in the G2/M phase as well. Although this issue will require further investigation, it is tempting to speculate that the reassociation of GSTP1-1 with TRAF2 may be required for the cell to exit from the G2/M phase and re-enter the G0/G1 phase.
Indeed, the findings described in this paper may lay the basis for new future studies aimed at optimizing the antitumor efficacy of therapies directed toward members of the TNF receptor superfamily through the combination with molecules targeting GSTP1-1.
Materials and Methods
Chemicals
GSH, DNSC, CDNB, PI, CQ, SRB, thymidine, deoxycytidine and nocodazole were from Sigma-Aldrich (St Louis, MO, USA); PLA was purchased from Olink Bioscience (Uppsala, Sweden); NBDHEX was synthesized as reported by Ricci et al.26
Construction of the TRAF2 expression plasmid
The sequence coding for the C-terminal domain of human TRAF2 (residues 310–501)4 was obtained through gene synthesis from Geneart (Regensburg, Germany) and cloned into a pET28a(+) (Novagen, Darmstadt, Germany) expression vector with NdeI and BamHI restriction sites at the 5′ and 3′ ends, respectively. The resulting plasmid was used to transform E. coli BL21 (DE3) cells to express the protein with a thrombin cleavable N-terminal 6-histidine tag.
Expression and purification of TRAF2 and GSTP1-1
E. coli BL21 (DE3) cells, transformed with the His-tagged TRAF2 C-terminal domain construct, were grown in Luria broth medium containing 30 μg/ml kanamicin sulfate. The expression of TRAF2 was induced by the addition of 1 mM isopropyl-1-thio-β galactopyranoside when the absorbance at 600 nm was 0.5. Cells were grown for 18 h at 25 °C, thereafter harvested by centrifugation and resuspended in lysis buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 20 mM imidazole, 10% glycerol, 1 mM DTT and an EDTA-free inhibitor of protease). The cell suspension was sonicated and clarified by centrifugation. The cellular extract was loaded on a 10-ml Ni-NTA column pre-equilibrated with lysis buffer, then the column was washed with 200 ml of lysis buffer and the protein was eluted using a linear gradient consisting of 50 ml of lysis buffer and 50 ml of the same buffer containing 500 mM imidazole. Imidazole was then removed from the TRAF2 sample by filtration through a Sephadex G-25 column (GE Healthcare Life Science, Chalfont St. Giles, UK) pre-equilibrated with 20 mM Tris-HCl pH 7.6 containing 150 mM NaCl and 10% glycerol. The TRAF2 content and its purity were analyzed in the eluted fractions by SDS-PAGE. GSTP1-1 was expressed and purified as previously described.26 The protein concentration was determined by measuring the absorbance at 280 nm and using an extinction coefficient of 17 780 and 25 460 M−1 cm−1 for TRAF2 and GSTP1-1 monomers, respectively. Proteins were stored at −80 °C.
Spectroscopic characterization of TRAF2
The purified protein was diluted at 2 μM in 20 mM Tris-HCl buffer, pH 7.6, containing 150 mM NaCl and 10% glycerol. CD spectra were recorded on a JASCO J-710 spectropolarimeter (Easton, MD, USA) at 20 °C, using a 0.1-cm quartz cuvette. The average values were calculated after accumulating at least six consecutive runs, to get a good signal-to-noise ratio. The TRAF2 α-helix and beta structure contents were estimated fitting the measured spectrum by a linear combination of reference spectra obtained from a set of proteins with known secondary and tertiary structures.20, 35 The steady-state fluorescence spectrum of the TRAF2 C-terminal domain was recorded at 20 °C on a PC1-ISS photon counting fluorometer (ISS, Champain, IL, USA) equipped with Glan Thompson polarizers. Excitation was set at 292 nm.
GSTP1-1 oxidative inactivation
Purified GSTP1-1 was extensively incubated at room temperature in 0.1 M K-phosphate buffer, pH 8.0. At different times, aliquots of the protein solution were assayed for GST activity and cysteine reactivity, as previously described.24 In brief, GST activity was assayed spectrophotometrically at 340 nm by measuring the rate of CDNB conjugation with GSH as a function of time, according to the method by Habig et al.36 The assay mixture contained 1 mM GSH and 1 mM CDNB in 0.1 M K-phosphate buffer, pH 6.5. The SA of the enzyme was expressed as μmol/min/mg protein. The reactivity of Cys-47 and Cys-101 toward 2,2′-dithiobis(2-nitrobenzoic acid) (DTNB) was assayed at 25 °C by following the increase of absorbance at 412 nm, where the thionitrobenzoate ion absorbs (ε412 nm=13.6 mM−1 cm−1). A typical incubation mixture contained 4 μM GSTP1-1 (the concentration refers to the monomeric protein) and 0.1 mM DTNB in 0.1 M K-phosphate buffer, pH 8.0. The reduction of the fully oxidized GSTP1-1 was performed at 37 °C in K-phosphate buffer pH 8.0 containing 100 mM DTT and 1 mM GSH. To remove DTT and GSH, the mixture was subjected to partition chromatography on a column of Sephadex G-25 (GE Healthcare Life Science), then GST activity and cysteine reactivity were evaluated as described above.
ELISA for protein–protein interaction analysis
Two hundred microliters of His-tagged TRAF2 C-terminal domain (0.005 μM in 20 mM Tris-HCl, pH 7.6 containing 150 mM NaCl and 10% glycerol) were added to each well of a 96-well His-Sorb plate (Qiagen, Hilden, Germany) and incubated overnight at 4 °C on a rocking platform. Afterward, wells were washed three times with PBS and incubated for 30 min with GSTP1-1 (concentration range of the monomeric protein from 0.1 to 2 μM) in 10 mM K-phosphate buffer, pH 7.0 containing 0.1 mM EDTA. Incubation with GSTP1-1 was also performed in the presence of different GSH concentrations (ranging from 1/3 to 10 times the Km of GSTP1-1 for GSH)22 both in the absence and presence of NBDHEX, at a concentration (8 μM) that is 10 times its IC50 value for GSTP1-1.26 Alternatively, TRAF2 was incubated with increasing amounts of oxidized GSTP1-1. At the end of incubation, the wells were washed with PBS and then filled with 200 μl of a mouse anti-GSTP1-1 antibody (Cell Signaling, Beverly, MA, USA; 1 : 1000 in TBS containing 0.1% Tween and 5% nonfat dry milk) for 2 h at room temperature. Subsequently, wells were washed with PBS and incubated with an anti-mouse IgG antibody (1 : 1000 in TBS containing 0.1% Tween and 5% nonfat dry milk) (Cell Signaling) for 45 min at room temperature. The immunocomplexes were detected by the addiction of 200 μl per well of the 1-Step-Turbo TMB substrate solution (Pierce, Rockford, IL, USA). The reaction was stopped after 45 min by the addition of 50 μl of 2 M H2SO4, and the absorbance was measured at 450 nm. The equilibrium dissociation constants (Kd) of the complexes were calculated by assuming a 1 : 1 interaction between one TRAF2 monomer and one GSTP1-1 subunit, the binding sites being equal and independent. The most generally valid equation to analyze our data, on formation of a 1 : 1 complex, is given by equation (1),37 where ν is the percentage of the saturated binding sites; [P]t and [L]t are the total concentrations of monomeric GSTP1-1 and TRAF2, respectively.
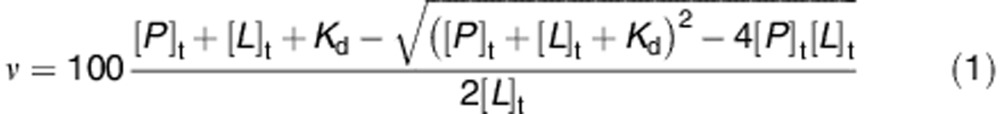 |
TRAF2 dansylation
TRAF2 (40 μM in 0.1 M K-phosphate buffer, pH 8.4) was incubated with 200 μM Dansyl Chloride (DNSC, Sigma-Aldrich) for 3 h at 4 °C in the dark with continuous stirring. After labeling, the sample was passed through gel filtration chromatography (Sephadex G-25, GE Healthcare Life Sciences) to remove the unreacted probe and secondary products. The stoichiometry of the dansylation was determined by measuring the protein and probe concentrations in labeled samples. The dye concentration was calculated using ɛ=5700 M−1 cm−1 at 340 nm. The mole ratio of dansyl/TRAF2 was determined as 2 : 1.
Fluorometric analysis
The interaction of dansyl/TRAF2 with GSTP1-1 was monitored by steady-state fluorescence. In particular, an average emission wavelength (actually the spectral center of mass, λ) of dansylated TRAF2 (2.5 μM) was obtained at increasing GSTP1-1 concentrations (from 0.2 to 9 μM, the concentration refers to the dimeric protein). The interaction of dansyl/TRAF2 (5.9 μM) with GSTP1-1 was also analyzed in the presence of saturating GSH (1 mM).22 The spectral shift recorded was fitted to both the general equations (1) and (2):
where λo and λ∞ are the average fluorescence wavelengths of dansylated TRAF2 in the absence or presence of saturating concentrations of GSTP1-1 and [L]t is the total TRAF2 concentration.
This second model analyzes the case where one TRAF2 (P1) or two TRAF2 monomers (P2) are bound to the dimeric GSTP1-1, as described by the following equilibria:
 |
The dependence of the [P]1 and [P]2 species on the total concentration of dimeric GSTP1-1 ([P]t) is expressed by the equations (3) and (4), obtained from the equilibria reported above.
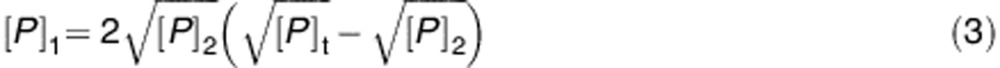 |
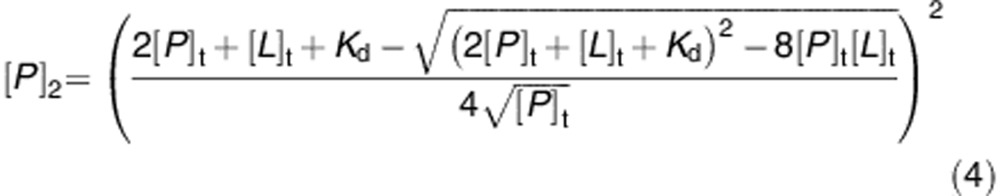 |
We assumed, also in this case, that the binding sites of GSTP1-1 are equal and independent.
Cell culture conditions
U-2OS human osteosarcoma cells were purchased from the American Type Culture Collection (ATCC) and grown in Iscove's modified Dulbecco's medium (IMDM, EuroClone, Milan, Italy) supplemented with 10% fetal bovine serum (Lonza, Basel, Switzerland), 2 mM L-glutamine and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin).
Cell synchronization
U-2OS cells were seeded at 50–60% of confluence and incubated in a medium containing 2 mM thymidine for 16 h, to obtain cells arrested in the G0/G1 phase. The cells were then extensively washed with PBS and incubated for 8 h in fresh medium containing 8 μM deoxycytidine to promote entry into the S phase. Moreover, to induce G2 and mitotic arrest, after deoxycytidine treatment, cells were incubated with fresh medium plus 50 ng/ml nocodazole. Mitotic cells were harvested by shaking off cells, whereas the G2 cells were obtained by harvesting the remaining adherent cells. Floating mitotic cells and adherent G2-arrested cells were separately collected.38, 39, 40
PLA and confocal laser scanning microscopy analysis of the TRAF2–GSTP1-1 complex, GSTP1-1, TRAF2 and P-JNK in U-2OS cells
U-2OS cells were seeded at 15 000 cells/cm2 on 12 mm poly-L-lysine-coated glass coverslips (Beckton & Dickinson, Franklin Lakes, NJ, USA) and cultured as reported above. Cells were stained for the TRAF2–GSTP1-1 complex, GSTP1-1 or TRAF2 24 h after plating and in each phase of the cell cycle. Alternatively, U-2OS cells were treated with 5 μM NBDHEX 48 h after plating and stained for the TRAF2–GSTP1-1 complex and P-JNK after 1, 3 or 6 h of treatment. The TRAF2–GSTP1-1 complex was visualized by PLA as follows: U-2OS cells were fixed in 4% formaldehyde, rinsed three times in PBS and permeabilized with ice-cold 100% methanol for 10 min, at −20 °C. After washing with PBS the slides were incubated for 1 h at room temperature with blocking buffer (5% normal goat serum, 0.3% Triton X-100 in PBS) and then overnight at 4 °C with the combination of a mouse anti-GSTP1-1 (1 : 100) and a rabbit anti-TRAF2 (1 : 25) antibodies (Cell Signaling) diluted in antibody dilution buffer. After washing, samples were incubated with Duolink PLA Rabbit MINUS and PLA Mouse PLUS proximity probes and proximity ligation was performed using the Duolink Far-Red detection reagent kit (Olink Bioscences), according to the manufacturer's protocol. Finally, the slides were extensively washed and mounted on a coverslip with the DAPI Mounting Medium (OLink Bioscences) to stain the nuclei. At the end of this procedure, each TRAF2–GSTP1-1 complex generated a fluorescent red spot. In parallel, U-2OS cells grown, fixed and permeabilized as reported above, were incubated for 1 h with blocking buffer and successively overnight with the mouse anti-GSTP1-1, the rabbit anti-TRAF2 or the rabbit anti-P-JNK, followed by an anti-mouse or anti-rabbit Alexa 647-conjugated secondary antibody (Cell Signaling, 1 : 1000). Fluorescence was detected using a Fluoview 1000 Olympus (Opera Zerbo, Milan, Italy) system equipped with an Olympus IX-81 inverted microscope. The acquisitions were performed with a × 60 magnification oil immersion objective (NA 1.42, WD 0.15 mm). Duolink Far-Red detection reagent and Alexa Fluor 647-conjugated antibodies were excited using the 635 nm diodo laser, DAPI was excited using the 405 nm diodo laser and NBDHEX, when present, was excited with the 488 nm argon ion laser. The emission apertures for fluorescence detection were as follows: 430–460 nm for DAPI, 655–685 nm for Alexa Fluor 647 and Duolink Far-Red detection reagent and 507–537 nm for NBDHEX. The Z-optical section series, obtained beginning from the nuclear apex and progressing down in 0.48 μM (at least 12 planes), were converted to maximum projection images to avoid subjectivity in the choice of the plane to be analyzed. The distributions of Far-Red Alexa Fluor 647-conjugated antibodies fluorescence (GSTP1-1 and TRAF2) and blue DAPI fluorescence (nuclei) were analyzed by area as follows: perimeters were drawn around the nucleus (according to the area marked with DAPI) and around the entire cell excluding the nucleus area, after subtraction of the background, using the ImageJ software (US National Institutes of Health, Bethesda, MD, USA). The BlobFinder software (Olink Bioscience) was used for PLA image analysis. Fluorescent signals from rolling circle amplification products were defined and counted per cell.
Assessment of GSH intracellular levels
Lysates from ∼1 × 106 unsynchronized U-2OS cells were subjected to HPLC determination of GSH, as reported previously.41 Briefly, 100 μl of 12% sulfosalicylic acid was added to 50 μl of cell lysate, and the GSH content was determined in the acid-soluble fraction. The molar concentrations of GSH was obtained using the U-2OS-cell volume of 4000 μm3.42
Western blot analysis
For the assessment of GSTP1-1 and TRAF2 intracellular levels, a fixed number of synchronized U-2OS cells (∼7.5 × 104 cells) were lysed as previously reported18 and loaded on a 12% SDS-polyacrylamide gel. Proteins were then transferred to PVDF membranes (Millipore, Billerica, MA, USA). For the assessment of LC3-II levels, cell lysates from U-2OS cultures, untreated or treated with CQ (2.5–40.0 μM dose range) for 24 h, were subjected to immunoblot analysis, as previously described.30 A monoclonal anti-GSTP1-1, a polyclonal anti-TRAF2 (Cell Signaling), a polyclonal anti-LC3 (Novus Biologicals, Littleton, CO, USA) and a monoclonal anti-β-actin (Sigma-Aldrich) were used as primary antibodies. Anti-rabbit or anti-mouse secondary antibodies (Cell Signaling) were revealed with the ECL LiteAblot Extend (EuroClone). ImageJ software was used to analyze the band intensities.
Flow cytometric analysis
The percentage of cells in every phase of the cell cycle was determined to verify cell synchronization and the effect of NBDHEX treatment. Cells were fixed with 70% ethanol overnight, stained with the PI staining buffer (50 μg/ml PI, 10 μg/ml RNAsi and 1% Triton X-100) and analyzed by a FACSCalibur instrument (BD Bioscence, San Jose, CA, USA) acquiring for 30 000 individual cells. Flow cytometric data were statistically analyzed by FlowJo 8.8.6 software (Tree Stare, Inc, Ashland, OR, USA).
Cell viability studies
U-2OS (2 × 104 cells per well) were seeded in 96-well plates and, after 24 h at 37 °C, were exposed to NBDHEX (5 μM), CQ (2.5–10.0 μM) or NBDHEX-CQ combinations and allowed to incubate for 48 h. After incubation, cell survival was evaluated by the SRB assay, as previously described.43
Statistical analysis
All the experiments were repeated at least three times; results are presented as means±S.E.M. Statistical evaluation was done using the Student's t-test. The criterion for statistical significance was P<0.05.
Acknowledgments
We thank the Centre for Advanced Microscopy ‘Patrizia Albertano' and Dr. Elena Romano of University of Rome ‘Tor Vergata' for imaging acquisition. This study was supported by a grant from Associazione Italiana per la Ricerca sul Cancro (AIRC, project number 10598).
Glossary
- CDNB
1-chloro-2,4-dinitrobenzene
- CQ
chloroquine diphosphate
- DTNB
2,2′-dithiobis(2-nitrobenzoic acid)
- GSTP1-1
glutathione transferase P1-1
- NBDHEX
6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol
- GSH
reduced glutathione
- PI
propidium iodide
- SRB
sulforhodamine B
- PLA
proximity ligation assay
- TRAF2
TNF receptor-associated factor 2
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Bradley JR, Pober JS. Tumor necrosis factor receptor-associated factors (TRAFs) Oncogene. 2001;20:6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- Chung JY, Park YC, Ye H, Wu H. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J Cell Sci. 2002;115 (Pt 4:679–688. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- Xie P. TRAF molecules in cell signaling and in human diseases. J Mol Signal. 2013;8:7. doi: 10.1186/1750-2187-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YC, Burkitt V, Villa AR, Tong L, Wu H. Structural basis for self-association and receptor recognition of human TRAF2. Nature. 1999;398:533–538. doi: 10.1038/19110. [DOI] [PubMed] [Google Scholar]
- Kyriakis JM, Banerjee P, Nikolakaki E, Dai T, Rubie EA, Ahmad MF, et al. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- Zhang L, Blackwell K, Altaeva A, Shi Z, Habelhah H. TRAF2 phosphorylation promotes NF-kappaB-dependent gene expression and inhibits oxidative stress-induced cell death. Mol Biol Cell. 2011;22:128–140. doi: 10.1091/mbc.E10-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HY, Régnier CH, Kirschning CJ, Goeddel DV, Rothe M. Tumor necrosis factor (TNF)-mediated kinase cascades: bifurcation of nuclear factor-kappaB and c-jun N-terminal kinase (JNK/SAPK) pathways at TNF receptor-associated factor 2. Proc Natl Acad Sci USA. 1997;94:9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich KP, Yeh WC, Yao Z, Mak TW, Woodgett JR. Mediation of TNF receptor-associated factor effector functions by apoptosis signal-regulating kinase-1 (ASK1) Oncogene. 1999;18:5814–5820. doi: 10.1038/sj.onc.1202975. [DOI] [PubMed] [Google Scholar]
- Nishitoh H, Saitoh M, Mochida Y, Takeda K, Nakano H, Rothe M, et al. ASK1 is essential for JNK/SAPK activation by TRAF2. Mol Cell. 1998;2:389–395. doi: 10.1016/s1097-2765(00)80283-x. [DOI] [PubMed] [Google Scholar]
- Liu H, Nishitoh H, Ichijo H, Kyriakis JM. Activation of apoptosis signal-regulating kinase 1 (ASK1) by tumor necrosis factor receptor-associated factor 2 requires prior dissociation of the ASK1 inhibitor thioredoxin. Mol Cell Biol. 2000;20:2198–2208. doi: 10.1128/mcb.20.6.2198-2208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS, Schumacker PT, Arch RH. Reactive oxygen species are downstream products of TRAF-mediated signal transduction. J Biol Chem. 2001;276:42728–42736. doi: 10.1074/jbc.M103074200. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Takeda K, Matsuzawa A, Saegusa K, Nakano H, Gohda J, et al. Recruitment of tumor necrosis factor receptor-associated factor family proteins to apoptosis signal-regulating kinase 1 signalosome is essential for oxidative stress-induced cell death. J Biol Chem. 2005;280:37033–37040. doi: 10.1074/jbc.M506771200. [DOI] [PubMed] [Google Scholar]
- Tobiume K, Saitoh M, Ichijo H. Activation of apoptosis signal-regulating kinase 1 by the stress-induced activating phosphorylation of pre-formed oligomer. J Cell Physiol. 2002;191:95–104. doi: 10.1002/jcp.10080. [DOI] [PubMed] [Google Scholar]
- De Luca A, Federici L, De Canio M, Stella L, Caccuri AM. New insights into the mechanism of JNK1 inhibition by glutathione transferase P1-1. Biochemistry. 2012;51:7304–7312. doi: 10.1021/bi300559m. [DOI] [PubMed] [Google Scholar]
- Sau A, Pellizzari Tregno F, Valentino F, Federici G, Caccuri AM. Glutathione transferases and development of new principles to overcome drug resistance. Arch Biochem Biophys. 2010;500:116–122. doi: 10.1016/j.abb.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Wu Y, Fan Y, Xue B, Luo L, Shen J, Zhang S, et al. Human glutathione S-transferase P1-1 interacts with TRAF2 and regulates TRAF2-ASK1 signals. Oncogene. 2006;25:5787–5800. doi: 10.1038/sj.onc.1209576. [DOI] [PubMed] [Google Scholar]
- Sau A, Filomeni G, Pezzola S, Serra M, D'Aguanno S, Tregno FP, et al. Targeting GSTP1-1 induces JNK activation and leads to apoptosis in cisplatin-sensitive and -resistant human osteosarcoma cell lines. Mol Biosyst. 2012;8:994. doi: 10.1039/c1mb05295k. [DOI] [PubMed] [Google Scholar]
- Xia Y, Makris C, Su B, Li E, Yang J, Nemerow GR, et al. MEK kinase 1 is critically required for c-Jun N-terminal kinase activation by proinflammatory stimuli and growth factor-induced cell migration. Proc Natl Acad Sci USA. 2000;97:5243–5248. doi: 10.1073/pnas.97.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venyaminov SYu, Baikalov IA, Shen ZM, Wu CS, Yang JT. Circular dichroic analysis of denatured proteins: inclusion of denatured proteins in the reference set. Anal Biochem. 1993;214:17–24. doi: 10.1006/abio.1993.1450. [DOI] [PubMed] [Google Scholar]
- Masters BR, Lakowicz JR. Principles of fluorescence spectroscopy, third edition. J Biomed Opt. 2008;13:029901. [Google Scholar]
- Ivanetich KM, Goold RD. A rapid equilibrium random sequential bi-bi mechanism for human placental glutathione S-transferase. Biochim Biophys Acta. 1989;998:7–13. doi: 10.1016/0167-4838(89)90111-8. [DOI] [PubMed] [Google Scholar]
- Adler V, Yin Z, Fuchs SY, Benezra M, Rosario L, Tew KD, et al. Regulation of JNK signaling by GSTp. EMBO J. 1999;18:1321–1334. doi: 10.1093/emboj/18.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci G, Del Boccio G, Pennelli A, Lo Bello M, Petruzzelli R, Caccuri AM, et al. Redox forms of human placenta glutathione transferase. J Biol Chem. 1991;266:21409–21415. [PubMed] [Google Scholar]
- Turella P, Cerella C, Filomeni G, Bullo A, De Maria F, Ghibelli L, et al. Proapoptotic activity of new glutathione S-transferase inhibitors. Cancer Res. 2005;65:3751–3761. doi: 10.1158/0008-5472.CAN-04-3903. [DOI] [PubMed] [Google Scholar]
- Ricci G, De Maria F, Antonini G, Turella P, Bullo A, Stella L, et al. 7-Nitro-2,1,3-benzoxadiazole derivatives, a new class of suicide inhibitors for glutathione S-transferases. Mechanism of action of potential anticancer drugs. J Biol Chem. 2005;280:26397–26405. doi: 10.1074/jbc.M503295200. [DOI] [PubMed] [Google Scholar]
- Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius K-J, Jarvius J, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Sundqvist A, Zieba A, Vasilaki E, Herrera Hidalgo C, Soderberg O, Koinuma D, et al. Specific interactions between Smad proteins and AP-1 components determine TGFβ-induced breast cancer cell invasion. Oncogene. 2012;32:3606–3615. doi: 10.1038/onc.2012.370. [DOI] [PubMed] [Google Scholar]
- Pellizzari Tregno F, Sau A, Pezzola S, Geroni C, Lapenta C, Spada M, et al. In vitro and in vivo efficacy of 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) on human melanoma. Eur J Cancer. 2009;45:2606–2617. doi: 10.1016/j.ejca.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Battisti S, Valente D, Albonici L, Bei R, Modesti A, Palumbo C. Nutritional stress and arginine auxotrophy confer high sensitivity to chloroquine toxicity in mesothelioma cells. Am J Respir Cell Mol Biol. 2012;46:498–506. doi: 10.1165/rcmb.2011-0195OC. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella L, Caccuri AM, Rosato N, Nicotra M, Lo Bello M, De Matteis F, et al. Flexibility of helix 2 in the human glutathione transferase P1-1. time-resolved fluorescence spectroscopy. J Biol Chem. 1998;273:23267–23273. doi: 10.1074/jbc.273.36.23267. [DOI] [PubMed] [Google Scholar]
- Federici L, Lo Sterzo C, Pezzola S, Di Matteo A, Scaloni F, Federici G, et al. Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases. Cancer Res. 2009;69:8025–8034. doi: 10.1158/0008-5472.CAN-09-1314. [DOI] [PubMed] [Google Scholar]
- Tew KD, Monks A, Barone L, Rosser D, Akerman G, Montali JA, et al. Glutathione-associated enzymes in the human cell lines of the National Cancer Institute Drug Screening Program. Mol Pharmacol. 1996;50:149–159. [PubMed] [Google Scholar]
- Yang JT, Wu CS, Martinez HM. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- Habig WH, Pabst MJ, Jakoby WB. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- Stella L, van de Weert M. Fluorescence quenching and ligand binding: a critical discussion of a popular methodology. J Mol Struct. 2011;998:144–150. [Google Scholar]
- Sánchez del Pino M, Farràs R, Ferrero M, Valero L, Orlando L, Font de Mora J, et al. Phosphorylation of AIB1 at mitosis is regulated by CDK1/CYCLIN B. PLoS One. 2011;6:e28602. doi: 10.1371/journal.pone.0028602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-S, Park Y-Y, Park S-Y, Cho H, Kang D, Cho H. The auto-ubiquitylation of E3 ubiquitin-protein ligase Chfr at G2 phase is required for accumulation of polo-like kinase 1 and mitotic entry in mammalian cells. J Biol Chem. 2011;286:30615–30623. doi: 10.1074/jbc.M111.231803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini D, Colaluca IN, Vodermaier HC, Biamonti G, Giacca M, Falaschi A, et al. Early mitotic degradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression. EMBO J. 2003;22:3715–3724. doi: 10.1093/emboj/cdg340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore A, Tozzi G, Gaeta LM, Bertini E, Serafini V, Di Cesare S, et al. Actin glutathionylation increases in fibroblasts of patients with Friedreich's ataxia: a potential role in the pathogenesis of the disease. J Biol Chem. 2003;278:42588–42595. doi: 10.1074/jbc.M301872200. [DOI] [PubMed] [Google Scholar]
- Aebersold R, Claassen M, Ellenberg J, Rinner O, Schmidt A, Ori A, et al. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]