Abstract
Telomere shortening can lead to chromosome instability, replicative senescence, and apoptosis in both somatic and male germ cells. To study roles for mammalian telomeres in homologous pairing and recombination, we characterized effects of telomere shortening on spermatogenesis and oogenesis in late-generation telomerase-deficient mice. We show that shortened telomeres of late-generation telomerase-deficient mice impair meiotic synapsis and decrease recombination, in particular, in females. In response to telomere shortening, male germ cells mostly undergo apoptosis, whereas female germ cells preferentially arrest in early meiosis, suggesting sexually dimorphic surveillance mechanisms for telomere dysfunction during meiosis in mice. Further, meiocytes of late-generation telomerase-deficient females with shortened telomeres, bred with early-generation males harboring relatively long telomeres, exhibit severely impaired chromosome pairing and synapsis and reduced meiotic recombination. These findings imply that functional telomeres are important in mammalian meiotic synapsis and recombination.
Keywords: meiosis, mouse, oocytes
Telomeres are repetitive DNA sequences composed of (TTAGGG)n repeats that cap chromosome ends and maintain chromosomal stability in somatic cells (1, 2). Comparatively, much less is known regarding telomere function in mammalian germ cells and meiosis, although telomere shortening and dysfunction result in sterility in mice (3). In the fission yeast and Caenorhabditis elegans, loss of telomere function decreases meiotic recombination and increases chromosome missegregation and loss (4–6). Telomere-mediated chromosome movement and/or telomere clustering in lower eukaryotes and plants promote homologue pairing (7–11). Telomeres have been postulated to function similarly in mammalian meiosis (12).
Telomere localization during mammalian gametogenesis recently has been reported, but only during spermatogenesis (for review, see ref. 13). The function of telomeres in mammalian meiotic pairing and recombination remains unclear, because male germ cells with telomere dysfunction from late-generation telomerase-deficient (TR-/-) mice undergo apoptosis (3, 14), with striking depletion of spermatogenic and meiotic cells in the seminiferous tubules (15), which may mask the role of telomeres in meiotic pairing and recombination in these cells. Moreover, meiosis differs between the two sexes in mammals (16). Female meiosis begins during fetal development, then arrests at prophase I stage for weeks or years, whereas spermatogenesis starts at 2–3 weeks in male mice and continues throughout the life of males. In addition, female meiosis lacks an effective checkpoint control at the metaphase/anaphase transition, in contrast to male meiosis (17, 18).
In meiosis, homologous chromosomes pair and recombine during prophase I, and synaptonemal complexes (SC) form along paired meiotic chromosomes. The synaptonemal complex protein 3 (SCP3), a structural component in the axial/lateral element of the SC, colocalizes with the sister chromatids of each homologue and connects along their entire length by forming a central element at pachytene stage of meiotic prophase I (19–22). Thus, the structure and number of SCP3 elements provide a key marker for evaluating synapsis between homologues. We analyzed effects of telomere shortening in late-generation TR-/- female mice on meiotic chromosome synapsis and recombination and compared them with males. Because very late-generation (G6 or G7) TR-/- male mice exhibit extensive apoptosis in germ cells and sterility, we focused on intermediate generation (e.g., G4) TR-/- mice, because these mice show telomere shortening, but less apoptosis and decreased fertility (3, 14).
Materials and Methods
Mice and Gonads. Two- to four-month-old WT and telomerase deficient (TR-/-) mice (23) were used in the experiments. For analysis of female meiosis, fetal ovaries were collected at day 17.5 of pregnancy (the day with mating plug was designated as day 0.5). For analysis of male meiosis, testes were collected from adult males at 20 days after birth or 2–4 months of age.
Immunofluorescence Microscopy of Cryosections. Fetal ovaries and adult testis were freshly collected and snap-frozen in OCT compound, cut in 6-μm sections, fixed in cold acetone for 2 min, and stained for SCP3, as described (15), except that rabbit anti hamster COR1/SCP3 (24) was used in this study. Images were captured with a Zeiss Axioplan 2 imaging microscope, equipped with epifluorescence, axiocam and axiovision 3.0 software. For some experiments, immunostaining of SCP3 was followed by fluorescence in situ hybridization of telomere probes, as described below.
Immunostaining and Fluorescence Microscopy of Meiocyte Spreads. Surface spreading of meiocytes was prepared by a drying-down technique and stained for synaptonemal complexes, according to the procedure described (19, 25, 26). In brief, dissected ovaries or testes were placed in hypoextraction buffer, fixed in 1% paraformaldehyde, washed in PhotoFlo, air-dried, and stored at -20°C. Slides with meiocytes were washed and blocked in PBS containing goat serum, BSA, and Triton-X, incubated with anti-COR1/SCP3 (1:1,000) for 1 h at 37°C, washed, then incubated with FITC or Texas red-conjugated goat anti-rabbit IgG (1:100), washed, and mounted in Vectashield mounting medium (Vector Laboratories) added with 0.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). For detection of MLH1 foci, mouse anti-human MLH1 antibody (BD PharMingen) at 1:50 dilution and Texas red-conjugated goat anti-mouse IgG (Jackson Immunoresearch) as secondary antibody were used. MLH1 foci were counted as described (27, 28).
Quantitative Fluorescence in Situ Hybridization (FISH). Telomere FISH was performed on cryosections of testis and fetal ovaries or on meiocyte spreads, as described (29). Telomeres were denatured at 80°C for 3 min and hybridized with FITC-labeled (CCCTAA)3 peptide nucleic acid probe (Applied Biosystems), washed, and mounted in Vectashield mounting medium added with 0.5 μg/ml DAPI. For quantitative measurement of telomere length in the pachytene meiotic cells, telomere fluorescence intensity was integrated by using the tfl-telo program (30), kindly provided by P. Lansdorp (Terry Fox Laboratory, British Columbia Cancer Agency, Vancouver, Canada), and calibrated with fluorescence beads. Telomere length (kb) was estimated based on the relative fluorescence intensity by using standard cell lines with known telomere length (31).
Terminal Deoxynucleotidyltransferase dUTP Nick-End Labeling (TUNEL) Assay. Testis and fetal ovaries were fixed in Bouins, paraffin-embedded, and sectioned. Sections were deparaffinized and pretreated for TUNEL assay, as described (14). Apoptosis was detected with the TUNEL assay by using the In Situ Cell Death Detection Kit (Roche Diagnostics), according to manufacturer's instructions, and nuclei counterstained with 50 μg/ml propidium iodide (Molecular Probes). Slides were mounted in Vectashield mounting medium. Images were captured with the Zeiss fluorescence microscopy.
Statistical Analysis. We analyzed data by one-way ANOVA with statview software (SAS Institute, Cary, NC). Individual statistical differences were determined by Fisher's probable least-significant difference comparison test.
Results
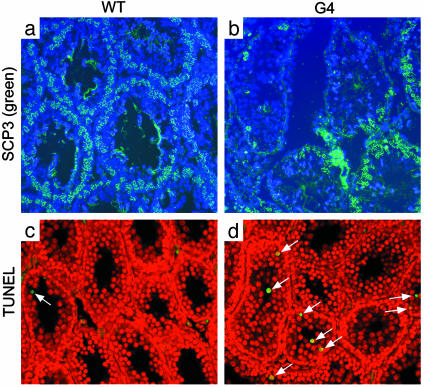
We observed that the number of spermatocytes with COR1/SCP3 lateral elements was markedly decreased in G4 TR-/- compared with WT controls, consistent with a recent report (15). Nearly 100% of seminiferous tubules (n = 400) of WT mice show spermatocytes with SCP3 elements, whereas 44% of 318 seminiferous tubules of G4 TR-/- mice exhibited decreased number of spermatocytes with SCP3 elements (Fig. 1 a and b). We also found that apoptosis, as demonstrated by TUNEL-positive nuclei, was increased 5-fold in seminiferous tubules from G4 TR-/- mice compared with WT mice (0.86 vs. 0.18 nuclei per tubule, respectively; Fig. 1 c and d). Furthermore, apoptosis was found in the outer layers of spermatocytes of G4 TR-/- mice (Fig. 1d, arrows), whereas the inner layer of spermatogonia mostly remained intact. The basic hematoxylin/eosin histological staining is provided in Fig. 7a, which is published as supporting information on the PNAS web site.
Fig. 1.
Axial/lateral elements of synaptonemal complexes and apoptosis of seminiferous tubules. (a) WT tubules show continuous peripheral layer of meiocytes with SCP3 staining. (b) Nearly 50% of G4 TR-/- tubules exhibit complete absence of or very few meiocytes with SCP3 staining. Green, SCP3 elements; blue, nuclear chromosomes stained with DAPI. (c) Rare apoptosis of WT seminiferous tubules. (d) Extensive apoptosis, indicated by green or yellow, in G4 TR-/- seminiferous tubules. Red, nuclear staining with propidium iodide.
To investigate effects of telomere shortening on female meiosis, we collected fetal ovaries at day 17.5 of gestation and examined SCs in both ovarian sections and meiocyte spreads. Late-generation TR-/- pregnant females had fewer fetuses as compared with the WT (9.3 WT and 8 G2 TR-/- fetuses from WT and G1 TR-/- pregnant females, respectively, vs. 5.8 G4 TR-/- fetuses from G3 TR-/- females). Furthermore, all WT fetuses were normal and alive, but only an average of 4.3 G4 TR-/- fetuses (74%) were alive and 41% of these showed reduced size (Table 2, which is published as supporting information on the PNAS web site). This finding is consistent with a previous finding that shortened telomeres impair embryonic viability and fetal development (32).
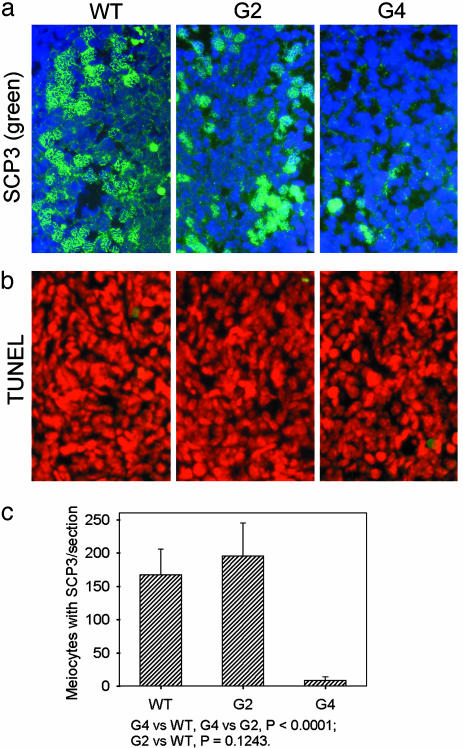
The number of primary oocytes stained with SCP3 elements was severely decreased in G4 TR-/-, compared with WT and G2 TR-/- fetal ovaries (Fig. 2a; hematoxylin/eosin histological staining in Fig. 7b). To quantify the degree to which oogenesis was disrupted in TR-/- female fetus, we scored the number of oocytes that had formed SCs per ovarian section. More than 150 meiocytes with SCP3 elements per section were found in WT and G2 TR-/- fetal ovaries, compared with <10 meiocytes with SCP3 elements per section in G4 TR-/- fetal ovaries (Fig. 2c), suggesting increased apoptosis or meiotic arrest with telomere shortening. Nonetheless, apoptosis appeared at low frequency in fetal ovaries at this stage (Fig. 2b) and did not differ among WT, G2, and G4 TR-/- ovaries (in the range of four to eight apoptotic nuclei per section). It has been shown that, during normal oogenesis in the mouse, germ-cell apoptosis does not become evident during fetal development, until 1–2 days after birth, when germ-cell numbers decline (33). Thus, it is likely that meiotic arrest or defective germ-cell differentiation before zygotene/pachytene stage underlies meiotic defects in late-generation TR-/- female mice.
Fig. 2.
Axial/lateral elements of synaptonemal complexes and apoptosis in WT, G2, and G4 TR-/- fetal ovaries. (a) Meiocytes with SCP3 elements (green) and nuclear chromosomes (blue). (b) Very few apoptotic cells (green) in WT, G2, and G4 TR-/- fetal ovaries. Red, nuclear staining with propidium iodide. (c) Number of meiocytes with SCP3 elements per section of fetal ovaries, showing significantly reduced SCP3 meiocytes in G4 TR-/- fetal ovaries, compared with WT and G2 TR-/- fetal ovaries.
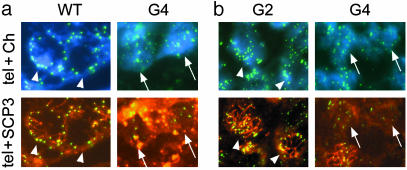
We further observed perinuclear distribution of telomeres at the ends of SCP3 elements (15, 34), from WT germ cells and meiocytes, but rarely from G4 TR-/- germ cells. Shortened telomeres and lack of perinuclear distribution of telomeres from G4 TR-/- mice, may contribute to impaired assembly of the synaptonemal complex (Fig. 3, arrows). These results imply that in mice, as in yeast, functional telomeres are involved in chromosome pairing and synapsis.
Fig. 3.
Immunostaining of SCP3 and FISH with telomere probe of cryosections. (a) WT meiocytes (arrowheads) show perinuclear distribution of telomeres (green), which are associated with both ends of SCP3 (red), whereas G4 TR-/- meiocytes exhibit disrupted perinuclear distribution of telomeres, coincident with lack of SCP3 elements (arrows). Blue, nuclear chromosomes stained with DAPI. (b)G2 TR-/- fetal ovaries show meiocytes with perinuclear distribution of telomeres (green; arrowheads), colocalized with both ends of SCP3 (red), but lack of perinuclear distribution of telomeres is seen in G4 TR-/- fetal ovaries (arrows). tel, telomere; Ch, chromosome.
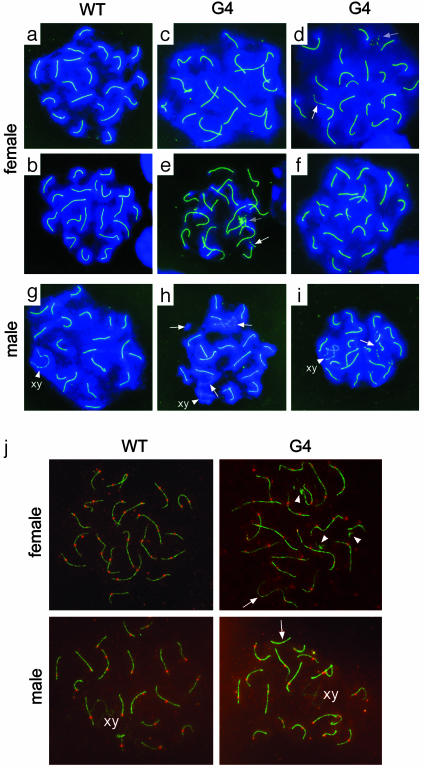
To examine whether homologous synapsis is disrupted in the few meiocytes that do reach pachytene stage in G4 TR-/- mice, we analyzed in detail the structure and number of SCs by immunostaining with anti-COR1/SCP3 and compared them with WT pachytene meiocytes. SCP3 elements were altered in G4 TR-/- female meiotic cells, which showed an increase (19%) in abnormal numbers of SCP3 elements (19, 21, or 22 vs. the normal 20 SCP3 elements) (Fig. 4 a–i and Table 1). Fragmented SCP3 also was increased (37%) in G4 TR-/- pachytema compared with WT (4%). These defects can arise from a delay in synapsis of homologues or from disrupted pairing. Fragmented elements also may indicate chromosome fragmentation and damage. Comparatively, the number and structure of SCP3 elements were less affected in males than in females. This difference occurs perhaps because male germ cells use apoptosis to deplete cells with shortened telomeres (14). By contrast, female meiosis lacks an effective mechanism to eliminate defective germ cells (e.g., those with telomere dysfunction). At least in fetal ovary, germ cells with short telomeres continue to develop, despite being incapable of pairing or synapsis, however, at the cost of possibly developing disrupted recombination and thus increased risk of aneuploidy.
Fig. 4.
Immunostaining of SCP3 elements and MLH1 foci at pachytene stage from female fetal ovary (day 17.5 of gestation) and male testis (day 20 after birth). (a–i) SCP3 elements and structure. WT pachytene oocytes show 20 SCP3 elements (a and b), whereas G4 TR-/- pachytene oocytes show 19 (c), 21 (d), 22 (e), and 21 (f) SCP3 elements. WT pachytene spermatocytes show 20 SCP3 elements (g), but G4 T-/- pachytene spermatocytes show altered number of SCP3 elements (21) (h and i). Green, SCP3; blue, chromosomes counterstained with DAPI; white arrows, unpaired lateral SCP3; gray arrows, SCP3 fragments. (j) MLH1 foci and SCP3 elements. Only those MLH1 dots (red) unambiguously colocated with SCs (green) are counted as MLH1 foci crossover sites (27, 28). Some MLH1 foci are present on the diffuse chromatin surrounding SCs and are considered as MLH1 background staining (27). Arrows, lack of MLH1 foci in the SCP3 elements; arrowheads, SCP3 fragments.
Table 1. Comparison of the number of SCP3 elements and MLH1 foci of pachytene meiocytes.
| SCP3 elements, n
|
MLH1 foci, n
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Abnormal
|
|||||||||
| Sex | Animals | Cells analyzed, n | Normal, 20 | 19 | 21 | 22 | Total abnormal, n (%) | SCP3 fragments, n (%) | Mean ± SEM (cells analyzed, n) |
| Female | G4 TR-/- | 52 | 42 | 5 | 3 | 2 | 10 (19.2)a | 19 (36.5)a | 23.2 ± 1.8c (21) |
| WT | 73 | 72 | 1 | 1 (1.4)b | 3 (4.1)b | 28.1 ± 0.6d (44) | |||
| Male | G4 TR-/- | 60 | 55 | 5 | 5 (8.3) | 0 | 19.8 ± 0.6e (37) | ||
| (XY) | WT | 70 | 70 | 0 | 0 | 21.5 ± 0.3f (36) | |||
Superscript letters: a vs. b, c vs. d, e vs. f; P < 0.01.
One proposed function for SCs is in the control of meiotic recombination or crossover along chromosomes (35). Defective pairing and/or synapsis may therefore compromise meiotic recombination of homologues. MLH1, a mismatch repair protein necessary for meiotic recombination, also has been used to detect crossover sites and to estimate chiasmata frequency (21, 27, 36). MLH1 foci were consistently found on SCs during pachytene, a stage of maximum homologous chromosome pairing (27, 28). MLH1 foci were significantly reduced in G4 TR-/- pachytene oocytes, compared with WT oocytes (Fig. 4j and Table 1). MLH1 foci also were decreased in pachytene spermatocytes of G4 TR-/- adult mice, but to a lesser extent than in oocytes. G4 TR-/- pachynema showed absence of crossover in some synapsis (Fig. 4j, arrows). In general, WT female meiocytes have one or more crossovers per homologous synapsis, whereas most male SCs have only one crossover, typically near the distal end of SCs. Oocytes have more crossovers than spermatocytes, demonstrating a difference in recombination frequency and variation in the distribution of crossovers between male and female meiosis, as has been shown in humans (28). Fewer crossovers in G4 TR-/- pachynema are consistent with the altered number of SCP3 elements reported above. Considering that absence of SCP3 has only subtle effects on recombination (21), we suggest that functional telomeres themselves are involved in the meiotic recombination processes.
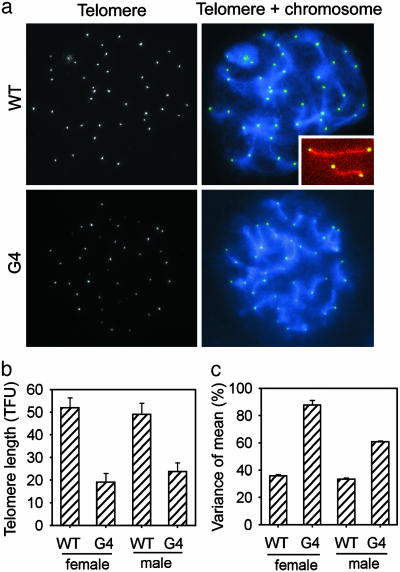
To correlate defects of pairing, synapsis, and crossovers with telomere shortening, we used both immunocytochemistry and quantitative FISH with a telomere probe (Fig. 5a). Telomeres of homologues paired at the end of SCP3 and were visually indistinguishable from each other (Figs. 5a Inset and 3). Telomeres were significantly shorter, on average, and their variance significantly larger, indicative of telomere dysfunction in G4 TR-/- pachynema of both males and females, compared with WT pachynema (Fig. 5 b and c). Moreover, some chromosomes had severely shortened telomeres (Fig. 5a). Pachytene meiocytes with altered SCP3 elements also exhibited telomere shortening at the ends of SCP3 elements. These findings associate telomere dysfunction with defective synapsis and recombination at the pachytene stage. Thus, telomere loss and dysfunction disrupt the onset of meiosis, chromosome pairing or synapsis, and recombination of homologues.
Fig. 5.
Telomere quantitative FISH in pachytene meiocytes. (a) Representative images of WT and G4 TR-/- male meiocytes (n > 20), showing telomeres (white/green dots) and chromosomes (blue). (Inset) Magnification (×2) of SCs, shown by SCP3 (red), flanked by telomeres (green/yellow) at both ends of homologues. Each telomere dot represents two homologous telomeres, which are too close to be visually distinguishable. (b) Quantitative telomere length (mean ± SEM) of homologous chromosomes. Telomere fluorescence unit (TFU) corresponds to 1 kb of TTAGGG repeats. Telomere length is significantly reduced (P < 0.0001) in both G4 TR-/- male and female meiocytes, compared with WT meiocytes. (c) The variance of the telomere length of G4 TR-/- meiocytes is significantly higher (P < 0.0001) than WT meiocytes of both males and females, indicating heterogeneous shortening of telomeres preferentially at some ends.
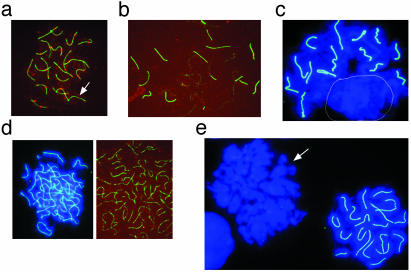
It is expected that unequal telomere lengths in homologues may further compromise meiotic pairing and/or synapsis and recombination. To test this hypothesis, we examined SCP3 elements and MLH1 distribution of both ovarian sections and meiocytes derived from day 17.5 G4/G2 TR-/- hetero-fetuses derived from G3 TR-/- adult females bred with G1 TR-/- adult males. These parents exhibit a telomere length difference of ≈10 kb. An average of 57 ± 7 (n = 3) meiocytes with SCP3 elements per section was observed in the G4/G2 TR-/- hetero-fetal ovaries. This number is significantly less than that of WT and G2 TR-/- fetal ovaries, but a clear increase compared with G4 TR-/- fetal ovaries (see Fig. 2c). In 45 analyzed meiocyte spreads of G4/G2 TR-/- hetero-fetal ovaries, 34 meiocytes showed staining of SCP3, but 14 (41%) of them exhibited an altered number of SCP3 elements (Fig. 6 a–d). Six meiocytes had SCP3 elements in the number of 14, 18, 19, and 21. MLH1 foci (19.6 ± 8.9, n = 26) of pachytene oocytes were reduced, compared with G4 TR-/- and WT oocytes (compare with Table 1). Eight (23.5%) meiocytes showed nearly 40 SCP3 elements, indicating absence of homologous pairing or synapsis. Moreover, these meiocytes lacked MLH1 foci on SCP3 elements (Fig. 6d), which is consistent with abortive chromosome pairing. Other meiocytes were deficient in SCP3 formation (Fig. 6e) or had a few discrete staining of SCP3 spots. Clearly, chromosome pairing, synapsis, and recombination were severely impaired in meiocytes with irregular telomeres. These results further support the notion that appropriate telomere length is critical for homologous pairing and recombination.
Fig. 6.
SCP3 elements and MLH1 foci of meiocytes of G4/G2 TR-/- heterofetal ovaries (day 17.5) derived from G3 TR-/- adult females bred with 1 TR-/- males. (a) A pachytene meiocyte having 20 SCP3 elements with one lack of MLH1 foci (white arrow). (b) Partially synapsed SCP3 elements and lack of MLH1 foci. (c) Fourteen synapsed SCP3 elements and the absence of synapsis in other chromosomes (circled). (d) Unsynapsed, nearly 40 SCP3 elements. (e) Absence of SCP3 staining in one meiocyte (arrow). Green, SCP3 synapsis; blue, chromosomes counterstained with DAPI; red, MLH1 staining and foci (crossover) on the SCP3 elements.
Discussion
We have shown that, in addition to compromised fetal development, telomere shortening and dysfunction result in fetuses with defects in chromosome synapsis, accompanied by increased apoptosis in male meiotic cells and early meiotic differentiation arrest in females. For cells capable of reaching pachytene, telomere shortening compromises chromosome synapsis and recombination. These multiple checkpoints might be set to ensure that only germ cells with minimum telomere length survive meiosis.
We noticed that SCs could form in some cells, even when telomeres had shortened to a critical length. However, we also observed shortening only of telomeres at one end of chromosomes, in particular, the p-arm, with no loss of telomeres at both ends of the synapsis of homologous chromosomes. Moreover, telomere shortening was found in both meiocytes with normal synapsis (numbered 20) and with abnormal synapsis (numbered 19, 21, and 22), suggesting that absolute length of telomeres may not be as important as concordance in telomere length to facilitate homologous pairing. This finding is supported by significantly increased impaired synapsis and recombination of germ cells derived from late-generation TR-/- females with shortened telomeres, which have been bred with early generation males that harbor relatively long telomeres. Mechanisms under-lying meiotic pairing and synapsis could be complex and involve multiple elements. It is also possible that telomeres themselves may be involved indirectly in homologous pairing. Barlow and Hulton (37) showed that synapsis started at the termini (most probably at the telomeres) of homologues. Genetic analysis of Saccharomyces cerevisiae strains has indicated that meiotic telomere clustering contributes to efficient homologue alignment and synaptic pairing telomeres. Telomere-associated meiotic protein (Ndj1) is required for tethering meiotic telomeres to the nuclear periphery (7, 38). Also, proteins Hop2 and Mnd1 work as a complex to promote meiotic chromosome pairing (39). Mammalian MutS homologue 5 and other proteins, such as FK506-binding protein (Fkbp6), also are required for chromosome pairing in meiosis (40, 41).
The dramatic reduction in crossovers of G4 TR-/- pachynema (with shortened telomeres) suggests that telomeres are involved in meiotic recombination. Telomeres have been implicated as promoters of meiotic recombination of mouse chromosome 8, where a recombination “hot spot” has been found (42). In this study, we directly associate telomere shortening with defective chromosome synapsis and recombination in the mouse.
We have shown that telomere dysfunction is associated with aberrant synapsis. By careful examination, Hemann et al. (14) found that apoptosis at the onset of meiotic prophase removes cells with dysfunctional telomeres from the germ-cell population in males. It is tempting to predict that apoptosis may have contributed to the decreased meiocytes with synapsis. Responses to telomere dysfunction appear to differ between male and female germ cells. Males can effectively trigger apoptosis and remove defective spermatocytes, whereas female germ cells undergo meiotic arrest, with loss of perinuclear distribution and disruption of synapsis formation. Indeed, deletion of p53, an important regulator for apoptosis, rescues spermatogenesis with shortened telomeres but only for a limited time. As telomere dysfunction worsens, germ cells undergo p53-independent apoptotic death associated with genetic catastrophe (43). Genetically induced SCP3 deficiency also leads to sexually dimorphic disruption of meiosis, a pattern similar to that observed with telomere dysfunction. SCP3 homozygous mutant males are sterile because of massive apoptotic cell death during meiotic prophase (20). SCP3-deficient male mice fail to form axial/lateral elements and SCs, and chromosomes in mutant spermatocytes do not synapse. By contrast, loss of SCP3 in female mice induces defective meiotic chromosome segregation and promotes aneuploidy in oocytes (21). Ineffective removal by apoptosis of oocytes with dysfunctional telomeres may contribute to increased rates of aneuploidy reported in female meiosis.
We observed few primary oocytes with SCs during the initiation of oogenesis in fetal ovaries of G4 TR-/- mice, coincident with markedly shortened telomeres in pachytene oocytes. Oocytes have been known to secret factors, such as GDF-9, which are crucial for stimulating growth and differentiation of surrounding granulosa cells and development of ovaries (44). Reduced numbers of oocytes could compromise development of the ovary. Indeed, G4 TR-/- adult females exhibit significant ovarian atrophy. Further, a limited number of germinal vesicle oocytes collected from these ovaries undergo aberrant meiotic division, specifically metaphase chromosome misalignment and/or spindle disruption (45), which could result, in part, from failed chromosome synapsis and reduced recombination arising early in prophase I of meiosis and lead to reduced fertility. Chromosome missegregation and nondisjunction increase with advancing age in women and are major causes of aneuploidy, pregnancy loss, and age-related infertility (16). Absent or reduced levels of recombination events, as observed in telomere-shortened mice, increase the likelihood of nondisjunction (16, 28). Trisomy 21 progeny could be derived from low levels of crossovers at telomeres and pericentromeric regions (46). Our findings of defective meiotic synapsis and recombination resulting from irregular telomeres may have implications in understanding nondisjunction, aneuploidy, and infertility in women.
Supplementary Material
Acknowledgments
We thank Lewis Kerr and Rudi Rottenfusser for help with cryosection and microscopic imaging; Peter Lansdorp for providing the tf l-telo software; Carla M. DiGirolamo for critical reading of the manuscript; and James Trimarchi for helpful discussion. This work was supported in part by Brown Faculty Funds (D.L.K.) and by the Ministry of Science and Technology, Spain, and the European Union (M.A.B).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TR-/-, telomerase-deficient; SC, synaptonemal complexes; SCP3, synaptonemal complex protein 3; DAPI, 4′,6-diamidino-2-phenylindole; FISH, fluorescence in situ hybridization; TUNEL, terminal deoxynucleotidyltransferase dUTP nick-end labeling.
References
- 1.Blackburn, E. H. (2000) Nature 408, 53-56. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn, E. H. (2001) Cell 106, 661-673. [DOI] [PubMed] [Google Scholar]
- 3.Lee, H. W., Blasco, M. A., Gottlieb, G. J., Horner, J. W., II, Greider, C. W. & DePinho, R. A. (1998) Nature 392, 569-574. [DOI] [PubMed] [Google Scholar]
- 4.Cooper, J. P., Watanabe, Y. & Nurse, P. (1998) Nature 392, 828-831. [DOI] [PubMed] [Google Scholar]
- 5.Nimmo, E. R., Pidoux, A. L., Perry, P. E. & Allshire, R. C. (1998) Nature 392, 825-828. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed, S. & Hodgkin, J. (2000) Nature 403, 159-164. [DOI] [PubMed] [Google Scholar]
- 7.Rockmill, B. & Roeder, G. S. (1998) Genes Dev. 12, 2574-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tease, C. & Fisher, G. (1998) Chromosome Res. 6, 269-276. [DOI] [PubMed] [Google Scholar]
- 9.Scherthan, H., Weich, S., Schwegler, H., Heyting, C., Harle, M. & Cremer, T. (1996) J. Cell Biol. 134, 1109-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scherthan, H., Jerratsch, M., Li, B., Smith, S., Hulten, M., Lock, T. & de Lange, T. (2000) Mol. Biol. Cell 11, 4189-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bass, H. W., Riera-Lizarazu, O., Ananiev, E. V., Bordoli, S. J., Rines, H. W., Phillips, R. L., Sedat, J. W., Agard, D. A. & Cande, W. Z. (2000) J. Cell Sci. 113, 1033-1042. [DOI] [PubMed] [Google Scholar]
- 12.de Lange, T. (1998) Nature 392, 753-754. [DOI] [PubMed] [Google Scholar]
- 13.Zalenskaya, I. A. & Zalensky, A. O. (2002) Int. Rev. Cytol. 218, 37-67. [DOI] [PubMed] [Google Scholar]
- 14.Hemann, M. T., Rudolph, K. L., Strong, M. A., DePinho, R. A., Chin, L. & Greider, C. W. (2001) Mol. Biol. Cell 12, 2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franco, S., Alsheimer, M., Herrera, E., Benavente, R. & Blasco, M. A. (2002) Eur. J. Cell Biol. 81, 335-340. [DOI] [PubMed] [Google Scholar]
- 16.Hassold, T. & Hunt, P. (2001) Nat. Rev. Genet. 2, 280-291. [DOI] [PubMed] [Google Scholar]
- 17.LeMaire-Adkins, R., Radke, K. & Hunt, P. A. (1997) J. Cell Biol. 139, 1611-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roeder, G. S. (1997) Genes Dev. 11, 2600-2621. [DOI] [PubMed] [Google Scholar]
- 19.Dobson, M. J., Pearlman, R. E., Karaiskakis, A., Spyropoulos, B. & Moens, P. B. (1994) J. Cell Sci. 107, 2749-2760. [DOI] [PubMed] [Google Scholar]
- 20.Yuan, L., Liu, J. G., Zhao, J., Brundell, E., Daneholt, B. & Hoog, C. (2000) Mol. Cell 5, 73-83. [DOI] [PubMed] [Google Scholar]
- 21.Yuan, L., Liu, J. G., Hoja, M. R., Wilbertz, J., Nordqvist, K. & Hoog, C. (2002) Science 296, 1115-1118. [DOI] [PubMed] [Google Scholar]
- 22.Pelttari, J., Hoja, M. R., Yuan, L., Liu, J. G., Brundell, E., Moens, P., Santucci-Darmanin, S., Jessberger, R., Barbero, J. L., Heyting, C. & Hoog, C. (2001) Mol. Cell. Biol. 21, 5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blasco, M. A., Lee, H. W., Hande, M. P., Samper, E., Lansdorp, P. M., DePinho, R. A. & Greider, C. W. (1997) Cell 91, 25-34. [DOI] [PubMed] [Google Scholar]
- 24.Tarsounas, M., Morita, T., Pearlman, R. E. & Moens, P. B. (1999) J. Cell Biol. 147, 207-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters, A. H., Plug, A. W., van Vugt, M. J. & de Boer, P. (1997) Chromosome Res. 5, 66-68. [DOI] [PubMed] [Google Scholar]
- 26.Spyropoulos, B. & Moens, P. B. (1994) Methods Mol. Biol. 33, 131-139. [DOI] [PubMed] [Google Scholar]
- 27.Anderson, L. K., Reeves, A., Webb, L. M. & Ashley, T. (1999) Genetics 151, 1569-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tease, C., Hartshorne, G. M. & Hulten, M. A. (2002) Am. J. Hum. Genet. 70, 1469-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hande, M. P., Samper, E., Lansdorp, P. & Blasco, M. A. (1999) J. Cell Biol. 144, 589-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poon, S. S., Martens, U. M., Ward, R. K. & Lansdorp, P. M. (1999) Cytometry 36, 267-278. [DOI] [PubMed] [Google Scholar]
- 31.McIlrath, J., Bouffler, S. D., Samper, E., Cuthbert, A., Wojcik, A., Szumiel, I., Bryant, P. E., Riches, A. C., Thompson, A., Blasco, M. A., et al. (2001) Cancer Res. 61, 912-915. [PubMed] [Google Scholar]
- 32.Herrera, E., Samper, E. & Blasco, M. A. (1999) EMBO J. 18, 1172-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pepling, M. E. & Spradling, A. C. (2001) Dev. Biol. 234, 339-351. [DOI] [PubMed] [Google Scholar]
- 34.Moens, P. B. & Pearlman, R. E. (1990) Chromosoma 100, 8-14. [DOI] [PubMed] [Google Scholar]
- 35.Zickler, D. & Kleckner, N. (1999) Annu. Rev. Genet. 33, 603-754. [DOI] [PubMed] [Google Scholar]
- 36.Moens, P. B., Kolas, N. K., Tarsounas, M., Marcon, E., Cohen, P. E. & Spyropoulos, B. (2002) J. Cell Sci. 115, 1611-1622. [DOI] [PubMed] [Google Scholar]
- 37.Barlow, A. L. & Hulten, M. A. (1996) Chromosome Res. 4, 562-573. [DOI] [PubMed] [Google Scholar]
- 38.Trelles-Sticken, E., Dresser, M. E. & Scherthan, H. (2000) J. Cell Biol. 151, 95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsubouchi, H. & Roeder, G. S. (2002) Mol. Cell. Biol. 22, 3078-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edelmann, W., Cohen, P. E., Kneitz, B., Winand, N., Lia, M., Heyer, J., Kolodner, R., Pollard, J. W. & Kucherlapati, R. (1999) Nat. Genet. 21, 123-127. [DOI] [PubMed] [Google Scholar]
- 41.Crackower, M. A., Kolas, N. K., Noguchi, J., Sarao, R., Kikuchi, K., Kaneko, H., Kobayashi, E., Kawai, Y., Kozieradzki, I., Landers, R., et al. (2003) Science 300, 1291-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashley, T., Cacheiro, N. L., Russell, L. B. & Ward, D. C. (1993) Chromosoma 102, 112-120. [DOI] [PubMed] [Google Scholar]
- 43.Chin, L., Artandi, S. E., Shen, Q., Tam, A., Lee, S. L., Gottlieb, G. J., Greider, C. W. & DePinho, R. A. (1999) Cell 97, 527-538. [DOI] [PubMed] [Google Scholar]
- 44.Elvin, J. A., Yan, C., Wang, P., Nishimori, K. & Matzuk, M. M. (1999) Mol. Endocrinol. 13, 1018-1034. [DOI] [PubMed] [Google Scholar]
- 45.Liu, L., Blasco, M. A. & Keefe, D. L. (2002) EMBO Rep. 3, 230-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanzi, R. E., Watkins, P. C., Stewart, G. D., Wexler, N. S., Gusella, J. F. & Haines, J. L. (1992) Am. J. Hum. Genet. 50, 551-558. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.