Abstract
Epithelial–mesenchymal transformation is a critical developmental process reiterated in multiple organs throughout embryogenesis. Formation of endocardial cushions, primordia of valves and septa, is a classic example of epithelial–mesenchymal transformation. Several gene mutations are known to affect cardiac valve formation. Sox9 is activated when endocardial endothelial cells undergo mesenchymal transformation and migrate into an extracellular matrix, called cardiac jelly, to form endocardial cushions. In Sox9-null mutants, endocardial cushions are markedly hypoplastic. In these mutants, Nfatc1 is ectopically expressed and no longer restricted to endothelial cells. Further, Sox9-deficient endocardial mesenchymal cells fail to express ErbB3, which is required for endocardial cushion cell differentiation and proliferation. Our results reveal a succession of molecular steps in the pathway of endocardial cushion development. We propose that loss of Sox9 inhibits epithelial–mesenchymal transformation after delamination and initial migration, but before definitive mesenchymal transformation.
Endocardial cushions are present within both the cardiac outflow tract and the atrioventricular (AV) canal. Some endocardial cushion cells in the distal part of the cardiac outflow tract originate from cardiac neural crest cells, whereas others originate from endothelial cells. All endocardial cushion cells in the AV canal are derived from endocardial endothelial cells that undergo epithelial–mesenchymal transformation (EMT). Both neural crest-derived and endothelial cell-derived mesenchymal cells proliferate and invade a thick layer of acellular extracellular matrix, known as the cardiac jelly, to form the endocardial cushions. During embryogenesis, Sox9, which encodes a high-mobility group transcription factor, is expressed in several embryonic tissues, including chondroprogenitors and differentiated chondrocytes, male gonads, endocardial cushions, notochord, and neural crest (1, 2). Heterozygous SOX9 mutations are the cause of the human disease campomelic dysplasia (3, 4), which is characterized by generalized hypoplasia of endochondral bones, XY sex reversal, and occasional anomalies of pancreas and/or kidney. In addition, ventricular septa defects and tetralogy of Fallot (a cardiac malformation involving outflow tract defects) have also been reported in patients with this disease (5). The perinatal lethality of Sox9 heterozygous mutant mice precludes the generation of Sox9 homozygous null mutants and, hence, studies of the functions of Sox9 during embryonic development by using conventional genetic methods (6, 7).
We have recently shown that the conditional inactivation of Sox9 in limbs before the appearance of chondrogenic mesenchymal condensations prevents the formation of these condensations without changing the extent of cell proliferation or cell death (8). Inactivation of Sox9 after the condensations are established largely blocks further progression of these cells through the normal pathway of chondrogenic differentiation. Furthermore, in the absence of Sox9, there is no expression of Sox5 and Sox6, whose proteins are also required for chondrogenic differentiation (9). Thus, although Sox9-null cells are specified in limb buds, these cells are unable to form chondrogenic mesenchymal condensations and to proceed into subsequent chondrocyte differentiation. Experiments by others showed that inactivation of the same conditional alleles of Sox9 in neural stem cells results in defects in specification of oligodendrocytes and astrocytes, suggesting that the switch from neurogenesis to gliogenesis fails to take place (10).
To examine whether Sox9 had additional roles during embryogenesis, we conditionally inactivated the Sox9 gene in male and female germ lines by using the Cre/loxP recombination system and generated embryos completely lacking Sox9. These Sox9-null mutant embryos provide the genetic basis to examine Sox9 functions during embryogenesis. Our results, which demonstrate that Sox9 has an essential role in EMT in the heart, establish a sequence of steps in the pathway of endocardial cushion formation.
Materials and Methods
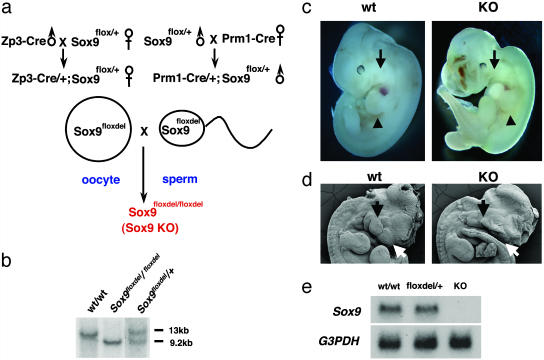
Generation of Sox9-Null Embryos and Sox9flox/flox;Cre Embryos. Introduction of loxP sites into the Sox9 gene and the generation of Sox9 floxed heterozygous and homozygous mice were described previously (8). In a first cross, Zona pellucida 3 (Zp3)-Cre or Protamine 1 (Prm1)-Cre transgenic mice (11, 12) were mated with mice heterozygous for the Sox9 floxed allele. The offspring inheriting Zp3-Cre or Prm1-Cre and a floxed allele were then mated with each other to obtain embryos in which two Sox9 floxed alleles were deleted. Routine mouse genotyping was performed by PCR as previously described (8). For generation of Sox9lacZ/floxdel embryos, Zp3-Cre male transgenic mice were mated with female mice heterozygous for the Sox9 floxed allele. The female offspring inheriting Zp3-Cre and a floxed allele were then mated with Sox9lacZ/+ chimera mice (6) to obtain embryos harboring the lacZ gene together with a Sox9 floxdel allele (Sox9lacZ/floxdel). To generate the Sox9 conditional null mice harboring the Wnt1-Cre transgene, Wnt1-Cre transgenic mice were mated with mice heterozygous for the Sox9 floxed allele. The offspring inheriting Wnt1-Cre and a floxed allele were then mated with Sox9 floxed heterozygous mice to obtain embryos harboring the Wnt1-Cre transgene together with two Sox9 floxed alleles.
Histological Analysis. 5-Bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) staining of embryos and scanning electron microscope analysis were performed as described previously (8). For the histological analysis, embryos were fixed with 4% paraformaldehyde and embedded in paraffin. Sections of 7 μm were stained with hematoxylin and Treosin (Statlab, Lewisville, TX), or alcian blue and nuclear fast red. Immunohistochemical staining of sections was performed by using peroxidase chromogens (Zymed)/TrueBlue substrate (Kirkegaard and Perry Laboratories, Gaithersburg, MD) with rabbit polyclonal anti-fibulin-2 antibody (1:150) (13, 14) and rabbit polyclonal anti-cleaved caspase-3 and caspase-9 (Cell Signaling Technology, Beverly, MA; 1:50 and 1:200, respectively). Immunofluorescent staining of sections was performed as previously described with monoclonal anti-Nfatc1 antibody 7A6 (1:50, Pharmingen) (15). Cell proliferation was evaluated by BrdUrd pulse-labeling. BrdUrd was injected i.p. into pregnant mice 4 h before killing and was detected by a Zymed BrdUrd staining kit following the manufacturer's protocol. Cell proliferation was also examined by using mouse monoclonal anti-phosphohistone H3 (Cell Signaling Technology; 1:1,000). Terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) analysis was performed on paraffin-embedded sections by using the ApopTag Plus peroxidase in situ apoptosis detection kit (Intergen, Purchase, NY) following the manufacturer's protocol. RNA in situ hybridization analysis was carried out as previously described (16). Pictures of hybridization signals were superimposed with blue fluorescence images of cell nuclei stained with Hoechst 33258 dye.
AV Canal Endocardial Cushion Explant Culture. The AV canal tissues from wild-type and mutant embryos at 9.5 days postcoitum (dpc) were placed on type I collagen gel and cultured for 48 h. Transformation of endothelium to mesenchyme and invasion into the collagen gel was scored as previously described (17).
Results
Sox9-Null Mutant Embryos Die During Midgestation with Heart Failure. Using the mating scheme shown in Fig. 1a, we produced animals that were heterozygous for Sox9flox carrying either the Zp3-Cre transgene, specifically expressed in oocytes, or the spermatid-specific Prm1-Cre transgene, in which one Sox9 allele was deleted (floxdel) in either oocytes or haploid spermatids (11, 12). These mice were then mated with each other to obtain embryos lacking both Sox9 alleles. Mice were genotyped by Southern hybridization (Fig. 1b). Sox9-null mutant embryos died between 11.5 and 12.0 dpc, presumably from congestive heart failure, as the major blood vessels were severely dilated. These embryos exhibited severe hypoplasia of the frontonasal mesenchyme, otic vesicles, and branchial arches (Fig. 1 c and d). The shape and size of trunk and limb buds were, however, normal. The absence of Sox9 RNA and protein in Northern hybridization (Fig. 1e) and immunohistochemistry (data not shown) of homozygous mutant embryos at 11.5 dpc indicated that the mutation was a null mutation.
Fig. 1.
Targeting strategy for Sox9-null embryos. (a) The mating scheme used to generate Sox9-null embryos. (b) Southern blot analysis of fetal genomic DNA. Genomic DNA isolated from the yolk sac was digested with BamHI and then hybridized with the 3′ probe. The wild-type and floxdel alleles were detected as 13-kb and 9.2-kb fragments, respectively. (c) The gross appearance of wild-type (wt) and Sox9-null (KO) embryos at 11.5 dpc. The arrows indicate the first branchial arch. The arrowheads indicate the forelimb buds. (d) Morphological analysis of craniofacial regions of wild-type and Sox9-null embryos at 11.5 dpc by using a scanning electron microscope. The white and black arrows indicate frontonasal mass and branchial arches, respectively. (e) Northern blot analysis of Sox9 mRNA expression in wild-type and Sox9-null embryos at 11.5 dpc.
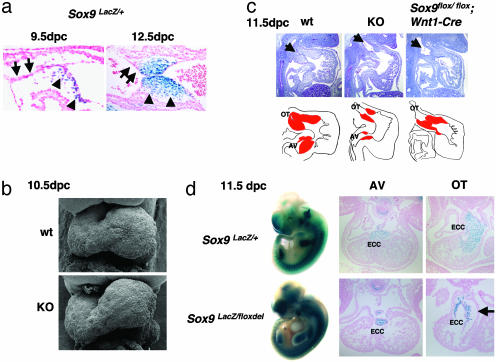
Endocardial Cushion Hypoplasia in Sox9-Null Mutant Embryos. In Sox9 heterozygous mutant embryos, in which a lacZ reporter gene was inserted in one Sox9 allele (6), all mesenchymal cells of the endocardial cushions expressed lacZ at 9.5 and 12.5 dpc. However, the single layer of endothelial cells lining the surface of the endocardial cushions is lacZ-negative (Fig. 2a). This suggests that the Sox9 gene is activated when the endothelial cells are transformed into mesenchymal cells after migration into the cardiac jelly. Scanning electron microscopy shows that the looping of the heart tubes in the mutants occurs normally (Fig. 2b). The aorticopulmonary spiral septum, which is derived from cardiac neural crest cells (18, 19), is also formed normally (data not shown), indicating that Sox9-null cardiac neural crest cells migrate to the heart. However, both proximal and distal portions of the endocardial cushions in the outflow tract as well as the endocardial cushions in the AV canal are severely underdeveloped in the mutant embryos (Fig. 2c). To examine the extent of endocardial cushion hypoplasia due to inactivation of Sox9 in cardiac neural crest cells, we generated embryos containing Sox9flox/flox and harboring the Wnt1-Cre transgene, in which Cre is expressed in cardiac neural crest cells (18). In these embryos, the abnormality of the endocardial cushions appears only in the distal part of the outflow tract. The proximal portion of the outflow tract and the AV cushions are normal (Fig. 2c and data not shown). Thus, Sox9 is required for the development of endocardial cushions derived from both neural crest cells and endothelial cells of the heart.
Fig. 2.
Hypoplastic endocardial cushions in Sox9-null embryos. (a) Sox9 expression in endocardial cushions of Sox9 heterozygous embryos at 9.5 and 12.5 dpc. The arrows and the arrowheads indicate endothelial cells and mesenchymal cells, respectively. (b) Morphological analysis of the heart of wild-type and Sox9-null embryos at 10.5 dpc by using scanning electron microscopy. (c) Hematoxylin and Treosin staining of Sox9-null embryos and the Sox9 conditional null mutant embryos at 11.5 dpc. The arrows indicate the outflow tract of the heart. In graphical drawings, red denotes endocardial cushions. Five independent experiments were performed and gave similar results. (d) Distribution of Sox9-null cells. Whole-mount X-Gal staining of Sox9 heterozygous (Sox9lacZ/+) and Sox9-null (Sox9lacZ/floxdel) embryos at 11.5 dpc, and transverse sections of Sox9 heterozygous (Sox9lacZ/+) and Sox9-null (Sox9lacZ/floxdel) embryos at 11.5 dpc stained with X-Gal. The arrow indicates the acellular cardiac jelly in Sox9-null embryos. ECC, endocardial cushion; OT, outflow tract. Three independent experiments were performed and gave similar results.
Sox9-Null Mutant Cells Are Specified in Endocardial Cushions. To follow the fate of Sox9 mutant cells, we also generated Sox9-null mutant embryos with a lacZ gene inserted in one Sox9-null allele (Sox9lacZ/floxdel), which enables the visualization of lacZ expression by X-Gal staining. In heterozygous embryos containing this allele, expression of lacZ reproduces expression of Sox9 (6). At 11.5 dpc, the overall pattern of lacZ expression in Sox9-null mutants was similar to that in Sox9 heterozygous mutant embryos except in the region of severe craniofacial anomalies (Fig. 2d). Transverse sections of Sox9lacZ/floxdel embryos at 11.5 dpc also confirm that the overall location of Sox9-null mutant cells is normal (Fig. 2d). However, consistent with the histological analysis of the embryos, the number of β-galactosidase-positive cells is markedly reduced in endocardial cushions in mutants (Fig. 2d).
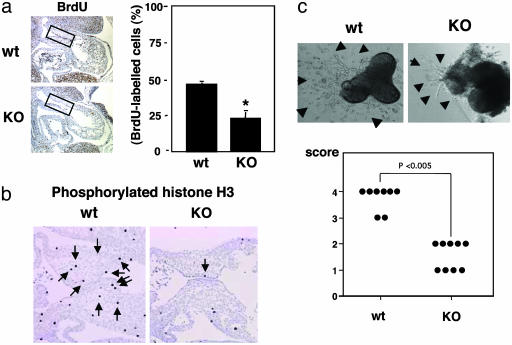
Reduced EMT and Decreased Cell Proliferation in Sox9-Null Mutant Endocardial Cells. The severe reduction in size of the endocardial cushions in the mutant embryos could have been due to a decrease in cell proliferation and/or an increase in apoptosis within the population of the endocardial cushion cells. To distinguish among these possibilities, we compared the levels of BrdUrd incorporation and the number of phosphorylated histone H3-positive cells, TUNEL-positive cells, and caspases-positive cells in wild-type and mutant endocardial cushions at 11.5 dpc. BrdUrd labeling and phosphorylated histone H3 are markers of the S phase and the mitotic phase, respectively. BrdUrd in vivo pulse labeling and immunohistochemistry of phosphorylated histone H3 revealed a marked reduction in cell proliferation of the endocardial cushions in mutant embryos compared with their wild-type siblings (Fig. 3 a and b). In contrast, no significant difference in the number of TUNEL-positive cells or in cleaved caspase-3 and caspase-9-positive cells between the wild-type and the mutant endocardial cushions was detected (data not shown). Histological analysis from X-Gal-stained sections was consistent with the notion that there was a severe reduction in the number of labeled cells in mutant embryos compared with wild-type within the cardiac jelly of the outflow tract (Fig. 2d). Indeed, in the mutants, an acellular space was seen adjacent to a thin layer of β-galactosidase-positive cells, in contrast to the abundance of cells in wild-type endocardial cushions (Fig. 2d). Thus, our results strongly suggest that Sox9 regulates proliferation of endocardial cushion cells. EMT and invasion of the cardiac jelly can be mimicked ex vivo by explanting endocardial cushion tissue, including endothelium and overlying myocardium, on collagen gels. AV canal explants from 9.5-dpc wild-type embryos exhibited numerous endothelial-like cells that first migrated over the surface of a collagen gel, underwent transformation, and invaded the gel matrix (Fig. 3c). In contrast, explants from Sox9-null embryos indicated that some cells were able to invade the collagen gel, but the number of transformed mesenchymal-like cells within the collagen gel was severely reduced. Although we did not directly prove that in these explants the cells migrating on and invading collagen gels were delaminating from endothelial cells, we hypothesize that Sox9 also controls differentiation of endocardial cushion cells during the critical process of EMT.
Fig. 3.
Cell proliferation and morphogenesis of AV canal endocardial cushion cells. (a) BrdUrd incorporation was reduced in endocardial cushion cells (boxed regions) in Sox9-null embryos. Statistical significance was assessed by one-way analysis of variance and unpaired Student's t test. Values shown are the mean + SD of four embryos. *, Statistically significant difference between wild-type and Sox9-null embryos at P < 0.001. (b) Immunohistochemistry of phosphorylated histone H3 revealed marked reduction of the number of phosphorylated histone H3-positive cells in endocardial cushion of Sox9-null embryos. These results were confirmed with two wild-type (wt) and two Sox9-null mutant (KO) embryos. (c) AV canal explant cultures showed markedly reduced transformation of endothelial cells into mesenchyme. The arrowheads indicate migrating endothelial cells and transformed mesenchymal cells. Eight wild-type and nine Sox9-null mutant explant cultures were examined. Statistical analysis used the Wilcoxon rank sum test (17).
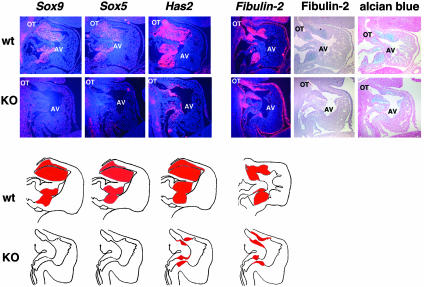
Ectopic Expression of Nfatc1 and Abolition of ErbB3 Expression in Sox9-Null Endocardial Mesenchymal Cells. To support this hypothesis, we examined the expression of a series of genes characteristic of endocardial cushion mesenchyme. In situ hybridization showed that expression of Sox5 and Sox6 was abolished in the mutant endocardial cushions (Fig. 4 and data not shown). Expression of Has2 and Fibulin-2, which are markers of migrating endocardial cushion mesenchymal cells, was reduced approximately in proportion to the reduction in cells present in the mutant endocardial cushions (Fig. 4). A similar reduction was seen in the accumulation of alcian blue-stainable proteoglycan and of fibulin-2 (Fig. 4). Fibulin-2 is normally up-regulated when endocardial endothelial cells undergo transformation into mesenchymal cells (20).
Fig. 4.
Histological analysis in endocardial cushions of Sox9-null embryos. Hearts of wild-type (wt) and Sox9-null (KO) embryos at 11.5 dpc were hybridized with probes for Sox9, Sox5, Has2, and Fibulin-2. In graphical drawings, red denotes the expression domains of these genes in endocardial cushions. The expression of Sox5 was abolished. Alcian blue staining and immunohistochemistry of fibulin-2 showed marked reduction of matrix accumulation in endocardial cushion of Sox9-null embryos. OT, outflow tract. Three independent experiments were performed and gave similar results.
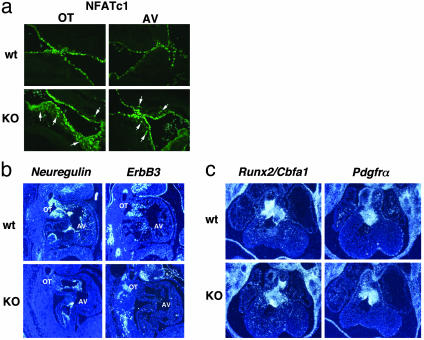
Nfatc1 is a transcription factor and is involved in the initial step of EMT (21, 22). In wild-type embryos at 11.5 dpc, the expression of Nfatc1 was detected in endocardial endothelial cells, not in mesenchymal cushion cells (Fig. 5a). Thus, Nfatc1 and Sox9 have reciprocal expression patterns in endocardial cushions because Sox9 is expressed in mesenchymal cells, not in endothelial cells. In contrast, in Sox9-null mutants, the expression of Nfatc1 was detectable in both endothelial cells and mesenchymal cells migrating into the cardiac jelly (Fig. 5a). Thus, the abnormal expression of Nfatc1 strongly suggests that Sox9-null mutant cells initiate EMT, but that their differentiation into mesenchymal cushion cells is markedly defective. ErbB3 is a member of the family of epidermal growth factor receptor tyrosine kinases and is activated by neuregulins, which are epidermal growth factor-like domain-containing glycoproteins (23). ErbB3 is expressed in endocardial cushion mesenchyme, whereas neuregulin is expressed by endocardium (24). ErbB3-null mutant embryos died around 13.5 dpc, exhibiting severe hypoplasia of endocardial cushions, leading to congestive heart failure (25). The similarities in expression patterns and phenotypes of null mutant embryos of both Sox9 and ErbB3 raise the possibility of their genetic interaction. In situ hybridization showed that expression of ErbB3 was abolished in the endocardial cushion mesenchyme of Sox9-null mutants at 11.5 dpc (Fig. 5b), and Neuregulin expression is down-regulated during EMT (Fig. 5b). Other markers of cushion mesenchyme were expressed normally, indicating that the defects in NFATc1 and ErbB3 expression were specific. Runx2/Cbfa1, which codes for a transcription factor required for osteoblast differentiation and bone formation (26, 27), is also expressed in endocardial cushions, although its function in these tissues is unknown (Fig. 5c). In Sox9-null mutants, Runx2/Cbfa1 expression is unchanged (Fig. 5c). Patch mutants, in which the platelet-derived growth factor receptor α subunit (PDGFRα) gene is deleted (28), develop cardiac outflow tract defects, and PDGFRα is expressed by endocardial cushion mesenchyme (29). However, expression of PDGFRα in Sox9-null mutants is similar to that in wild-type embryos (Fig. 5c).
Fig. 5.
Histological analysis in endocardial cushions of Sox9-null embryos. (a) Immunofluorescence of Nfatc1 in sections of wild-type (wt) and Sox9-null (KO) embryos at 11.5 dpc. The arrows indicate expression of Nfatc1 in mesenchymal cells of endocardial cushions in Sox9-null mutants. (b and c) Hearts of wild-type and Sox9-null embryos at 11.5 dpc were hybridized with probes for Neuregulin, ErbB3, Runx2/Cbfa1, and Pdgfrα. The expression of ErbB3 was abolished. OT, outflow tract. Three independent experiments were performed and gave similar results.
These results indicate that the consequences of Sox9 deficiency during endocardial cushion formation are due to a failure of endothelial cells to complete mesenchymal transformation. The endothelial marker, Nfatc1, is expressed in endothelial cells but fails to be appropriately down-regulated during the transformation process, and ErbB3 is not activated, leading to a deficiency of mesenchymal proliferation.
Discussion
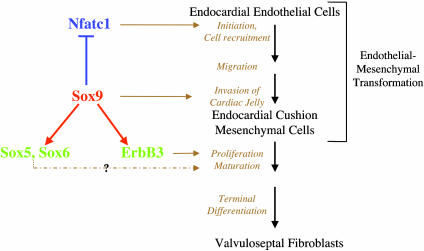
We have recently shown that the conditional inactivation of Sox9 in limbs before and after chondrogenic mesenchymal condensations reveals its essential roles in successive steps of chondrocyte differentiation (8). The results presented here indicate an essential role for Sox9 in a multistep pathway of endocardial cushion cell differentiation outlined in Fig. 6. Nfatc1, whose expression is restricted to endocardial endothelial cells, has a critical role in the initiation of EMT by these cells. Expression of Sox9, which is activated when these cells start to migrate into the cardiac jelly, is required for Nfatc1 down-regulation in endocardial mesenchymal cells after initiation of EMT. Indeed, in wild-type embryos, Nfatc1 expression is abolished once the cushion cells start migrating into the jelly. The finding that Nfatc1 continues to be expressed in Sox9 mutant mesenchymal cells indicates that EMT is disrupted subsequent to delamination and initial migration. We speculate that the complete inhibition of Nfatc1 expression once cushion cells enter the cardiac jelly is a critical step in the differentiation of these cells. Sox9 is also required for expression of ErbB3, which itself is needed for proliferation and differentiation of endocardial cushion mesenchymal cells. ErbB3 can be activated both by heregulin and by hyaluronan (30). The marked reduction in cushion cell proliferation in Sox9 mutants is in agreement with the abolition of ErbB3 expression in this tissue. As in the chondrogenic differentiation pathway, Sox9 is needed for expression of Sox5 and Sox6 in endocardial cushion cells. It is very unlikely, however, that the effects of Sox9 in endocardial cushions are mediated solely by Sox5 and Sox6 because Sox5;Sox6 double null mutants die at 16.5–17.5 dpc, because of increased abdominal pressure caused by failure in endochondral bone formation (9).
Fig. 6.
Model of the functions of Sox9 in the differentiation of endocardial cushion cells. Sox9 is expressed in mesenchymal cells, not in endothelial cells of endocardial cushions, prevents expression in mesenchymal cells of Nfatc1, which is expressed in endocardial endothelial cells, and is needed at an early step for endocardial cushion formation. Sox9 is required for expression of ErbB3, which is needed for proliferation and differentiation of endocardial cushion cells. Sox9 also activates Sox5 and Sox6.
A growing number of single-gene mutations have been associated with abnormal development of cardiac valves and with defects of endocardial cushion formation. These include mutations of Nfatc1 (21, 22), Neuregulin (31), ErbB3 (25), type 1 neurofibromatosis (Nf1) (15), PDGFRα (29), Shp2 (32), BMPs and TGFβs and their receptors (33–36), and others (37, 38). However, to date, no progress has been made in assembling these individual genes into genetic or molecular cascades that control EMT or subsequent valve remodeling. In several cases, molecules implicated in endocardial cushion formation are also active in EMT processes that take place elsewhere in the embryo, suggesting that molecular pathways may be conserved. The analysis of Sox9-deficient embryos indicates that the processes of delamination/migration and mesenchymal transformation can be distinguished. Sox9 does not appear to be necessary for delamination and initial migration, and some mesenchymal markers are activated correctly (e.g., PDGFα and Runx2/Cbfa1). Also, at least one endothelial marker, Neuregulin, is down-regulated in the Sox9-null mutants. However, persistent expression of Nfatc1 and failure to activate ErbB3 expression indicate that mesenchymal differentiation is inhibited and allow us to place Sox9 function upstream of ErbB3 but downstream or independent of PDGFRα and Runx2/Cbfa1.
Based on the symptoms of campomelic dysplasia, which is caused by heterozygous mutations in Sox9 (3, 4), and on the results of other experiments (10, 39, 40), Sox9 must have important roles in additional tissues including Sertoli cells of the testis, cranial neural crest cells, precursor cells in the kidney and the pancreas, and neural stem cells. It seems likely that the function of Sox9 during EMT in the heart will have analogies in other organs.
Acknowledgments
We thank Kenneth Dunner, Jr., for scanning electron microscopy; Andrew Spicer for advice with AV canal explant culture; Xiaohong Yang, Zhaoping Zhang, Min Min Lu, and Danilo Landrock for technical support; and Rupert Timpl, Mon-Li Chu, and Andrew Spicer for plasmids, probes, and antibodies. We also thank Janie Finch for editorial assistance. This work was supported by the National Institutes of Health (B.d.C. and R.R.B.) and the G. Harold and Leila Y. Mathers Charitable Foundation (B.d.C.). We also acknowledge National Institutes of Health Grant CA16672 for DNA sequence analysis, scanning electron microscope analysis, and veterinary services.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EMT, epithelial–mesenchymal transformation; AV, atrioventricular; X-Gal, 5-bromo-4-chloro-3-indolyl β-d-galactoside; dpc, days postcoitum.
References
- 1.Zhao, Q., Eberspaecher, H., Lefebvre, V. & De Crombrugghe, B. (1997) Dev. Dyn. 209, 377-386. [DOI] [PubMed] [Google Scholar]
- 2.Ng, L. J., Wheatley, S., Muscat, G. E., Conway-Campbell, J., Bowles, J., Wright, E., Bell, D. M., Tam, P. P., Cheah, K. S. & Koopman, P. (1997) Dev. Biol. 183, 108-121. [DOI] [PubMed] [Google Scholar]
- 3.Wagner, T., Wirth, J., Meyer, J., Zabel, B., Held, M., Zimmer, J., Pasantes, J., Bricarelli, F. D., Keutel, J., Hustert, E., et al. (1994) Cell 79, 1111-1120. [DOI] [PubMed] [Google Scholar]
- 4.Foster, J. W., Dominguez-Steglich, M. A., Guioli, S., Kowk, G., Weller, P. A., Stevanovic, M., Weissenbach, J., Mansour, S., Young, I. D., Goodfellow, P. N., et al. (1994) Nature 372, 525-530. [DOI] [PubMed] [Google Scholar]
- 5.Houston, C. S., Opitz, J. M., Spranger, J. W., Macpherson, R. I., Reed, M. H., Gilbert, E. F., Herrmann, J. & Schinzel, A. (1983) Am. J. Med. Genet. 15, 3-28. [DOI] [PubMed] [Google Scholar]
- 6.Bi, W., Deng, J. M., Zhang, Z., Behringer, R. R. & de Crombrugghe, B. (1999) Nat. Genet. 22, 85-89. [DOI] [PubMed] [Google Scholar]
- 7.Bi, W., Huang, W., Whitworth, D. J., Deng, J. M., Zhang, Z., Behringer, R. R. & de Crombrugghe, B. (2001) Proc. Natl. Acad. Sci. USA 98, 6698-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akiyama, H., Chaboissier, M. C., Martin, J. F., Schedl, A. & de Crombrugghe, B. (2002) Genes Dev. 16, 2813-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smits, P., Li, P., Mandel, J., Zhang, Z., Deng, J. M., Behringer, R. R., de Croumbrugghe, B. & Lefebvre, V. (2001) Dev. Cell 1, 277-290. [DOI] [PubMed] [Google Scholar]
- 10.Stolt, C. C., Lommes, P., Sock, E., Chaboissier, M. C., Schedl, A. & Wegner, M. (2003) Genes Dev. 17, 1677-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries, W. N., Binns, L. T., Fancher, K. S., Dean, J., Moore, R., Kemler, R. & Knowles, B. B. (2000) Genesis 26, 110-112. [PubMed] [Google Scholar]
- 12.O'Gorman, S., Dagenais, N. A., Qian, M. & Marchuk, Y. (1997) Proc. Natl. Acad. Sci. USA 94, 14602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan, T. C., Sasaki, T., Zhang, R. Z., Fassler, R., Timpl, R. & Chu, M. L. (1993) J. Cell Biol. 123, 1269-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, H. Y., Chu, M. L., Pan, T. C., Sasaki, T., Timpl, R. & Ekblom, P. (1995) Dev. Biol. 167, 18-26. [DOI] [PubMed] [Google Scholar]
- 15.Gitler, A. D., Zhu, Y., Ismat, F. A., Lu, M. M., Yamauchi, Y., Parada, L. F. & Epstein, J. A. (2003) Nat. Genet. 33, 75-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albrecht, U., Eichele, G., Helms, J. A. & Lu, H. C. (1997) in Molecular and Cellular Methods in Developmental Toxicology, ed. Daston, G. P. (CRC, Boca Raton, FL), pp. 23-48.
- 17.Camenisch, T. D., Spicer, A. P., Brehm-Gibson, T., Biesterfeldt, J., Augustine, M. L., Calabro, A., Jr., Kubalak, S., Klewer, S. E. & McDonald, J. A. (2000) J. Clin. Invest. 106, 349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, X., Rowitch, D. H., Soriano, P., McMahon, A. P. & Sucov, H. M. (2000) Development (Cambridge, U.K.) 127, 1607-1616. [DOI] [PubMed] [Google Scholar]
- 19.Epstein, J. A., Li, J., Lang, D., Chen, F., Brown, C. B., Jin, F., Lu, M. M., Thomas, M., Liu, E., Wessels, A. & Lo, C. W. (2000) Development (Cambridge, U.K.) 127, 1869-1878. [DOI] [PubMed] [Google Scholar]
- 20.Tsuda, T., Wang, H., Timpl, R. & Chu, M. L. (2001) Dev. Dyn. 222, 89-100. [DOI] [PubMed] [Google Scholar]
- 21.de la Pompa, J. L., Timmerman, L. A., Takimoto, H., Yoshida, H., Elia, A. J., Samper, E., Potter, J., Wakeham, A., Marengere, L., Langille, B. L., et al. (1998) Nature 392, 182-186. [DOI] [PubMed] [Google Scholar]
- 22.Ranger, A. M., Grusby, M. J., Hodge, M. R., Gravallese, E. M., de la Brousse, F. C., Hoey, T., Mickanin, C., Baldwin, H. S. & Glimcher, L. H. (1998) Nature 392, 186-190. [DOI] [PubMed] [Google Scholar]
- 23.Riese, D. J., II, & Stern, D. F. (1998) BioEssays 20, 41-48. [DOI] [PubMed] [Google Scholar]
- 24.Lakkis, M. M. & Epstein, J. A. (1998) Development (Cambridge, U.K.) 125, 4359-4367. [DOI] [PubMed] [Google Scholar]
- 25.Erickson, S. L., O'Shea, K. S., Ghaboosi, N., Loverro, L., Frantz, G., Bauer, M., Lu, L. H. & Moore, M. W. (1997) Development (Cambridge, U.K.) 124, 4999-5011. [DOI] [PubMed] [Google Scholar]
- 26.Komori, T., Yagi, H., Nomura, S., Yamaguchi, A., Sasaki, K., Deguchi, K., Shimizu, Y., Bronson, R. T., Gao, Y. H., Inada, M., et al. (1997) Cell 89, 755-764. [DOI] [PubMed] [Google Scholar]
- 27.Otto, F., Thornell, A. P., Crompton, T., Denzel, A., Gilmour, K. C., Rosewell, I. R., Stamp, G. W., Beddington, R. S., Mundlos, S., Olsen, B. R., et al. (1997) Cell 89, 765-771. [DOI] [PubMed] [Google Scholar]
- 28.Stephenson, D. A., Mercola, M., Anderson, E., Wang, C. Y., Stiles, C. D., Bowen-Pope, D. F. & Chapman, V. M. (1991) Proc. Natl. Acad. Sci. USA 88, 6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison-Graham, K., Schatteman, G. C., Bork, T., Bowen-Pope, D. F. & Weston, J. A. (1992) Development (Cambridge, U.K.) 115, 133-142. [DOI] [PubMed] [Google Scholar]
- 30.Camenisch, T. D., Schroeder, J. A., Bradley, J., Klewer, S. E. & McDonald, J. A. (2002) Nat. Med. 8, 850-855. [DOI] [PubMed] [Google Scholar]
- 31.Meyer, D. & Birchmeier, C. (1995) Nature 378, 386-390. [DOI] [PubMed] [Google Scholar]
- 32.Chen, B., Bronson, R. T., Klaman, L. D., Hampton, T. G., Wang, J. F., Green, P. J., Magnuson, T., Douglas, P. S., Morgan, J. P. & Neel, B. G. (2000) Nat. Genet. 24, 296-299. [DOI] [PubMed] [Google Scholar]
- 33.Boyer, A. S., Ayerinskas, I. I., Vincent, E. B., McKinney, L. A., Weeks, D. L. & Runyan, R. B. (1999) Dev. Biol. 208, 530-545. [DOI] [PubMed] [Google Scholar]
- 34.Brown, C. B., Boyer, A. S., Runyan, R. B. & Barnett, J. V. (1999) Science 283, 2080-2082. [DOI] [PubMed] [Google Scholar]
- 35.Kim, R. Y., Robertson, E. J. & Solloway, M. J. (2001) Dev. Biol. 235, 449-466. [DOI] [PubMed] [Google Scholar]
- 36.Gaussin, V., Van de Putte, T., Mishina, Y., Hanks, M. C., Zwijsen, A., Huylebroeck, D., Behringer, R. R. & Schneider, M. D. (2002) Proc. Natl. Acad. Sci. USA 99, 2878-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galvin, K. M., Donovan, M. J., Lynch, C. A., Meyer, R. I., Paul, R. J., Lorenz, J. N., Fairchild-Huntress, V., Dixon, K. L., Dunmore, J. H., Gimbrone, M. A., Jr., et al. (2000) Nat. Genet. 24, 171-174. [DOI] [PubMed] [Google Scholar]
- 38.Kumai, M., Nishii, K., Nakamura, K., Takeda, N., Suzuki, M. & Shibata, Y. (2000) Development (Cambridge, U.K.) 127, 3501-3512. [DOI] [PubMed] [Google Scholar]
- 39.Kent, J., Wheatley, S. C., Andrews, J. E., Sinclair, A. H. & Koopman, P. (1996) Development (Cambridge, U.K.) 122, 2813-2822. [DOI] [PubMed] [Google Scholar]
- 40.Piper, K., Ball, S. G., Keeling, J. W., Mansoor, S., Wilson, D. I. & Hanley, N. A. (2002) Mech. Dev. 116, 223-226. [DOI] [PubMed] [Google Scholar]