Abstract
The hematopoietic system offers many advantages as a model for understanding general aspects of lineage choice and specification. Using oligonucleotide microarrays, we compared gene expression patterns of multiple purified hematopoietic cell populations, including neutrophils, monocytes, macrophages, resting, centrocytic, and centroblastic B lymphocytes, dendritic cells, and hematopoietic stem cells. Some of these cells were studied under both resting and stimulated conditions. We studied the collective behavior of subsets of genes derived from the Biocarta database of functional pathways, hand-tuned groupings of genes into broad functional categories based on the Gene Ontology database, and the metabolic pathways in the Kyoto Encyclopedia of Genes and Genomes database. Principal component analysis revealed strikingly pervasive differences in relative levels of gene expression among cell lineages that involve most of the subsets examined. These results indicate that many processes in these cells behave differently in different lineages. Much of the variation among lineages was captured by the first few principal components. Principal components biplots were found to provide a convenient visual display of the contributions of the various genes within the subsets in lineage discrimination. Moreover, by applying tree-constructing methodologies borrowed from phylogenetics to the expression data from differentiated cells and stem cells, we reconstructed a tree of relationships that resembled the established hematopoietic program of lineage development. Thus, the mRNA expression data implicitly contained information about developmental relationships among cell types.
The hematopoietic system contains cells of more than a dozen mature types, derived from a single stem cell. Individual lineages and their dedicated precursor cells are stably determined and resistant to a variety of changes in the milieu of a cell or exhibit a narrow range of developmental choices that are selected through some combination of stochastic and instructive processes under precise conditions (1, 2). Instead of focusing on a many individual, unrelated, differentially expressed genes, we sought to investigate the differences among multiple hematopoietic cell types from a global viewpoint.
To study the transcriptome composition of hematopoietic cells, one could apply standard methods to identify single genes that are expressed at levels that are different for each lineage. This approach is limited in explaining the overall differences among lineages because we did not find even a small number of individual genes whose expression levels distinguished among all cell types, although there are many genes that significantly differentiate between two or more cell types.
Another analytical approach toward the goal of learning about how the cell types differ in gene expression would be to search in our data for a set of genes, together with a particular discriminant function of the expression levels of those genes that optimally distinguishes the cell types. Such a “supervised learning” approach is common in pattern recognition problems in which a large number of examples of patterns from the different classes are given (3, 4). In our case, we have thousands of variables (genes) to use in discriminating among the types of cells, but for each type, we have only a very small number of examples of that type. In a case like this, “overfitting” in pattern recognition becomes a particularly serious concern; we can always find many functions that discriminate among sets of a handful of points in a space of thousands of dimensions, but then we would have little confidence that we have learned anything real that would generalize beyond the few given data points.
To avoid this problem, we investigated whether an unsupervised approach to dimension reduction would also lead to discrimination among the cell types. Because we do not make use of information about the cell types in reducing the data to a few dimensions, worries about overfitting are ameliorated. When we project the data onto two dimensions by using principal components and then see that the different cell types project onto different regions of the plane (as in Figs. 1 and 2), we can trust that the discrimination we are witnessing is genuine. Because the projection used no information about the identities of the cell types, we know we have not overfit the data; indeed the projection is simply a view of the data without “fitting” the cell-type labels at all.
Fig. 1.
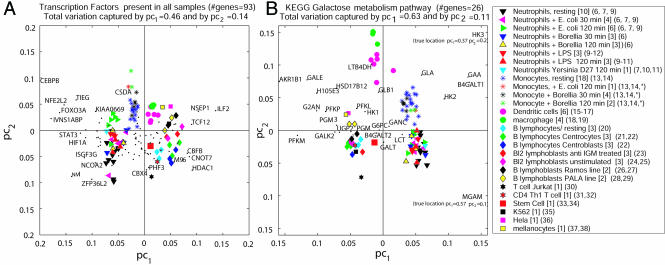
(A) PCA and biplot. Shown is the projection of 92 samples collected from 27 cell types onto the two leading principal components of a submatrix consisting of expression profiles of 93 transcription factors present in all samples. Clustering of samples is evident, even though the identities of the samples were not used in performing the projection. Before PCA the genome-wide data matrix was preprocessed by taking the logarithm of each entry in the matrix followed by subtraction of the row and column means of this entry and addition of the overall mean, leading to a normalized matrix having all row sums and all column sums equal to zero. Each cell type is represented by a distinct symbol (numbers of samples are in brackets and method of preparation is referenced). Overlaying a 2D scatter plot, representing the contribution of the genes (represented by dots) to the first and second principal components on top of the 2D sample scatter plot, forms a biplot. An inner product between a gene and a sample (which is equal to the product of the length of a vector pointing from the origin to the sample, the length of a vector pointing from the origin to the gene, and the cosine between these vectors) approximates the normalized expression value of the gene at this sample. The accumulated variation captured by the first and second principal components is 60% of the total variation. (B) A biplot for the KEGG galactose metabolism pathway. This biplot represents samples as in A for 26 genes from the galactose metabolism pathway, which was expressed in at least one sample. The HK3 and MGAM genes were located out of the frame; therefore the true location is indicated in parentheses. The biplot can be used to read approximated normalized expression levels. For example, normalized expression levels of HK3 are elevated in the monocyte samples in comparison with the B cell samples. Similarly, expression levels of MGAM are highly elevated in neutrophils in comparison with the rest of the cells. The accumulated variation captured by the first and second principal components is 74%.
Fig. 2.
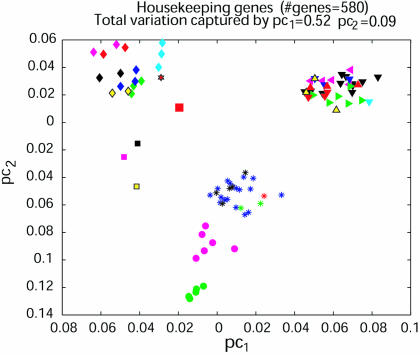
PCA as in Fig. 1 but for 580 housekeeping genes reported in the literature that were expressed in at least one of the 92 samples. (Although the literature refers to 575 housekeeping genes, a few of the sequences match more than one gene so that the total number of matching genes is slightly greater.) The Jurkat cell is an outlier of this set of genes and therefore it was excluded in this analysis. The accumulated variation captured by the first and second principal components is 61%.
Reduction in the number of variables could have been achieved by representing gene clusters obtained in an unsupervised clustering analysis by their leading principal components (5). However, our analysis was constructed to represent the system by using variables that are directly related to biological processes (or functions), which would allow us to investigate differences and similarities in activity of each of these processes among the cell types.
We attempted to identify functional categories of genes whose expression levels vary between the different lineages more extensively than within any single lineage, but we found that a large number of broadly expressed genes have relative levels of expression that distinguish among various types of cells. A similar distinction is also apparent in the expression levels of genes limited to specific metabolic pathways. Lineage-specific patterns of expression were seen even with “housekeeping” genes that are expressed very broadly in experiments reported in the literature. Most individual genes show significant lineage-specific differences in their relative levels of expression, and each lineage is characterized by a pervasive distinct pattern of gene expression. However, these patterns are constrained such that the first few principal components account for a large part of the variation. The gene expression data also contain information about the developmental histories of these cells, as shown by a tree resembling the standard hematopoietic program that we constructed by using methodology borrowed from phylogenetics.
Materials and Methods
Cell Isolation, RNA Extraction, and Probe Preparation. The methods for preparation and treatment of individual cell types are indicated in the legend of Fig. 1. RNA from each cell type was extracted most often by use of commercial kits (RNeasy, Qiagen, Valencia, CA), except for neutrophils, from which total RNA was extracted by a guanidine·HCl method, as described (6, 7). RNA quality was evaluated by measuring the ratio of 28S to 18S rRNA on agarose gel electrophoresis and by determining the ratio of optical absorbance at 260 and 280 nm. Samples were rejected if the A280:A260 or the 18S:28S rRNA ratio was >0.6 or if samples showed any traces of degradation or DNA contamination on gel electrophoresis. In general, samples were rejected if <10 mg of total RNA was obtained from a single preparation. Each preparation was from a single patient, and for the most part, replicate samples of the same cell type were from different donors. RNA analysis was performed on the Affymetrix HGU133 chip set (Santa Clara, CA), using standard techniques for probe preparation and the Affymetrix mas 5.0 normalization procedure. The human cell population was derived from peripheral blood stem cells from a healthy donor mobilized by granulocyte colony-stimulating factor (G-CSF) (8). We used a two-step positive–negative selection technique in which CD34+ cells are isolated and then depleted of CD38 by using immunimagnetic beads. The resultant population was 93% CD34+ and had <1% CD38+ cells.
In the present work, we have included only one stem cell sample because of the expense of obtaining enough human cells of this type to perform RNA analysis without previous amplification and, therefore, to keep the results strictly comparable to those from the other samples. However, we have data from a second human stem cell preparation analyzed with an earlier version of the oligonucleotide chip (Affymetrix HGU95) that gave similar results (data not shown).
Simultaneous Array and Gene Normalization. Recently (39) we developed a normalization procedure based on the concept that two genes (and likewise two samples) whose expression profiles differ only by a multiplicative constant of proportionality are really behaving in the same way. After taking logarithms of each element in the data matrix, we considered two genes (each represented by a row of the matrix) or two samples (represented by the columns of the matrix) to be equivalent if their expression profiles differ by an additive constant. The resulting normalization, which involves taking logarithms of the entries in the data matrix followed by subtraction of the row and column means and addition of the overall mean at each entry, can be thought as a two-way ANOVA-like procedure, which removes first-order effects and extracts the desired gene–sample interaction effects. Previously (39) this choice of normalization led to better separation between distinct cell types.
Filtering. Before further analysis, we discarded the genes labeled by the Affymetrix mas software as absent in all samples. This procedure maximizes the number of remaining genes so that the collective expression profiles we derive for each pathway will have a sufficient number of representative genes. Alternatively, we used filtering in which we kept only genes that were flagged as present in every sample by the Affymetrix software.
Principal Component Analysis (PCA). Our dataset is tabulated in a matrix consisting of tens of thousands of genes (rows) and 92 samples (columns) that span 27 different cell types. The number of samples is two orders of magnitude smaller than the number of variables (genes). Reduction in the number of variables is useful for visualizing major trends and structure inherent in the data. PCA is an unsupervised dimension reduction method that generates a new set of decorrelated variables (principal components) as linear combinations of the original variables (genes). For an introductory explanation see www.statsoftinc.com/textbook/stfacan.html. The majority of the variation of microarray datasets (samples) can be captured by the most dominant principal components. An additional advantage of expressing the data in terms of the leading principal components is their robustness to noise. The projections of the samples onto the leading principal components are computed by applying the singular value decomposition to the data matrix (after preprocessing as described above).
Data Balancing. The number of replicate samples in our data repository of 27 cell types varies from 1 (for the hematopoietic stem cell) to 18 (for the monocytes). This imbalance should be taken into account before performing PCA. We did this by forming a matrix for each cell type containing 18 columns. For cell types with <18 measurements, we replicated existing measurements. When 18 was not exactly divisible by the number of measurements, we added columns with median expression levels across the existing samples of the particular cell type. For example, we had four samples of macrophages; therefore we concatenated their median profile twice together with four replicates of each sample. The resulting balanced data matrix contained 18 × 26 columns.
Biplot. We used principal component biplots to display the expression profiles of the genes (rows of the data matrix) and the cell types (columns of the data matrix) simultaneously as points in 2D space (40). The biplot provides an optimal approximation of the data matrix by such a 2D structure, in the sense that it displays the singular value decomposition, which gives the rank-two approximation to the data matrix having the smallest mean-squared error. The expression of a given gene in a given sample is approximated by the projection of the gene vector onto the direction of the sample vector, multiplied by the length of the sample vector. Thus, in this rank-two approximation, for a given gene and for sample vectors of a given length, the gene is expressed at a higher (or lower) level in samples whose vector points in nearly the same (or opposite) direction as the gene. A gene is not differentially expressed between samples that are located on a line orthogonal to the gene vector.
Linear Discriminant Analysis. One of our primary methods for visualizing data corresponding to a given subset of genes was to project the data onto the first two principal component directions and then inspect the resulting 2D scatter plot to see how well the various cell types were separated. We used linear discriminant analysis and cross validation to construct a descriptive statistic to quantify the extent to which the cell type groups are separated in a given projection. The starting point is a scatter plot containing, for each of the n samples in the data set, the 2D coordinates for that sample together with the corresponding class label, i.e., the cell type of that sample. For each of the n samples in turn, we would remove that sample from the data, find the optimal linear discriminant boundaries for the resulting data set of n - 1 samples, and then check whether the resulting discriminant boundaries correctly classify the held-out data point. (This procedure was carried out by using the function “lda” from the MASS library of the statistical computing package r.) We took the fraction of samples that were classified correctly after being left out of the data in this way as a measure of how well the classes were separated in the 2D projection. This measure is used in Fig. 3.
Fig. 3.
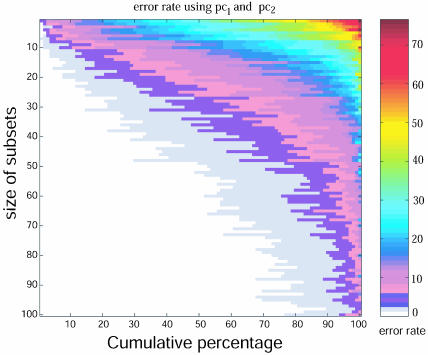
Map of error rates as a function of a gene subset sizes. For each subset size we randomly selected 100 subsets of genes out of the annotated 17,460 genes printed on the U133 Affymetrix chip. We then derived the first and second principal components for each of these gene subsets. These principal components were used as predictors in linear discriminant analysis to classify the different lineages. The vertical axis of the map represents the subset size. Each subset size has 100 samples sorted along the horizontal axis according their misclassification error rate. As we increase the subset size, the fraction of subsets with zero error rate increases. Thus the potential to partition the samples by using the information stored in the first two principal components increases as the gene subset size increases.
Phylogenetic Analysis. In typical phylogenetic studies, the goal is to recover the tree that represents the evolutionary history of species by using a collection of biological sequences. Instead of different species of organisms, our taxa are the different cell types in our study. Our data are also not typical for current phylogenetic studies because we are using gene expression measurements potentially taking a continuum of values, rather than sequences chosen from a small alphabet such as nucleotides or amino acids.
The class of phylogenetic methods that seemed most straight-forward to adapt to our problem are the distance-based methods, such as neighbor joining and the unweighted pair group method using arithmetic averages (UPGMA), which take as their input a matrix of estimated distances between pairs of taxa. Applying these methods to our data requires us to choose a method of calculating a distance between two gene expression profiles. The most important property desired for a measure of distance in this context is that it be additive, or nearly additive, in the following sense. A distance measure is additive on a tree if the distance between any pair of observed taxa is the sum of the lengths of all branches on the path joining those two species in the tree, where each branch length is the result of applying the same distance measure to the (possibly unobserved and hypothetical) taxa at the two ends of the branch. Consideration of the stochastic processes by which the measured characteristics change as the tree is traversed can suggest additive distance measures, e.g., Markov process models have led to various measures of distance in standard phylogenetic studies (41, 42). In our problem, as a first rough model, we could postulate that the logarithms of the gene expression values change according to a random-walk-like process in development. After a bifurcation at which one lineage splits into two, the two resulting lineages would be imagined to develop independently. Because the variance is additive for independent random variables, the requirement of additivity in such a model suggested that we use the variance of the differences in logarithm of gene expression values as our distance measure.
Results
Pathway Analysis. To study cell type specificity of various biological processes, we computed the leading principal components for each of the pathways annotated in Biocarta or the Kyoto Encyclopedia of Genes and Genomes (KEGG), or for functionally defined families of genes obtained from Gene Ontology. Each such set of principal components gives a low-dimensional summary of the expression measurements for the corresponding set of genes. More specifically, the complete expression information of any given pathway is stored in a matrix whose rows represent the genes of this pathway and whose columns represent the multiple samples from the various cell types. This information can be represented more simply by computing a relatively small number of specific linear combinations of the gene profiles of the pathway. The coefficients in the linear combination are principal component directions. For a given pathway, the expression profiles of the samples (cell types) are summarized by their projections onto a small number of principal components directions. For many pathways, the projection of the expression profiles of the samples onto two principal component directions reveals clear separation between the different cell types that is not observed by inspecting the expression patterns of individual genes. Studying the system in terms of a few hundred collective variables representing biologically defined pathways, instead of tens of thousands of variables corresponding to individual genes, allows us to have a more coarse-grained global picture of the cell type characteristics.
As primary data, we used expression analyses of 92 samples representing the 27 cell types detailed in Fig. 1. To examine the differences in pathways and functional activities among these 27 cell types, we performed a PCA of gene expression data normalized as described in Materials and Methods, to determine whether well separated cell type clusters could be identified. We projected the multidimensional data onto a 2D space spanned by the two leading principal components, represented by the two axes of the graph.
A major regulator of the pattern of gene transcription in any cell is the abundance and activity of protein complexes including the sequence specific factors that interact with DNA. We therefore initially studied the patterns of transcripts for genes encoding known transcription factors. To investigate the specificity and stability of lineage-specific gene expression, we included expression patterns of several continuous cell lines including Epstein–Barr virus-transformed lymphoblasts, T and B cell lymphomas, and control melanocyte cultures. We used Gene Ontology to select genes related to transcriptional activity. The list from Gene Ontology was curated by hand to eliminate obvious misclassifications and add missing components. To aid in this process, Locus Link was used to collect genes whose description included relevant terms, and these genes were examined individually for appropriateness. In addition, the transcriptional activity list was curated to remove subunits of core polymerases, general transcriptional components, and elongation or termination factors.
We derived principal components by using either all of the genes that were found to be present in at least one sample or, more stringently, only those genes listed as present in every sample. As shown in the scatter plot in Fig. 1a, even in the latter case most of the samples of the different cell types are separable. The projection of the samples onto the first principal component explains 46% of the variability of the data and is sufficient for partitioning the major groups of lineages. Moreover, the second principal component captures 14% of the data variability. The transcription factors contributing most to the first two principal components are labeled on the figure. The factors whose projection along the first two principal components was largest include some, such as CEBPB (CCAAT/enhancer-binding protein β), that would have been expected to vary in levels between cell types, based on a number of studies of their role in lineage-specific gene expression. A number of other factors, such as HDAC1, a common histone deacetylase, would not a priori have been predicted to be important in distinguishing among cell lineages.
These studies were extended to genes related to apoptosis, translation, receptor activity, cytokine activity, proteolysis, or protein kinase activity. Broadly speaking, the subsets of genes fell into two categories. In the case of cytokines and receptors, most of the genes that were present in some samples were not detected in other samples. In these cases, a large number of genes were listed as present in at least one sample, and classification of lineages by using all these genes gave a clear separation according to lineage by use of the first two principal components. On the other hand, if only genes listed as present in all samples were included, very few genes were left, and the separation of lineages by using these genes was incomplete. For other gene sets, such as transcription factors or proteolytic genes, the number of genes whose expression was detected in all samples was relatively high, with correspondingly good lineage separation by the first principal components. Generally, the first two principal components accounted for a major fraction of the total variation of samples and revealed extensive separation between cell lineages.
The KEGG and Biocarta databases present catalogs of groups of genes, classified according to their linkage in known pathways including those for small molecules. We examined the principal component analyses for each of the 137 pathways described in KEGG and the 245 networks listed in Biocarta. The number of genes per pathway varies, and this variation contributes to the ability of each gene set to separate lineages. However some small pathways clearly had greater power to separate lineages. As an example of lineage discrimination by a KEGG pathway, Fig. 1b shows the separation of lineages obtained by use of the first two principal components from the galactose pathway, together with the genes listed for the pathway. The bias in expression of MGAM (α-glucosidase), and HK3 (hexokinase 3) is particularly striking. The physiology underlying this bias is not immediately evident, although one might speculate, for example, about effects on innate immunity of the membrane-associated glucosidase in removing terminal glucose residues from cell surface complex carbohydrates. These initial results show that, without supervision, the data clearly separated samples according to the lineage of cells from which they were derived.
Because most subsets of genes seemed to give good lineage separation, we then asked whether a less discriminating set of housekeeping genes could be identified as a subset of genes expressed commonly in all lineages. To do this, we used data reported for genes whose expression had been detected in each of 47 types of cells or tissues reported at various times in the literature (9). As shown in Fig. 2, of ≈580 of the housekeeping genes listed in the literature, only 260 were recorded as expressed in all of our experiments. When either the 580 or the 260 genes in this group were used, the first two principal components again separated most or all lineages.
Having found that sets of genes that we expected to behave similarly across the cell lines have the ability to distinguish lineages, we speculated that even randomly chosen subsets of genes might exhibit some ability to discriminate the cell types. To test this conjecture, we applied multiclass linear discriminant analysis to randomly selected gene subsets of variable size. We found that for the majority of these subsets, the first and second principal components were sufficient to predict the cell type with very good accuracy (Fig. 3). Furthermore, for most pathways containing more than ≈20 genes, there was a range of the number of principal components that led to optimal separation of the cell types. This result indicates that the differences among the cell types are distributed among a very large set of genes and remain consistently evident in sufficiently large randomly chosen subsets of genes.
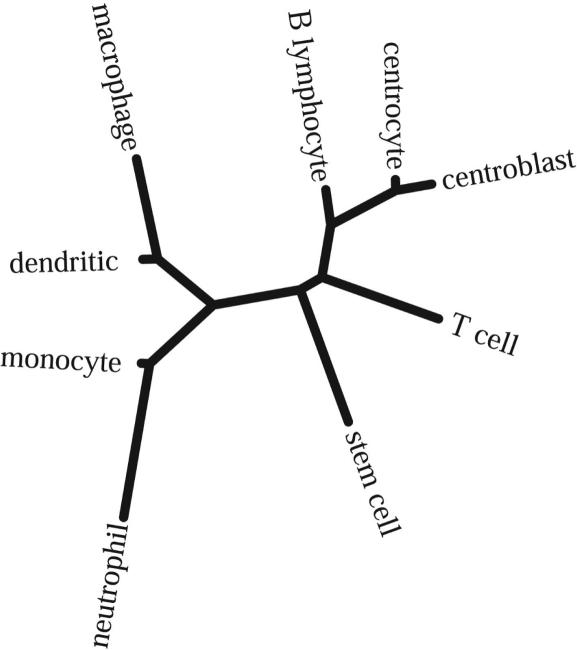
Development Tree. We next examined whether cell-type expression profiles provide sufficient information that may be used to infer a development tree in the same fashion that DNA sequences from different species are used to infer phylogenetic trees. Fig. 4 illustrates a developmental tree generated by using the neighbor-joining algorithm (43) together with the a measure of distance between expression profiles as described in Materials and Methods. We note that the stem cell is positioned between the myeloid and lymphoid lineages. Moreover, the first split in the lymphoid branch separates the B cells from the T cell sample. Down the B lymphocyte branch, the next split separates the resting cells from the centrocytes and centroblasts. The first split in the myeloid branch separates the neutrophil group and monocytes from the dendritic cells and macrophages that were generated by stimulating monocytes. We would expect the monocytes to branch off from the edge leading to the dendritic cells and macrophages, instead of the edge leading to the neutrophils. Aside from this single discrepancy, the topology of this tree matches our current knowledge of hematopoietic development. Thus, cell-type expression profiles containing the expression values of all transcription factors (and other functional groups of genes) are sufficient not only to differentiate between the cell lineages but also to capture developmental relationships between these cells. We note that using UPGMA, an alternative distance-based approach for building trees that implicitly relies on a molecular clock type of assumption, reconstructed an incorrect tree.
Fig. 4.
Development tree: An unrooted tree with undetermined direction of time was derived by using the neighbor-joining tree reconstruction algorithm as implemented in phylip (43). The input distance matrix between the different cell types was derived by using the variance between the logarithms of the mean cell-type profiles. The lymphoid and myeloid branches and their subbranches are separated in accordance with the established hematopoietic program.
Discussion
In the present study, we compared expression profiles of a large number of genes in a number of terminally differentiated cells that all derive from a common precursor. To examine the expression data, we used a normalization procedure that set the geometric mean of expression values of the particular group of genes to be the same for each sample and also set the geometric mean of expression values of each separate gene across all samples to be the same. This process loses some information. However, the procedure focuses on the differences in patterns of gene expression within a particular group. Also, the relative levels of expression of a given gene may more accurately reflect its contribution to a process in different cell types than does its absolute level of expression. The high quality of lineage separations obtained in this study indicates that, regardless of rationale, the normalization approach retained important biologic information.
As expected, the amount of total variation contributed by the first two principal components decreased as the number of genes in a group increased. However, it was striking that the first two principal components contributed a relatively large fraction, generally more than one-third, of the total variation in almost all cases. These data indicate considerable structure to the patterns of variation of lineage-specific expression.
Graphing the position of specific genes on the same principal component plot as that showing the cell lineages is a convenient way of visualizing the potential contribution of specific factors to the separation of the various lineages. The projection of the vector from the origin to the factor onto the vector pointing to each lineage is an estimate of the contribution of that factor to the specificity of that lineage. It is noteworthy that most vectors for individual genes were not parallel to the vector for any lineage, consistent with the expectation that most factors are used in different relative amounts and in a combinatorial fashion to specify lineages.
Describing the cells in terms of principal component variables summarizing the collective behavior of genes in various pathways may be a suitable framework for studying statistical relationships among the pathways. For example, one can inspect the level of correlations between the pathway variables or investigate the extent to which these variables interact with each other in multivariate statistical models designed to discriminate between the various cell types. Identifying strong pathway–pathway statistical associations is useful for designing experiments to explore biological inter-relationships among pathways. These collective variables are expected to be more robust to microarray experimental noise than the individual gene variables and therefore could be useful for future system biology studies.
One unanticipated result of the present set of analyses was the pervasiveness of lineage specificity of the relative levels of expression of genes. The number of products whose level was relatively constant (<1.5-fold variation in median expression) was so small as to be almost statistically insignificant. To a greater degree than anticipated, the lineage specificity of differential expression of known genes is not readily interpreted, as shown here for the enzymes in the KEGG galactose metabolism group. Among other things, this observation suggests the difficulty in predicting cell type-specific effects of pharmacologic agents or even transcription factors. More generally, the present approach affords a convenient exploratory tool for investigating lineage differences among cells with respect to biologically defined functions and gene subsets.
The position of the stem cells was often somewhat closer to B cells than the other lineages, but stem cells occupied a relatively central position in the principal component projections for almost all of the gene sets examined. Morphologically, stem cells resemble lymphocytes, and the genes contributing to this appearance might be one factor in the positioning of the stem cells. However, the central position could be due to several more important factors. Technically, the stem cell population is a very small subset of cells, and their definition by surface markers is rather indirect, so that the preparations could be heterogeneous in ways that are not obvious from fluorescence-activated cell sorter analysis. Low levels of mRNA for some B cell-specific genes were present in the stem cell sample, and this result could represent either contamination or expression of some lineage-specific genes in true stem cells. The stem cells do express a somewhat larger number of genes than the cells of the various differentiated lineages. They take an intermediate position in the 2D space spanned by the first two principal components but have a projection on the remaining principal components that is larger than most of the other cell types. Our analyses also suggest that each lineage specification represents a different perturbation from a generalized stem cell rather than a progressive addition of layers of differentiation on some other lineage.
In summary, we have analyzed gene expression in differentiated cells of multiple hematopoietic lineages, experimentally and mathematically. The results show an impressively pervasive lineage specificity in gene expression, extending across many pathways and gene types. The hematopoietic cell types studied represent terminally differentiated cells, some of which are postmitotic. This may heighten the differences between lineages. It will be of considerable interest to extend the analyses across other well defined cell types and to compare the results with more undifferentiated cells and cells of nonhematopoietic lineages.
Acknowledgments
This work was supported by National Institutes of Health Grant POL HL 633571.
Abbreviations: PCA, principal component analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes.
References
- 1.Payne, K. J. & Crooks, G. M. (2002) Immunol. Rev. 187, 48-64. [DOI] [PubMed] [Google Scholar]
- 2.Cantor, A. B. & Orkin, S. H. (2001) Curr. Opin. Genet. Dev. 11, 513-519. [DOI] [PubMed] [Google Scholar]
- 3.Pavlidis, P., Lewis, D. P. & Noble, W. M. (2002) Pacific Symp. Biocomput. 7, 474-485. [PubMed] [Google Scholar]
- 4.Mateos, A., Dopazo, J., Jansen, R., Tu, Y., Gerstein, M. & Stolovitzky, G. (2002) Genome Res. 12, 1703-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang, E., Cheng, S. H., Dressman, H., Pittman, J., Tsou, M. H., Horng, C. F., Bild, A., Iversen, E. S., Liao, M., Chen, C. M., et al. (2003) Lancet 361, 1590-1596. [DOI] [PubMed] [Google Scholar]
- 6.Subrahmanyam, Y. V., Baskaran, N., Newburger, P. E. & Weissman, S. M. (1999) Methods Enzymol. 303, 272-297. [DOI] [PubMed] [Google Scholar]
- 7.Subrahmanyam, Y. V., Yamaga, S., Prashar, Y., Lee, H. H., Hoe, N. P., Kluger, Y., Gerstein, M., Goguen, J. D., Newburger, P. E. & Weissman, S. M. (2001) Blood 97, 2457-2468. [DOI] [PubMed] [Google Scholar]
- 8.Debelak, J., Shlomchik, M. J., Snyder, E. L., Cooper, D., Seropian, S., McGuirk, J., Smith, B. & Krause, D. S. (2000) Transfusion 40, 1475-1481. [DOI] [PubMed] [Google Scholar]
- 9.Tsukahara, Y., Lian, Z., Zhang, X., Whitney, C., Kluger, Y., Tuck, D., Yamaga, S., Nakayama, Y., Weissman, S. M. & Newburger, P. E. (2003) J. Cell. Biochem. 89, 848-861. [DOI] [PubMed] [Google Scholar]
- 10.Lian, Z., Wang, L., Yamaga, S., Bonds, W., Beazer-Barclay, Y., Kluger, Y., Gerstein, M., Newburger, P. E., Berliner, N. & Weissman, S. M. (2001) Blood 98, 513-524. [DOI] [PubMed] [Google Scholar]
- 11.Lian, Z., Kluger, Y., Greenbaum, D. S., Tuck, D., Gerstein, M., Berliner, N., Weissman, S. M. & Newburger, P. E. (2002) Blood 100, 3209-3220. [DOI] [PubMed] [Google Scholar]
- 12.Zhang, X., Kluger, Y., Poddar, R., Whitney, W., DeTora, A., Weissman, S. M. & Newburger, P. E. (2004) J. Leukocyte Biol. 75, 358-372. [DOI] [PubMed] [Google Scholar]
- 13.Kumagai, K., Itoh, K., Hinuma, S. & Tada, M. (1979) J. Immunol. Methods 29, 17-25. [DOI] [PubMed] [Google Scholar]
- 14.Cathcart, M. K., Morel, D. W. & Chisolm, G. M., III 1985) J. Leukocyte Biol. 38, 341-350. [DOI] [PubMed] [Google Scholar]
- 15.Williams, L. A., Egner, W. & Hart, D. N. (1994) Int. Rev. Cytol. 153, 41-103. [DOI] [PubMed] [Google Scholar]
- 16.Fearnley, D. B., McLellan, A. D., Mannering, S. I., Hock, B. D. & Hart, D. N. (1997) Blood 89, 3708-3716. [PubMed] [Google Scholar]
- 17.Markowicz, S. & Engleman, E. G. (1990) J. Clin. Invest. 85, 955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sallusto, F. & Lanzavecchia, A. (1994) J. Exp. Med. 179, 1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelchen-Matthews, A., Kramer, B. & Marsh, M. (2003) J. Cell Biol. 162, 443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chalouni, C., Banchereau, J., Vogt, A. B., Pascual, V. & Davoust, J. (2003) Int. Immunol. 15, 457-466. [DOI] [PubMed] [Google Scholar]
- 21.Sims-Mourtada, J. C., Guzman-Rojas, L., Rangel, R., Nghiem, D. X., Ullrich, S. E., Guret, C., Cain, K. & Martinez-Valdez, H. (2003) Immunology 110, 296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pascual, V., Liu, Y. J., Magalski, A., de Bouteiller, O., Banchereau, J. & Capra, J. D. (1994) J. Exp. Med. 180, 329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denepoux, S., Fournier, N., Peronne, C., Banchereau, J. & Lebecque, S. (2000) J. Immunol. 164, 1306-1313. [DOI] [PubMed] [Google Scholar]
- 24.Poltoratsky, V., Woo, C. J., Tippin, B., Martin, A., Goodman, M. F. & Scharff, M. D. (2001) Proc. Natl. Acad. Sci. USA 98, 7976-7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakayama, Y., Iwamoto, Y., Maher, S. E., Tanaka, Y. & Bothwell, A. L. (2000) Biochem. Biophys. Res. Commun. 277, 124-127. [DOI] [PubMed] [Google Scholar]
- 26.Sale, J. E. & Neuberger, M. S. (1998) Immunity 9, 859-869. [DOI] [PubMed] [Google Scholar]
- 27.Papavasiliou, F. N. & Schatz, D. G. (2000) Nature 408, 216-221. [DOI] [PubMed] [Google Scholar]
- 28.Chicz, R. M., Urban, R. G., Gorga, J. C., Vignali, D. A., Lane, W. S. & Strominger, J. L. (1993) J. Exp. Med. 178, 27-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arunachalam, B., Pan, M. & Cresswell, P. (1998) J. Immunol. 160, 5797-5806. [PubMed] [Google Scholar]
- 30.Schneider, U., Schwenk, H. U. & Bornkamm, G. (1977) Int. J. Cancer 19, 621-626. [DOI] [PubMed] [Google Scholar]
- 31.Brosterhus, H., Brings, S., Leyendeckers, H., Manz, R. A., Miltenyi, S., Radbruch, A., Assenmacher, M. & Schmitz, J. (1999) Eur. J. Immunol. 29, 4053-4059. [DOI] [PubMed] [Google Scholar]
- 32.Dagna, L., Iellem, A., Biswas, P., Resta, D., Tantardini, F., Fortis, C., Sabbadini, M. G., D'Ambrosio, D., Manfredi, A. A. & Ferrarini, M. (2002) Eur. J. Immunol. 32, 2934-2943. [DOI] [PubMed] [Google Scholar]
- 33.Angelopoulou, M., Novelli, E., Grove, J. E., Rinder, H. M., Civin, C., Cheng, L. & Krause, D. S. (2003) Exp. Hematol. 31, 413-420. [DOI] [PubMed] [Google Scholar]
- 34.Donnelly, D. S. & Krause, D. S. (2001) Leuk. Lymphoma 40, 221-234. [DOI] [PubMed] [Google Scholar]
- 35.Lozzio, C. B. & Lozzio, B. B. (1975) Blood 45, 321-334. [PubMed] [Google Scholar]
- 36.Yee, C., Krishnan-Hewlett, I., Baker, C. C., Schlegel, R. & Howley, P. M. (1985) Am. J. Pathol. 119, 361-366. [PMC free article] [PubMed] [Google Scholar]
- 37.Bejar, J., Hong, Y. & Schartl, M. (2003) Development (Cambridge, U.K.) 130, 6545-6553. [DOI] [PubMed] [Google Scholar]
- 38.Bohm, M., Moellmann, G., Cheng, E., Alvarez-Franco, M., Wagner, S., Sassone-Corsi, P. & Halaban, R. (1995) Cell Growth Differ. 6, 291-302. [PubMed] [Google Scholar]
- 39.Kluger, Y., Basri, R., Chang, J. T. & Gerstein, M. (2003) Genome Res. 13, 703-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox, T. F. & Cox, A. A. (2001) Multidimensional Scaling (Chapman & Hall, London).
- 41.Baake, E. & Haeseler, A. V. (1999) Theor. Popul. Biol. 55, 166-175. [DOI] [PubMed] [Google Scholar]
- 42.Chang, J. T. (1996) Math. Biosci. 137, 52-73. [Google Scholar]
- 43.Felsenstein, J. (1989) Cladistics 5, 164-166. [Google Scholar]