Abstract
Isolated oceanic archipelagos have played a major role in the development of evolutionary theory by offering a unique setting for studying spatial and temporal patterns of biological diversification. However, the evolutionary events that cause associations between genetic variation and geography in archipelago radiations are largely unknown. This finding is especially true in the Galápagos Islands, where molecular studies have revealed conflicting biogeographic patterns. Here, we elucidate the history of diversification of giant Galápagos tortoises by using mtDNA sequences from 802 individuals representing all known extant populations. We test biogeographic predictions based on geological history and assess the roles of volcano emergence and island formation in driving evolutionary diversification. Patterns of colonization and lineage sorting appear highly consistent with the chronological formation of the archipelago. Populations from older islands are composed exclusively of endemic haplotypes that define divergent monophyletic clades. Younger populations, although currently differentiated, exhibit patterns of colonization, demographic variation and genetic interchange shaped by recent volcanism. Colonization probably occurs shortly after a volcano emerges through range expansion from older volcanoes. Volcanism can also create temporal shifts from historical to recurrent events, such as promoting gene flow by creating land bridges between isolated volcanoes. The association of spatial and temporal patterns of genetic variation with geophysical aspects of the environment can best be attributed to the limited dispersal and migration of tortoises following an oceanographic current. The endangered giant Galápagos tortoises represent a rapid allopatric radiation and further exemplify evolutionary processes in one of the world's greatest natural laboratories of evolution.
Volcanic oceanic archipelagos, such as the Galápagos and Hawaiian Islands, contain some of the world's most remarkable biological radiations and offer unparalleled natural laboratories for studying spatial and temporal patterns of population diversification (1–7). These isolated islands have provided opportunities for rapid colonization and speciation and have a geological record that can be used to estimate the chronology of evolutionary events (2, 7, 8). Despite the prominent role of archipelagos in the advancement of evolutionary theory, our knowledge about population history on islands is still limited. Molecular studies of island radiations have generally relied on phylogenetic methods (2) that lack the power to discriminate among various evolutionary factors that might cause associations between genetic and geographic variation, such as range expansion, past fragmentation, and gene flow. One particularly germane topic for testing hypotheses about population history concerns the identification of evolutionary events chronologically associated with the process of volcano emergence and island formation. This information is needed to better understand patterns of population colonization, demographic variation, and the progression of lineage diversification in an archipelago.
For the Galápagos, molecular studies have detected conflicting relationships between population diversification and island formation because of secondary contact between species (e.g., Darwin's finches; ref. 3), sex-biased dispersal (e.g., marine iguanas; ref. 4), or multiple colonizations from the mainland (e.g., lava lizards; ref. 5). The Galápagos Islands are true oceanic volcanoes formed as a result of an eastward displacement of the Nazca plate over a “hot spot,” such that the easternmost islands are the oldest with progressively younger islands to the west (8–10). Thus, the historical biogeographic scenario expected is one promoting chronological colonizations and decreasing phylogeographic structure in an east-to-west direction (3–5, 11, 12). Yet, the archipelago is arranged as clusters of islands with similar age (8, 9), as opposed to the linear arrangement of the Hawaiian Islands (7). This factor, coupled with problems of obtaining reliable ages of formation for volcanoes within a single island (8), complicates the identification of general biogeographic patterns in the Galápagos.
Here, we elucidate the history of colonization and diversification of giant Galápagos tortoises by using control region mtDNA sequences from 802 individuals representing all known extant populations. Giant Galápagos tortoises are the largest terrestrial chelonians in the world and represent the only surviving group of giant tortoises where evolutionary divergence is evident among populations (13). The group is monophyletic and is derived from a mainland South American lineage (14). The radiation is comprised of 11 extant and four extinct taxa endemic to different islands, or is restricted to each of Isabela's five major volcanoes (ref. 13 and Fig. 1). These taxa have been treated either as different species or as subspecies of Geochelone nigra based on morphological differences among populations (6, 13). Nonetheless, morphological divergence in giant Galápagos tortoises is potentially a better indicator of present ecological conditions than of evolutionary relationships (6). These animals cannot dive, they barely swim, and have a distribution linked to vegetated areas of the slopes or tops of volcanoes (13). The apparent allopatric nature of the radiation of giant tortoises, coupled with their dependency on volcanic environments and limited dispersal capability, make them an ideal model for investigating evolutionary processes in volcanic archipelagos.
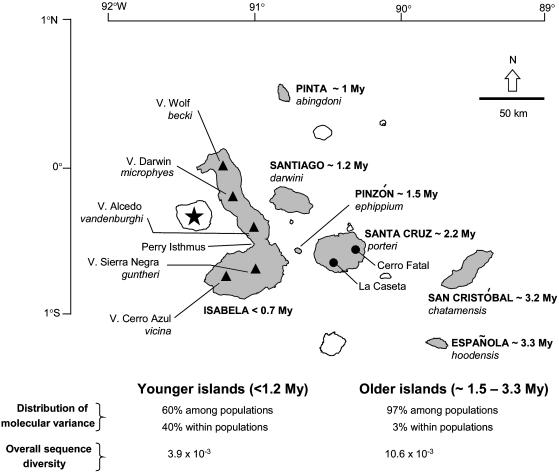
Fig. 1.
The Galápagos Islands with sampled populations, taxon name, and approximate island age (8, 9). Shaded islands have extant populations of tortoises (Pinta is represented by a single male kept in captivity). The star on the left represents the position of the archipelago's hot spot in the Island of Fernandina, ▴, The tops of volcanoes on Isabela.
Previous molecular studies (11, 14–16) of giant Galápagos tortoises revealed marked genetic differentiation among most populations and a general pattern of colonization from geologically older to younger islands. However, due to the recent age of the radiation and incomplete lineage sorting these surveys presented insufficient information about relationships among recently diverged lineages. In addition, because these studies used relatively small sample sizes and were mostly based on analyses of phylogenetic relationships and current gene flow, they provided no inferences about general evolutionary processes shaping population history in giant tortoises. An exception to this observation is a recently published coalescent-based analysis of the effects of a prehistoric volcano eruption in the history of one tortoise population (17). Here, we use a combination of a large sample size and phylogeographic and population genetic approaches to elucidate spatial and temporal dimensions of the tortoise radiation. Our results are used to (i)test biogeographic predictions based on the geological history of the Galápagos, and to (ii) asses the roles of volcano emergence and island formation in driving evolutionary diversification in archipelago radiations.
Materials and Methods
Samples and mtDNA Methods. Blood samples of giant tortoises were obtained from 22 sites on seven islands (Fig. 1). This sampling includes all extant taxa plus a genetically divergent population from Cerro Fatal, in Santa Cruz (11, 15). Islands were represented by a single population, except for Isabela (five volcano populations) and Santa Cruz (La Caseta and Cerro Fatal; Table 1). We also sampled one captive individual from a small group of ≈10 animals from Cerro Montura (Santa Cruz). This isolate population may represent an introduction from Pinzón (15) and therefore was excluded from the analysis. Samples from Wolf, Sierra Negra, and Cerro Azul are pools of individuals sampled from sites located near or on the slopes of each volcano's caldera (we did not detect substantial genetic structure between sites from each volcano). Methods used for DNA extraction, PCR amplification, and sequencing of an ≈700-bp fragment of the mtDNA control region (including its hypervariable section) are available elsewhere (15). We used single-stranded conformation polymorphism (SSCP) analysis to screen for sequence variants (18). The reliability of our SSCP protocol was confirmed by sequencing multiple individuals with the same SSCP gel band (see supporting information, which is published on the PNAS web site), and by comparing our data set with ≈200 tortoise sequences from other studies (11, 15) used as controls in all gels. Our protocol showed that individuals with the same SSCP phenotype as the control had the same sequence (the reverse was true for individuals with different sequences) and allowed unequivocal detection of haplotypes separated by as few as one base pair. Sequences were edited and aligned with sequencer 4.1.2 (Gene Codes, Ann Arbor, MI). Our aligned data set, based on >1.1 million bases of DNA (both strands were sequenced), has a length of 705 bp, and includes seven indels. We identified 115 variable sites that define 82 haplotypes.
Table 1. Populations studied and summary of statistical parameters for the mtDNA tortoise data.
| Island (volcano or site) | Estimated population size* | n | No. of haplotypes† | h | Percent of individuals with endemic haplotypes | Θ × 10-3 | Historical demographic inference‡ |
|---|---|---|---|---|---|---|---|
| San Cristobal | 500—700 | 27 | 1 | — | 1.0 | — | — |
| Española | 15 (native) | 16 | 1 | — | 1.0 | — | — |
| Santa Cruz (La Caseta) | 2,000—3,000 | 66 | 12 | 0.80 | 1.0 | 9.9 | Equilibrium |
| Santa Cruz (Cerro Fatal) | 20 | 16 | 1 | — | 1.0 | — | — |
| Pinzón | 150—200 | 53 | 8 | 0.76 | 1.0 | 3.1 | Expansion |
| Pinta | 1 | 1 | 1 | — | 1.0 | — | — |
| Santiago | 500—700 | 48 | 8 | 0.82 | 0.73 | 4.8 | Expansion |
| Isabela (V. Wolf) | 1,000—2,000 | 83 | 4 | 0.69 | 0.45 | 1.3 | Expansion |
| Isabela (V. Sierra Negra) | 100—300 | 168 | 13 | 0.83 | 0.68 | 2.2 | Expansion |
| Isabela (V. Cerro Azul) | 400—600 | 211 | 26 | 0.87 | 0.92 | 4.4 | Expansion |
| Isabela (V. Alcedo) | 3,000—5,000 | 84 | 5 | 0.17 | 0.09 | 1.6 | Expansion |
| Isabela (V. Darwin) | 500—1,000 | 29 | 2 | 0.50 | 1.0 | 0.3 | § |
n, number of individuals analyzed; h, haplotypic diversity; Θ, sequence diversity.
Ref. 35
Results for aliens (11) are in supporting information
Based on analysis of mismatch distribution
Inadequate variability for mismatch analysis
Genetic Diversity and Population Divergence. Sequence diversity (Θ) was calculated based on a maximum likelihood estimation that uses genealogy sampling, allows for fluctuating population sizes, and therefore reflects the influence of population history (19). We used arlequin 2.0 (20) to calculate haplotypic diversity (h) and to assess population differentiation with the fixation index ΦST (21), an estimator that includes information on haplotype frequency and molecular distance. The distance for ΦST, chosen by modeltest (22), was based on the Tamura–Nei model (23) with a proportion of invariable sites of 0.52 and a γ value of 0.47 determined in paup*, version 4b10 (24). In addition, ΦST was used to perform a hierarchical analysis of molecular variance (21) among and within populations grouped according to island age (Fig. 1 and refs. 8–10).
Interisland Divergence Time. The age of major interisland migrations was calculated by estimating the rate of mtDNA control region evolution on a maximum-likelihood tree by using the r8s program (25). Two different calibration points were used to allow scaling of rating and times to real units: we assigned the root of the tree at 3.3 million years (Myr; approximate age of the oldest island in Galápagos, Fig. 1) and also enforced a maximum age of 700,000 years (age of emergence of Isabela Island) for a clade comprised exclusively of Isabela populations (clade A). Both methods resulted in the rate of evolution of 3.4% per Myr.
Recent Demographic History. Demographic inferences were assessed by analysis of mismatch distributions, a method that explores signatures of demographic fluctuations in the DNA sequences of a population, and distinguishes between equilibrium and expanding situations (26). arlequin was used to obtain a test statistic for the expansion model and to estimate the age of inferred population expansions (27), such as those expected after a population colonizes a recently emerged island. Although the coalescent-based method implemented in fluctuate uses more information present in the data (28) and seems to be more adequate to estimate exponential growth (see ref. 17 for an example on one giant tortoise population), it tends to produce unclear results in more complex demographic situations (29). On the other hand, a large number of empirical studies (including one on two giant tortoise populations) have validated the usefulness of mismatch analysis when discriminating between contrasting demographic scenarios (15, 30), which relates closer to our study aims.
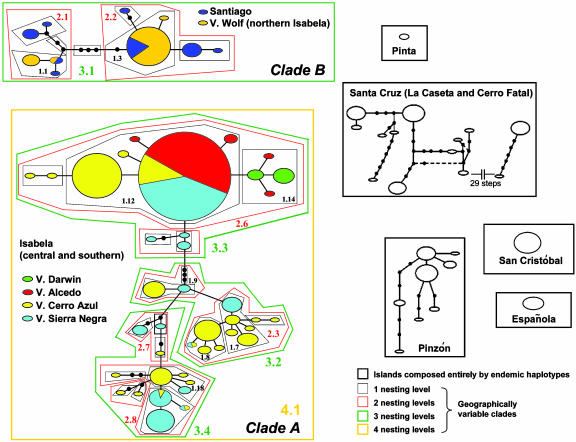
Haplotype Network and Nested Clade Analysis (NCA). Genealogical relationships were investigated by constructing a haplotype network with the statistical parsimony method implemented in tcs (31). This program established a 95% connection limit between haplotypes at 11 mutational steps. The haplotype network was converted into a nested series of clades (32) used to carry out nested clade phylogeographic analysis as implemented in geodis 2.0 (33). The NCA uses genealogical information to infer and geographically localize both historical events (such as range expansions) and recurrent forces (such as gene flow), influencing the distribution of genetic variation (32). It also estimates the dynamic structure and temporal juxtaposition of these evolutionary factors without regard to any prior model (32, 34). The NCA was conducted only when a clade showed geographic variation. The analysis is limited to situations where phylogeographic signal is based on sufficient sampling and genetic resolution and when the null hypothesis of no association between geography and the haplotype tree is rejected. Geographic distances incorporated into NCA were measured among sampling locations by using Global Positioning System coordinates collected in the field. Nested clades and NCA inferences are shown in Fig. 2 and Table 2, respectively. Biological patterns were interpreted with the revised version of Templeton's inference key available at http://zoology.byu.edu/crandall_lab/geodis.htm. The sequential interpretations of NCA results are in the supporting information.
Fig. 2.
Haplotype network showing relationships among geographic populations (taxa), based on statistical parsimony. Size of the ovals is proportional to haplotype frequency in each population. Each line between haplotypes indicates one mutational change, or step. Color boxes describe the nesting design of clades A and B. Only clades with significant geographic association are numbered (see Table 2). Thick black boxes delineate clades that have no genetic or geographic variation, and therefore cannot be included in the NCA (Santa Cruz populations showed insufficient variability for NCA). Minimum number of steps connecting boxed clades (data not shown) range between 13 and 31 (not supported by statistical parsimony).
Table 2. Inferences of the NCA for clades A and B and direction of colonization or gene flow.
| Clade A: Southern Isabela (V. Sierra Negra, V. Cerro Azul) and Central Isabela (V. Alcedo, V. Darwin) | Nested clade | Clade B: Santiago and Northern Isabela (V. Wolf) | Nested clade |
|---|---|---|---|
| LDC out of Sierra Negra to Alcedo, Darwin, Cerro Azul | 4.1 | ||
| LDC from southern to central volcanoes | 3.3 | IBD from Santiago to Wolf | 3.1 |
| LDC from Sierra Negra to Cerro Azul | 3.4 | ||
| IBD from Sierra Negra to Cerro Azul | 3.2 | ||
| IBD from Sierra Negra to Cerro Azul | 2.3 | LDC or LDD, origin ambiguous | 2.1 |
| IBD from Sierra Negra to Cerro Azul | 2.7 | LDC or LDD, origin ambiguous | 2.2 |
| LDC or LDD in southern volcanoes, origin ambiguous | 2.8 | ||
| PF into a Alcedo-Darwin and a Cerro Azul clade | 2.6 | ||
| LDC from Cerro Azul to Sierra Negra | 1.8 | IBD, origin ambiguous | 1.1 |
| LDC from Sierra Negra to Cerro Azul | 1.9 | IBD, origin ambiguous | 1.3 |
| LDC from Cerro Azul to Sierra Negra | 1.18 | ||
| LDC or LDD between southern and central, origin ambiguous | 1.12 | ||
| LDC between Darwin and Alcedo, origin ambiguous | 1.14 | ||
| LDC or LDD from Sierra Negra to Cerro Azul | 1.7 |
Only clades with statistically significant variation are included. Inferences are shown from oldest (top) to most recent (bottom); and are based on unambiguous relationships among clades and age estimates of islands and volcanoes. LDC, range expansion with long distance colonization; IBD, gene flow restricted by isolation by distance; PF, past fragmentation into two allopatric groups; LDD, restricted gene flow with long distance dispersal.
Results and Discussion
mtDNA Variability and Alien Lineages. Giant tortoise populations (or taxa) showed distinct levels of mtDNA variation, ranging from one control region haplotype in the Islands of Española and San Cristóbal to moderate and high variability in more westerly located islands (Table 1). Hypothetically, this disparity could be the result of a smaller evolutionary population size in arid and isolated islands (e.g., Española and San Cristóbal); as opposed to larger population sizes in younger islands, which have higher elevations and more ecological complexity (6, 13). Another potential cause is the unequal exploitation of tortoise populations by whalers in the 17th to 19th centuries. Whalers heavily collected tortoises in the more accessible islands such as Pinta, Española, and San Cristóbal, whereas exploitation was less intense or was nonexistent on islands with higher volcanoes (13).
A surprising finding was that 91% of individuals from the Volcano Alcedo, the largest extant population in the archipelago with ≈4,000 individuals (35), appear to be from the same maternal lineage (h = 0.17). We presented evidence elsewhere that the lower variability of Alcedo tortoises is probably related to a severe bottleneck caused by a sizeable and explosive prehistoric eruption ≈100,000 years ago (17). Although volcanic activity on young islands could have influenced the demographic history of other populations and obscured phylogeographic patterns, it seems that catastrophic eruptions are not the norm in the Galápagos. The Alcedo eruption is the only explosive eruption documented for the archipelago; otherwise, Galápagos volcanoes are constructed of basaltic lavas that erupted by nonexplosive effusion (36), and possibly did not severely impact populations living on the slopes of volcanoes.
We detected six “alien” haplotypes in our data set. Aliens have been previously defined as rare occurrences of tortoises with haplotypes that differ from others from the same location by 27–70 substitutions in 4.5 kb of mtDNA sequences, whereas they are identical or differ by very few substitutions (three on the average) from haplotypes found in other islands (11). Aliens have been found in northern Isabela (Wolf and Darwin) and in the adjacent island of Santiago. They are most abundant in Wolf, where they may have been deposited by whalers, a hypothesis supported by old logbooks from the whaling industry (11, 13). Ongoing work in the Wolf population using morphological, microsatellite, and mtDNA data has detected the only known example of genetic introgression between divergent lineages of giant tortoises (N.H., L.B.B., C. Cioffi, A. C., and J.R.P., unpublished work), a situation most likely related to human-mediated introductions. Importantly, data analyses including aliens (see supporting information) do not contradict the outcomes of this study. The only difference is the lack of support for population expansion in Wolf when alien haplotypes are included, which is an expected result due to increased genetic diversity (Θ, without and with aliens is 1.3 and 2.9, respectively). Details about the distribution and evolutionary relationships of aliens are available elsewhere (11).
General Biogeographic Consensus. Our data show a pattern of lineage sorting consistent with the temporal formation of the archipelago. Taxa from older islands (≈ 1.5–3.3 Myr) are composed exclusively of endemic haplotypes that define divergent monophyletic clades (overall Θ is 10.6 × 10-3). In contrast, taxa from younger islands (<1.2 Myr), although currently differentiated, have haplotypes that are not totally sorted among volcanoes and exhibit a recent history of coalescence (overall Θ is 3.9 × 10-3; Figs. 1 and 2 and Table 1). An analysis of molecular variance shows that in older islands differences among and within populations account for 97% and 3%, respectively, of total genetic variation, whereas these values are 60% and 40% in younger islands. This strong pattern of structure resulted in substantial genetic divergence among all populations (overall ΦST = 0.71), including those from young environments (see supporting information), an outcome consistent with analyses of microsatellite markers (15, 16).
We estimated divergence times for major interisland migrations and combined them with demographic analyses to provide an approximate time frame to evaluate events of colonization and diversification. A previous phylogenetic study (11) indicates that one of the earliest migrations was from either San Cristóbal or Española to Santa Cruz (founding La Caseta population). Our estimate for this event of ≈2 Myr is consistent with geological evidence (Fig. 1) and demographic history: La Caseta has the highest Θ (10 × 10-3) and is the only taxon with a mismatch distribution of a relatively old population with stable demography. All remaining populations for which demographic inference was obtained inhabit younger islands. Their haplotypes show much lower Θ (ranges from 0.3 to 4.8 × 10-3) and contain signals of demographic expansions (Table 1), which is probably associated with the onset of island colonization (except in Alcedo). For instance, one of the most recent interisland migrations in the Galápagos took place from Santa Cruz to one of the four volcanoes in southern and central Isabela (11). Divergence times between these taxa and estimates of population expansions in southern and central Isabela are very recent (0.05–0.57 and 0.2–0.7 Myr, respectively) and consistent with the geological age of the island (≈0.7 Myr; ref. 8). In general, we detected a correspondence between demographic variation and the structure of population genealogies linked to the geological history of major islands.
Volcano Emergence, Island Formation, and Population Diversification. By using NCA, we inferred specific evolutionary causes for phylogeographic associations in younger clades that appear related to aerial emergence and development of Isabela's volcanoes (Table 2 and Fig. 2). For clade A (southern and central Isabela), we detected 14 significant geographical associations, with the dominant biological event at various time scales being range expansion with long-distance colonization. Reconstructing the colonization in Isabela ideally requires two kinds of information. We need to understand phylogenetic relationships among recently diverged population groups, but these are not well resolved due to rapid diversification and incomplete lineage sorting in giant tortoises (Fig. 2 and ref. 11). In addition, we should use information about the relative ages of Isabela's five major volcanoes. This approach is also problematic because these volcanoes are perpendicular to the direction of plate motion (precluding a simple chain-like temporal progression for their origin), they are nearly contemporaneous, and display poor records about earliest subaerial activity (37). However, it appears that each volcano shows an evolutionary stage characterized by gradational differences in morphology and structure through which all volcanoes eventually pass and through which the oldest has already passed (10). This process enabled Nordlie (10) to propose a model for the systematic growth and development of Isabela's volcanoes that arranges them in a sequential formation where Sierra Negra is the oldest and Cerro Azul and Wolf are the youngest. Interestingly, Nordlie's model (10) is consistent with our finding that colonization apparently began at Sierra Negra, the population with the oldest estimated expansion (0.7 Myr), and with haplotypes occupying all interior positions of old nested clades (Fig. 2). Moreover, given that Sierra Negra is the most voluminous volcano in the Galápagos (37), it is most likely that this was the first landmass in Isabela to be colonized by drifting tortoises. Our inferences based on NCA (Table 2) support the idea that Sierra Negra acted as the source for subsequent range expansions to newly emerged neighboring islands (represented today by volcanoes Alcedo, Darwin, and Cerro Azul).
Once these islands emerged, lava flows promoted subaerial growth and establishment of land bridges between volcanoes (10). These land bridges, which eventually formed the island of Isabela, would theoretically promote movements of individual tortoises. Accordingly, NCA shows a temporal shift from the historical factors related to colonization of Sierra Negra and Cerro Azul to three events of gene flow between these two volcanoes (clades 3.2, 2.3, and 2.7). A more recent event was the past fragmentation inferred by NCA in a widespread clade (clade 2.6) separated into an Alcedo–Darwin and a Cerro Azul clade. This finding contrasts with the previous scenario of gene flow and is perhaps associated with the flooding of the Perry Isthmus (Fig. 1) during the interglacial period ≈130,000 years ago, which may have isolated southern populations from those in central volcanoes. The remaining expansion and migration events in clade A are very recent and are generally restricted to Sierra Negra and Cerro Azul. This result agrees with other studies that show reduced genetic differentiation between southern Isabela taxa (11, 16).
Finally, for clade B, NCA supports the shallow phylogeographic divergence between the Island of Santiago and volcano Wolf detected here (Fig. 2) and in previous surveys (11, 16) by inferring relatively recent episodes of gene flow and possible range expansions. Drifting events from Santiago to the recently formed Wolf (10) agree with prevailing current directions (12, 35), whereas dispersal from the south may have been impaired by the extensive lava fields between Wolf and Darwin. Although NCA was not conducted for clades from older islands because they show either no geographic or genetic variation (Fig. 2), the general agreement between estimated genealogical times of divergence with those based on geology (11) favors range expansion as the most likely explanation for the colonization of older islands.
Giant Tortoises: Linkages Between Volcanic and Population Histories. Our study of giant tortoises indicates an intimate association of temporal patterns of genetic variation with geophysical aspects of the environment. It contrasts to most reconstructions of archipelago radiations (2) because we have focused on population genetic parameters rather than merely interpreting phylogenetic relationships. By combining large sample size and statistical approaches that reflect the influence of population history, we hypothesize evolutionary events related with volcano emergence and island formation (but see ref. 38 for a phylogeny-based model for the development of island species in Hawaiian damselflies). In general, colonization probably occurs shortly after a volcano emerges by means of range expansion from older volcanoes, and allopatric diversification, both genetic and morphological (6), apparently proceeds very rapidly. Volcanism can also create temporal shifts from historical to recurrent events, such as promoting gene flow by creating land bridges between neighboring volcanoes.
We should also point out some limitations of our analysis of recent demographic history. One is the unavailability of data from extinct taxa, especially those from centrally located islands that could potentially be involved in interisland migrations. In addition, given that Galápagos tortoises are virtually invariable in 4 kb of noncoding nuclear DNA sequences (39), we were forced to base our analyses solely on the mtDNA locus. Finally, the identification of a specific population history that best fits to genetic data is an obviously difficult exercise, and for many situations (such as in Isabela), assessing error in phylogeographic inferences may simply not be feasible due to current methodological constraints (40). Despite these limitations, several relevant factors support the conclusions of our historical reconstruction. First, results of the NCA in Isabela are in agreement with estimates of demographic expansions based on other methods, and are concordant with the history of volcanism and geotectonics (8–10). Second, our inferences about recent gene flow, especially between southern volcanoes, agree well with population parameters estimated for giant tortoises based on multilocus microsatellite data (16). Furthermore, our reconstruction was conducted based on an extensive sampling across the island and using lineages endemic to a relatively recent environment (e.g., genetic variability was generated in situ and in <0.7 Myr). This finding is important because it reduces the risks that geographical gaps in sampling would result in NCA inferences of long-distance colonization. It also reduces the number of alternative historical scenarios to be considered and the problems commonly associated with ancient divergences, such as lineage extinctions, dramatic changes in ecology and distribution, and the stochastic nature of population genetic processes (17, 30, 41, 42).
We also describe a general scenario markedly consistent with biogeographic expectations of chronological colonization and decreasing phylogeographic structure in an east-to-west direction. Two major aspects accounting for this concordance are the unidirectional tortoise migration due to dependence on a predominantly Southeast-to-Northwest oceanographic current (12, 43) and the noticeable lack of natural movements between islands after population establishment. The latter is a remarkable finding given the tortoises' ability for drifting on currents, and thus raises the possibility that factors such as strong agonistic behavior between divergent populations (44), and morphological specializations for particular ecological resources (6), may limit the effectiveness of invasion by later migrants into an established population. Our study indicates that the endangered and long-lived giant tortoises represent a compelling example of a rapid allopatric radiation and are testimony to the complexity of biogeographic processes of one of the world's greatest natural laboratories of evolution. Although intricate, application of modern evolutionary techniques and concepts can nevertheless shed considerable light on the biological past. Galápagos tortoises continue to inspire and elucidate natural history processes.
Supplementary Material
Acknowledgments
We thank the staff from the Charles Darwin Research Station and Parque Nacional de Galápagos for their support with sample collection; D. Geist for helpful discussion on the geological history of the archipelago; and two anonymous reviewers for comments on earlier drafts. Samples were collected under CITES Permit 01/1S784934/9. This work was supported by the National Science Foundation, the National Geographic Society, and the Yale Institute for Biospheric Studies. L.B.B. acknowledges a Gaylord Donnelley Fellowship from the Yale Institute for Biospheric Studies.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NCA, nested clade analysis; Myr, million years.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF548204–AF548285).
References
- 1.Darwin, C. (1859) The Origin of Species by Means of Natural Selection (John Murray, London).
- 2.Emerson, B. C. (2002) Mol. Ecol. 11, 951-966. [DOI] [PubMed] [Google Scholar]
- 3.Grant, P. R. & Grant R. (1996) Philos. Trans. R. Soc. London B 351, 765-772. [Google Scholar]
- 4.Rassmann, K., Tautz, D., Trillmich, F. & Gliddon, C. (1997) Mol. Ecol. 6, 437-452. [Google Scholar]
- 5.Wright, J. W. (1983) in Patterns of Evolution in Galápagos Organisms, eds. Bowman, R. I., Berson, M. & Leviton, A. E. (Am. Assoc. Adv. Sci. Pacific Div., San Francisco), pp. 123-155.
- 6.Fritts, T. H. (1983) Biol. J. Linn. Soc. 21, 165-176. [Google Scholar]
- 7.Wagner, W. L. & Funk, V. A. (1995) Hawaiian Biogeography: Evolution on a Hot Spot Archipelago (Smithsonian Inst. Press, Washington, DC).
- 8.Cox, A. (1983) in Patterns of Evolution in Galápagos Organisms, eds. Bowman, R. I., Berson, M. & Leviton, A. E. (Am. Assoc. Adv. Sci. Pacific Div., San Francisco), pp. 11-23.
- 9.White, W. M., McBirney, A. R. & Duncan, R. A. (1993) J. Geophys. Res. 98, 533-563. [Google Scholar]
- 10.Nordlie, B. E. (1973) Geol. Soc. Am. Bull. 84, 2931-2956. [Google Scholar]
- 11.Caccone, A., Gentile, G., Gibbs, J. P., Fritts, T. H., Snell, H. L. & Powell, J. R. (2002) Evolution (Lawrence, Kans.) 56, 2052-2066. [DOI] [PubMed] [Google Scholar]
- 12.Harvey, L. E. (1994) J. R. Soc. N. Z. 24, 45-63. [Google Scholar]
- 13.Pritchard, P. C. H. (1996) Chelonian Res. Monographs 1, 1-85. [Google Scholar]
- 14.Caccone, A., Gibbs, J. P., Ketmaier, V., Suatoni, E. & Powell, J. R. (1999) Proc. Natl. Acad. Sci. USA 96, 13223-13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beheregaray, L. B., Ciofi, C., Caccone, A., Gibbs, J. P. & Powell, J. R. (2003) Conserv. Genet. 4, 31-46. [Google Scholar]
- 16.Ciofi, C., Milincovitch, M., Gibbs, J. P., Caccone, A. & Powell, J. R. (2002) Mol. Ecol. 11, 2265-2283. [DOI] [PubMed] [Google Scholar]
- 17.Beheregaray, L. B., Ciofi, C., Geist, D., Gibbs, J. P., Caccone, A. & Powell, J. R. (2003) Science 302, 75. [DOI] [PubMed] [Google Scholar]
- 18.Sunnucks, P., Wilson, A. C. C., Beheregaray, L. B., Zenger, K., French, J. & Taylor, A. C. (2000) Mol. Ecol. 9, 1699-1710. [DOI] [PubMed] [Google Scholar]
- 19.Kuhner, M. K., Yamato, J. & Felsenstein, J. (1995) Genetics 140, 1421-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider, S., Kueffer, J., Roessli, D. & Excoffier, L. (2000) arlequin: A Software for Population Genetic Data Analysis (Univ. of Geneva, Geneva), Version 2.000.
- 21.Excoffier, L., Smouse, P. E. & Quattro, J. M. (1992) Genetics 131, 479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 23.Tamura, K. & Nei, M. (1995) Mol. Biol. Evol. 10, 512-526. [DOI] [PubMed] [Google Scholar]
- 24.Swofford, D. L. (1998) paup*, Phylogenetic Analysis Using Parsimony (* and Other Methods) (Sinauer, Sunderland, MA), Version 4b10.
- 25.Sanderson, M. J. (1997) Mol. Biol. Evol. 14, 1218-1231. [Google Scholar]
- 26.Rogers, A. R. & Harpending, H. C. (1992) Mol. Biol. Evol. 9, 552-569. [DOI] [PubMed] [Google Scholar]
- 27.Schneider, S. & Excoffier, L. (1999) Genetics 152, 1079-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhner, M. K., Yamato, J. & Felsenstein, J. (1998) Genetics 149, 429-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lessa, E., Cook, J. A. & Patton, J. L. (2003) Proc. Natl. Acad. Sci. USA 100, 10331-10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernatchez, L. (1991) Evolution (Lawrence, Kans.) 55, 351-379. [Google Scholar]
- 31.Clement, M., Posada, D. & Crandall, K. A. (2000) Mol. Ecol. 9, 1657-1659. [DOI] [PubMed] [Google Scholar]
- 32.Templeton, A. R. (1998) Mol. Ecol. 7, 381-397. [DOI] [PubMed] [Google Scholar]
- 33.Posada, D., Crandall, K. A. & Templeton, A. R. (2000) Mol. Ecol. 9, 487-488. [DOI] [PubMed] [Google Scholar]
- 34.Templeton, A. R. (2002) Nature 416, 45-51. [DOI] [PubMed] [Google Scholar]
- 35.MacFarland, C. G., Villa, J. & Toro, B. (1974) Biol. Conserv. 6, 118-133. [Google Scholar]
- 36.Geist, D., Howard, K. A., Jellinek, A. M. & Rayder, S. (1994) Bull. Volcanol. 86, 243-260. [Google Scholar]
- 37.Naumann, T. & Geist, D. (2000) Bull. Volcanol. 61, 497-514. [Google Scholar]
- 38.Jordan S., Simon, C. & Polhemus, D. (2003) Syst. Biol. 52, 89-109. [DOI] [PubMed] [Google Scholar]
- 39.Caccone, A., Gentile, G., Burns, C., Sezzi, E., Bergman, W., Ruelle, M. & Powell, J. R. Mol. Phylogenet. Evol., in press. [DOI] [PubMed]
- 40.Knowles, L. L. & Maddison, W. P. (2002) Mol. Ecol. 11, 2623-2635. [DOI] [PubMed] [Google Scholar]
- 41.Masta, S. E., Laurent, N. M. & Routman, E. J. (2003) Mol. Ecol. 12, 1541-1554. [DOI] [PubMed] [Google Scholar]
- 42.Moritz, C., Patton, J. L., Schneider, C. J. & Smith, T. B. (2000) Annu. Rev. Ecol. Syst. 31, 533-563. [Google Scholar]
- 43.Pak, H. & Zaneveld, J. R. V. (1973) J. Geophys. Res. 20, 7845-7859. [Google Scholar]
- 44.Schafer, S. F. & Krekorian, C. O. (1983) Herpetologica 39, 448-456. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.