Abstract
Centromere repositioning provides a potentially powerful evolutionary force for reproductive isolation and speciation, but the underlying mechanisms remain ill-defined. An attractive model is through the simultaneous inactivation of a normal centromere and the formation of a new centromere at a hitherto noncentromeric chromosomal location with minimal detrimental effect. We report a two-generation family in which the centromeric activity of one chromosome 4 has been relocated to a euchromatic site at 4q21.3 through the epigenetic formation of a neocentromere in otherwise cytogenetically normal and mitotically stable karyotypes. Strong epigenetic inactivation of the original centromere is suggested by retention of 1.3 megabases of centromeric α-satellite DNA, absence of detectable molecular alteration in chromosome 4-centromereproximal p- and q-arm sequences, and failure of the inactive centromere to be reactivated through extensive culturing or treatment with histone deacetylase inhibitor trichostatin A. The neocentromere binds functionally essential centromere proteins (CENP-A, CENP-C, CENP-E, CENP-I, BUB1, and HP1), although a moderate reduction in CENP-A binding and sister-chromatid cohesion compared with the typical centromeres suggests possible underlying structural/functional differences. The stable mitotic and meiotic transmissibility of this pseudodicentric-neocentric chromosome in healthy individuals and the ability of the neocentric activity to form in a euchromatic site in preference to a preexisting alphoid domain provide direct evidence for an inherent mechanism of human centromere repositioning and karyotype evolution “in progress.” We discuss the wider implication of such a mechanism for meiotic drive and the evolution of primate and other species.
The evolutionary history of centromeres in humans and other primates is characterized by extraordinary plasticity, which has led to a substantial divergence in both the sequence and location of centromeric DNA on phylogenetically related chromosomes in different primate species (1, 2). Sequence divergence can be attributed to the fact that centromeric DNA is subjected to an unprecedented level of rearrangements including amplifications, duplications, transpositions, inversions, and deletions, compared with the bulk of euchromatic DNA (3). Divergence in the location of centromeres in phylogenetically related chromosomes also might be expected to result from chromosome rearrangements; however, recent studies suggest that centromere repositioning can occur in the absence of any alteration in the order of DNA markers along the chromosome (4, 5). These studies suggest that in some instances centromere repositioning occurs as a result of the emergence of a new centromere rather than by the relocation of an existing centromere from another genomic site.
One mechanism for the emergence of a centromere at a new site is by neocentromere formation. Neocentromeres are ectopic centromeres that originate occasionally from noncentromeric regions of chromosomes (6, 7). Despite the complete absence of normal centromeric α-satellite DNA, neocentromeres are able to form a primary constriction and assemble a functional kinetochore. The formation of neocentromeres is now a recognized phenomenon in humans, with >60 cases of constitutional neocentromeres described, in addition to the occurrence of neocentromeres in certain types of cancers (for review see ref. 7). Constitutional neocentromeres typically are detected in children with birth defects or developmental delay and occur in association with a chromosome rearrangement that generates an unbalanced karyotype and a chromosome fragment lacking α-satellite DNA. The emergence of neocentromeres in noncentromeric euchromatin therefore has been viewed as a mechanism for the “rescue” of analphoid chromosome fragments that otherwise would be lost, although the generally deleterious consequence of the karyotype rearrangement seems a priori incompatible as an evolutionary mechanism.
We describe a healthy family in which centromere repositioning has occurred on chromosome 4 via neocentromere formation with retention of α-satellite DNA but without any other detectable chromosomal alterations. The result is a mitotically and meiotically stable pseudodicentric chromosome that contains an active neocentromere and an inactive alphoid centromere. These findings provide direct evidence in support of neocentromere formation as an inherent mechanism for centromere repositioning and the potential for karyotype evolution.
Materials and Methods
Cell Lines. Epstein–Barr virus-transformed lymphocyte cell lines were established from all available family members and cultured in RPMI medium 1640 (Thermo Trace, Melbourne) supplemented with 10% FCS. Primary fibroblasts and simian virus 40 large T antigen-transformed fibroblasts from the 7-year-old female patient were maintained in DMEM supplemented with 10% FCS (Thermo Trace). Colcemid (GIBCO/BRL) was added to the medium at a concentration of 0.1 μg/ml for 1–2 h before harvesting.
Genotyping and Linkage Analysis. Genomic DNA was extracted from transformed lymphocytes of individuals I:1, II:1, II:2, III:1, and III:2. A chromosome 4 linkage screen was performed at the Australian Genome Research Facility (Parkville, Victoria, Australia) by using 45 microsatellite markers from the ABI Prism linkage mapping set, version 2 (PE Applied Biosystems) at a marker density of 5 centimorgans. Haplotypes were derived manually.
Bacterial Artificial Chromosome Probes and Fluorescence in Situ Hybridization. Bacterial artificial chromosome (BAC) clones were obtained from the human genomic BAC library, RP11. The mapping data were obtained from the University of California (Santa Cruz) human genome browser (http://genome.ucsc.edu) and the Celera Discovery System (www.celera.com). BAC DNA was labeled by nick translation with digoxigenin-11-dUTP or biotin-16-dUTP (Roche Diagnostics). Fluorescence in situ hybridization (FISH) at high or low (for some α-satellite probing experiments) stringency was performed as described (6).
Immunofluorescence and Immuno-FISH. Immunofluorescence was as described (8, 9), with modification for use in conjunction with FISH (immuno-FISH) (6). Immuno-FISH signals were measured by using iplab (Scanalytics, Billerica, MA) software. Background fluorescence was removed from each raw image before fluorescence quantitation. The integrated total fluorescence within a region immediately surrounding the signal of interest then was recorded. All signals of raw images were nonsaturating, and measurements for each of at least 40 different combined images were confirmed at different exposure levels.
Antibodies. Antisera used in this study included human anticentromere autoimmune serum CREST6 (specific for centromere proteins CENP-A and CENP-B) (6), anti-human CENP-A from Upstate Biotechnology (Lake Placid, NY), anti-HP1α (Santa Cruz Biotechnology), and anti-CENP-I (10). Antibodies to CENP-B, CENP-C, CENP-E, and hBUB1 were as described (11).
Cell Viability and Anaphase-Lag Assays. Cell viability in lymphoblastoid cell lines was determined by flow cytometry after propidium iodide (PI) staining. Briefly, 106 cells were subcultured 48 h before harvesting. Cells were washed twice and resuspended in 500 μl of buffer (PBS + 1% FCS) before staining with 10 μl of PI staining solution for 10 min. The number of PI-staining (dead) cells for 10,000 flow-sorted cells was determined.
To measure chromosome lag at anaphase, patient-transformed fibroblasts containing the pseudodicentric-neocentric chromosome 4 (PD-NC4) were arrested at anaphase by treatment overnight with nocodazole, a benzimidazole derivative that binds to tubulin dimers and inhibits microtubule assembly. After anaphase arrest, the cells were treated with cytochalasin B to prevent cytokinesis, suspending cells in anaphase or telophase. Chromosome segregation was monitored by using FISH to identify chromosome 4, comparing with other chromosomes visualized by 4′,6-diamidino-2-phenylindole staining. A total of 200 pairs of daughter cells were scored in anaphase or telophase. The rate of chromosome lagging was calculated as a percentage per chromosome per cell division.
Pulse-Field Gel Electrophoresis (PFGE). Agarose plugs of high molecular weight DNA were obtained from transformed lymphocytes of patient III:1 and digested overnight using restriction enzymes AccI, ApaI, BglII, SacI, or PvuII. The digested plugs were subjected to PFGE on 1% agarose/0.5× TBE gels by using a Chef Mapper (Bio-Rad). After the completion of PFGE, the gels were transferred to nylon membranes and Southern hybridization was performed by using standard methodology. α-Satellite sequences were detected by using the 32P-labeled Y chromosome probe pLAY5.5 (12), and fragment size was estimated by comparison with yeast chromosome pulse-field gel markers (New England Biolabs).
Trichostatin A Treatment. Cells (1 × 106) were seeded into small flasks with trichostatin A (TSA) (Wako Pure Chemical, Osaka) at concentrations of between 12.5 and 2,000 nmol. The medium was replaced 48 h after treatment. Cell samples for analysis by immunofluorescence and FISH were collected 0 and 5 days after removal of TSA.
Results
Identification of a Pseudodicentric Neocentric Human Chromosomal Variant. An unusual PD-NC4 was identified in three members of a family (Fig. 1A, II:1, III:1, and III:2). The family was ascertained after a routine karyotype was performed on a 7-year-old girl (III:2) after a diagnosis of mild cognitive impairment. Her karyotype was 46,XX psu dic (4)(q21.3;p10), with every cell examined containing one chromosome 4 with an abnormal centromeric constriction (attributable to the formation of a neocentromere; see below) in the interstitial long arm at q21.3 (Fig. 1B). We have designated this chromosome PD-NC4. Individual III:2 was otherwise healthy. The same PD-NC4 was subsequently detected in her 9-year-old brother (III:1) and 41-year-old father (II:1), both of whom were healthy and of normal intellect. The grandmother of the proband (I:2) had a normal karyotype (46,XX), but the grandfather (I:1) was not available for study. Linkage analysis performed on individuals I:2, II:1, II:2, III:1, and III:2 using 45 microsatellite markers at a 5-centimorgan interval along the length of chromosome 4 demonstrated the origin of PD-NC4 to be individual I:1, but it cannot be determined whether the 4q21.3 neocentromere was present in this individual or was a de novo meiotic event. The analysis also did not reveal evidence of a chromosome deletion along the length of chromosome 4, with two alleles being present at all informative loci including near the sites of the original centromere and the neocentromere (data not shown). There also was no evidence of meiotic recombination in the vicinity of either centromeric locus on the PD-NC4 (Fig. 1 A).
Fig. 1.
(A) Pedigree of family carrying PD-NC4, first detected in III:2. Haplotype bars derived from the microsatellite linkage screen indicate that the allele containing the neocentromere (black bar) was derived from the paternal grandfather (I:1) of the proband. (B) The PD-NC4 (Left) and normal chromosome 4 (Right) from G-banded karyotype of individual III:2. The primary constriction has relocated to 4q21.3 without any change in banding pattern. (C) BAC probes used to characterize the PD-NC4 neocentromere (NC) (Left) and alphoid centromere (Right). The neocentromere domain was defined by combined immunofluorescence and FISH to a region of 1.5 Mb bordered by BACs RP11–209g6 and RP11–204i22 (see also Fig. 2D). All 12 BACs flanking the alphoid centromere hybridized with equal intensity to the PD-NC4 and the normal chromosome 4, excluding a deletion of these regions.
Lymphoblast cell lines were established from individuals I:2, II:1, II:2, III:1, and III:2, and a fibroblast cell line was established from individual III:2. FISH using a chromosome 4-specific α-satellite probe (p4n1/4) at high stringency and a pan-α-satellite probe (pTRA7) under low-stringency conditions confirmed the absence of any α-satellite DNA at the neocentromere in individuals II:2, III:1, and III:2 and retention of a strong α-satellite signal at the original centromere position (4p10) (Fig. 2 A and B). Centromere-associated proteins CENP-A, CENP-C, CENP-E, CENP-I, and BUB1 all localized to the neocentromere but not the 4p10 α-satellite region, confirming the relocation of centromeric function to 4q21.3 (Fig. 2 B and C; and Fig. 5, which is published as supporting information on the PNAS web site). CENP-B, on the other hand, was retained at the 4p10 α-satellite and was not evident at the neocentromere site (Figs. 2C and 5), consistent with the known specificity of CENP-B for CENP-B-box-containing α-satellite irrespective of its centromere activity status (13). Heterochromatin protein HP1 was observed to bind to both the active and inactive centromeres (Fig. 2C), in keeping with previous observations in human pseudodicentric chromosomes (9, 14).
Fig. 2.
(A) FISH with chromosome 4-specific α-satellite probe (p4n1/4) showing hybridization to the centromere of the normal chromosome 4 (Left) and the inactive centromere (solid arrow) but not the neocentromere (open arrow) of PD-NC4 (Right). (B) FISH with pan-α-satellite probe (pTRA7) showing hybridization to the inactive centromere (solid arrow) of PD-NC4 and the centromere of all other chromosomes but not the PD-NC4 neocentromere (open arrow). (Inset) Combined immunofluorescence and FISH (immuno-FISH) on PD-NC4 using anti-CENP-A antibody (red) and FISH wish pTRA7 (green). (C) Immunofluorescence using anti-CENP-A (green) and anti-CENP-B (red). The CENP-A signal is reduced in strength compared with other centromeres (see also Fig. 3B). (Inset) Immunofluorescence on PD-NC4 using anti-HP1α antibody (red) followed by FISH using a BAC probe from the neocentromere site to identify PD-NC4 (picture not shown), showing the presence of HP1α at both the inactive alphoid centromere and the neocentromere. (D) Localization of the neocentromere site using BAC probes (see Fig. 1C; green) and anticentromere antibody (CREST6, red), showing colocalization with BAC RP11-113g13 and RP11-458j15 (not shown) bounded by BACs RP11-204i22 and RP11-209g6. A p-telomeric BAC, RP11-661f1, was used for orientation.
To further define the location of the neocentromere, we performed FISH on lymphoblasts from individual III:2 by using 11 BACs from 4q21.2–4q22.1 (Fig. 1C) followed by immunofluorescence using anticentromere antibody (CREST6). The CREST signal was observed to colocalize with BACs 113g13 and 458j15 and to be on the proximal q-arm and p-arm sides of BACs 209g6 and 204i22, respectively (Fig. 2D). Thus, a conservative estimate places the neocentromere in a region of 1.5 megabases (Mb) bounded by BACs 209g6 and 204i22. Bioinformatic analysis revealed that this region contains 11 known and 4 predicted genes and is enriched in AT content (61.2%), consistent with findings in other neocentromeres (15–17).
PD-NC4 Is Mitotically Stable and Nondeleterious to Cell Viability. Analysis of 100 cells from transformed lymphoblast lines from individuals II:1, III:1, and III:2 and a fibroblast sample from III:2 identified PD-NC4 in all of the cells. Similarly, analysis of 100 transformed lymphoblasts from these individuals after ≥50 passages in culture detected no loss of PD-NC4, which suggests that PD-NC4 is 100% stable, both in vivo and in long-term culture. In addition, no cells were observed in which the primary constriction on PD-NC4 had reverted to its original α-satellite location, indicating that the centromere repositioning is stable.
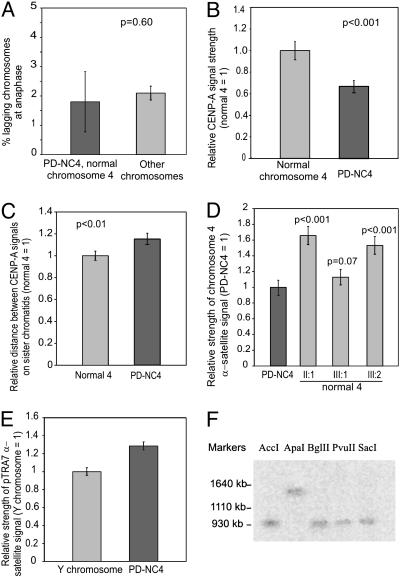
Mitotic stability was examined further by using nocodazole to arrest cells in anaphase and determine the prevalence of lagging. From 200 cell divisions, the anaphase-lag rate for PD-NC4 and chromosome 4 as a group was calculated as 1.8% per chromosome per cell division, which is not significantly different from the 2.1% for the collective anaphase-lag rate for all other chromosomes (P = 0.60) (Fig. 3A), providing evidence that the PD-NC4 kinetochore functions comparably with the conventional centromeres.
Fig. 3.
(A) Histogram comparing the rate of lagging for combined PD-NC4 and chromosome 4 at anaphase after nocodazole treatment with that for all other chromosomes. (B) Intensity of CENP-A signal on PD-NC4 (normalized to 1) as detected by immunofluorescence compared with that of the normal chromosome 4. (C) Distance between CENP-A signals on sister chromatids for PD-NC4 and normal chromosome 4 (normalized to 1). (D) Intensity of chromosome 4-specific α-satellite signal (p4n1/4) on PD-NC4 (normalized to 1) and normal chromosome 4 in each of the three PD-NC4 carriers. Variation between different chromosomes 4 represents normal polymorphic variation between homologous centromeres. (E) Relative intensity of pan-α-satellite signal (pTRA7) on PD-NC4 (normalized to 1) and the Y chromosomes in the two male individuals carrying the PD-NC4. (F) Sizing of Y-chromosome α-satellite DNA in individual III:1 using pTRA7 after PFGE. The α-satellite generates a single band of 950 kb when digested with enzymes AccI, BglII, PvuII, and SacI.
Cells viability was assayed by using flow cytometry after propidium iodide staining. The result for the three PD-NC4-containing patient cell lines (mean, 93.9%; range, 91.1–96.0%) was not significantly different compared with six control cell lines with no known cytogenetic abnormalities (mean, 93.2%; range, 91.6–96.4%), further indicating the absence of any detrimental effect of PD-NC4 on cell viability.
Reduced CENP-A Binding and Sister Cohesion at the Neocentromere. We compared the quantity of CENP-A at the PD-NC4 neocentromere with the normal chromosome 4 centromere by quantitative immunofluorescence using anti-CENP-A antibody on a lymphoblast cell line. Based on 40 metaphase spreads, the CENP-A signal at the neocentromere was significantly reduced, on average, to 67% of the normal chromosome 4 centromeric signal (P < 0.001) (Figs. 2C and 3B), a result that agrees with those we have obtained for a number of other neocentromeres (D. Irvine, D.J.A., R. Saffery, and K.H.A.C., unpublished data) and suggests that, despite its apparently normal centromeric function, the size of the neocentromere kinetochore is reduced.
Using the same 40 metaphases, we measured the relative distance between the paired CENP-A signals on sister chromatids at the PD-NC4 neocentromere. This result was found to be, on average, 15% greater than that of the normal chromosome 4 centromere (P < 0.01) (Figs. 2C and 3C), suggesting a reduction in sister-chromatid cohesion at the neocentromere. Cohesin is recruited to centromeric regions by heterochromatin, which itself may be present in minimal quantity at the neocentromere because of the absence of repetitive DNA, indicating a possible pathway to reduced cohesion. Alternatively, our observation may result directly from an altered chromatin conformation at the neocentromere.
No Detectable Pericentric Deletion at the Inactive Centromere. Twelve different BAC clones immediately flanking the proximal p- and q-arm regions of the chromosome 4 centromere were used in FISH to investigate the structural integrity of the chromosomal regions bordering the inactive centromere (Fig. 1C). All clones hybridized with equal intensity to the corresponding regions of the normal chromosome 4 and the PD-NC4, suggesting the intactness of these regions. The most proximal q-arm BAC (RP11-98b6) extends into the α-satellite DNA, ruling out a deletion of α-satellite from the q-arm DNA. The most proximal available p-arm BAC (RP11-1122f17) does not extend into the α-satellite DNA, and therefore the possibility that a deletion involving α-satellite and proximal p-arm DNA cannot be ruled out.
The Inactive Centromere Contains 1.3 Mb of α-Satellite. Quantitative FISH was performed with an α-satellite probe specific to chromosome 4 (p4n1/4) on lymphocytes from all three individuals carrying the PD-NC4. In each case, the fluorescence signal on the normal chromosome 4 was greater than that of the PD-NC4: by 66% in individual II:1 (P < 0.001); 13% in individual III:1 (P = 0.07); and 53% in individual III:2 (P < 0.001) (Fig. 3D). These results indicate that the α-satellite array on the PD-NC4 is slightly to moderately reduced compared with those of the normal chromosome 4.
The presence of a normal chromosome 4 in the patient cell lines made direct measurement of the size of the α-satellite DNA domain on PD-NC4 problematic. To overcome this problem, we used a two-step strategy involving comparing the α-satellite DNA on PD-NC4 with that of the Y chromosome (in the two male individuals II:1 and III:1) by using quantitative FISH, followed by PFGE to directly measure the size of the chromosome Y α-satellite DNA. For quantitative FISH, we used a pan-α-satellite probe pTRA7 (18), which under low-stringency conditions has been shown to hybridize to all α-satellite sequences (19). The combined results for the two individuals (who both carry the same Y chromosome) showed that the α-satellite signal on PD-NC4 was 29% greater than that of the Y chromosome (Fig. 3E). For PFGE, DNA from III:1 was digested with restriction enzymes that do not cut within the Y α-satellite DNA. Fragments of ≈950 kb were obtained by using AccI, BglII, PvuII, and SacI and of 1,350 kb by using ApaI (Fig. 3F), consistent with the Y α-satellite DNA in III:1 being ≈950 kb in size (20). This result suggests that the α-satellite domain on PD-NC4 is ≈1.3 Mb in size.
The Inactive Centromere Is Resilient to TSA Activation. The histone deacetylase inhibitor TSA was shown previously to induce centromere activity in α-satellite DNA that has been transfected into human cells and integrated at a noncentromeric site, resulting in functionally dicentric chromosomes and breakage of these chromosomes to form minichromosomes (21). We therefore treated a PD-NC4-containing lymphocyte cell line with TSA to determine whether the inactive alphoid centromere can be induced to reassemble a kinetochore. The results of Western blotting indicated that the level of histone H4 acetylation of histone H4 was increased by 62%, 166%, 448%, and 1,119% after TSA treatment for 48 h at 12.5, 25, 50 and 100 nmol, respectively. Analysis of chromosomes from up to 100 metaphases by immunofluorescence and FISH 0 and 5 days after removal of TSA showed no evidence of assembly of CENP-A at the inactive centromere or of minichromosome formation at any of the TSA concentrates tested, suggesting that the inactive centromere is resilient to TSA induction.
Discussion
Example of a PD-NC on a Human Autosome. Neocentromeres identified to date have typically occurred in association with chromosomal rearrangement, karyotypic imbalance, and/or mosaicism, which result in phenotypic abnormalities (7). We report here a family in which centromere activity on a chromosome 4 has relocated from its usual alphoid domain to an interstitial euchromatic site at 4q21.3. The result is a pseudodicentric chromosome, designated PD-NC4, containing an active neocentromere and an inactive alphoid centromere. This is an example of a neocentromere forming at an euchromatic site in the absence of any detectable chromosomal rearrangement and in conjunction with the inactivation and retention of what must have been a once-active alphoid centromere. The PD-NC4 is unlikely to be of phenotypic consequence, because cognitive impairment was observed in only one of three affected family members. Moreover, the absence of an aberrant phenotype relating to neocentromere formation at 4q21.3 can be explained by the finding of our previous study on a 10q25 neocentromere showing unaltered transcriptional competence of underlying genes at the neocentromeric chromatin (22).
Three examples of PD-NC have been described for the human Y chromosome (23–25). However, our PD-NC4 case differs from these PD-NCY cases in a number of significant ways. First, the PD-NC4 is 100% stable in mitosis, whereas mitotic instability of the three PD-NCY chromosomes was evidenced by their detection in 60–99% (23), 5% (24), and 84–95% (25) of cells examined. Second, PD-NCY formation seems to involve chromosomal rearrangement in the form of inversion or partial deletion of α-satellite DNA. Third, clinical phenotype is observed in some PD-NCY individuals, such as partial Turner syndrome (24) and short stature and cryptorchidism (23). Fourth, the PD-NCY neocentromeres all are located within the Yq heterochromatin containing an abundance of tandem repeat sequences, a feature that is known to support neocentromere formation (26, 27) but provides a more limited evolutionary scope for centromere repositioning and karyotype evolution (see below).
Mechanism of Origin of the PD-NC. The emergence of a PD-NC must involve two events, the loss of function at the alphoid centromere and the assembly of a kinetochore at the neocentromere site, in an undetermined order (see Fig. 4 and discussion below). Extensive studies using FISH and microsatellite markers suggest that centromere repositioning in the PD-NC4 has occurred in the absence of any detectable chromosomal deletion or rearrangement. Quantitative-FISH studies indicate a slight to moderate reduction in α-satellite on the PD-NC4 compared with the normal chromosome 4. Unfortunately the nonavailability of the progenitor chromosome 4 for investigation means that it is not possible to determine whether this reduction represents a deletion-linked inactivation of the PD-NC4 centromere or reflects the expected polymorphic heterogeneity of α-satellite array lengths among homologous chromosomes (19).
Fig. 4.
Models for the generation of PD-NC. (A) The formation of the PD-NC may result from a deletion of the kinetochore domain of the alphoid DNA. If the remaining α-satellite is not capable of assembling a new kinetochore (see text), the epigenetic formation of a neocentromere would enable the rescue of the otherwise acentric chromosome. Alternatively, neocentromere formation could be the initiating event, creating a functional dicentric-neocentric chromosome, with the alphoid centromere concurrently or subsequently being inactivated by a deletion. (B) Inactivation of the alphoid centromere and the formation of the neocentromere both occur solely as an epigenetic event, without sequence alteration in either the alphoid DNA or in the neocentromere DNA. As with that shown in A, the formation of the neocentromere might be the initiating event or occur concurrently with or after inactivation of the alphoid centromere.
Two possible models may be proposed for the origin of a PD-NC. The first model stipulates that inactivation of the alphoid centromere indeed results from the partial deletion of α-satellite DNA (Fig. 4A). The role of DNA sequence in centromere formation remains unclear, because no specific DNA sequence seems sufficient or necessary for centromere formation. In human cells, regular arrays of canonical α-satellite repeats are a preferred substrate for kinetochore assembly (28, 29). These arrays vary in size from <200 kb to >4 Mb without evidence of any impairment of centromeric function (19); however, studies of artificially created minichromosomes and naturally occurring deleted chromosomes suggest that centromeric function depends on a relatively small subdomain of α-satellite (30–32). Considerable evidence also suggests that not all α-satellite sequences are functionally equivalent. For example, the highly homogenized sequences and those containing CENP-B-box motif provide suitable substrates for kinetochore assembly but not the more diverged monomeric or oligomeric α-satellite units and those not containing CENP-B-boxes (29, 31, 33). Thus, any fortuitous deletion of a functionally essential minimal α-satellite domain, irrespective of the retention of other functionally inapt α-satellite sequences, would result in the temporary or permanent impairment of centromere function.
Alternatively, the nondeleted α-satellite DNA may be prevented from acquiring centromere function by epigenetic factors. In this explanation, the residual alphoid DNA would be capable of supporting centromere function under different circumstances (such as when transfected into cultured cells), but in vivo, such function is kept in check by epigenetic factors such as chromatin packaging that prevent the incorporation of CENP-A at this site (34). Studies in Drosophila have indicated that epigenetic factors may spread from a functional centromere to predispose centromere formation (35, 36); however, the range of such spreading is unclear and could fall within the deleted region in our model (Fig. 4A).
The second model of PD-NC formation stipulates that inactivation of the alphoid centromere results entirely from an epigenetic event, without any deletion or alteration to the DNA sequence of the alphoid centromere (Fig. 4B). In this model, inactivation of the alphoid centromere may result from the loss or gain of a hypothetical epigenetically controlled centromere activity-promoting or -suppressing factor.
Centromere Repositioning and Karyotype Evolution. Regardless of the mechanism of origin of a PD-NC, its successful meiotic transmission without obvious adverse karyotypic and phenotypic effect indicates that centromere repositioning via simultaneous centromere inactivation and neocentromere formation provides an inherent mechanism for karyotype evolution and speciation. The establishment of a new species is often accompanied by changes in chromosome morphology or number, with the probability of a given chromosome change becoming fixed within a population being influenced by factors such as genetic drift, selection in favor of individuals who carry the new variant, inbreeding, and meiotic drive (37). Although selection in favor of individuals carrying a neocentromere seems unlikely, given that most reported neocentromeres are detrimental to the individual (7), meiotic drive offers the greatest potential to establish a new neocentric chromosomal variant (38). Meiotic drive is the process whereby one of the four chromosomes in the meiotic tetrad is favored for transmission to the next generation as a consequence of the mechanics of meiotic divisions (39). In human female meiosis, any bias in the segregation of homologous chromosomes between the oocyte and the polar body would create such an opportunity, as long as a functional heterozygosity exists at the locus (centromere or neocentromere) that mediates attachment of the chromosome to the spindle (40). In fact, such an example has been described for nonhomologous Robertsonian translocations (which are typically functional dicentrics or pseudodicentrics), in which the segregation ratio in humans is distorted in favor of the translocation chromosome at female meiosis (41). Other examples demonstrating the specific role of neocentromeres in generating meiotic drive also have been reported in plant (see below).
One mechanism whereby a difference in DNA sequence at the centromere locus might lead to functional heterozygosity at the kinetochore is if it results in a quantitative change in CENP-A deposition (42). We have demonstrated that the neocentromere of the PD-NC4 chromosome binds less CENP-A than its homologous normal chromosome 4. Intuitively one might expect this arrangement to place the PD-NC4 at an evolutionary disadvantage, but that would be to (incorrectly) assume that a “stronger” centromere would preferentially segregate toward the oocyte side of the spindle rather than toward the polar body. In fact, in humans, the most efficient spindle is on the polar-body side of the meiotic division (40), and therefore a centromere or neocentromere binding less CENP-A might be selected preferentially at meiosis and transmitted to offspring.
The likelihood of any human PD-NC chromosome reaching fixation may be extremely slim; nonetheless, evidence from this study explicates this theoretical possibility. Such a possibility is expected to increase in lower organisms, in which the factors influencing fixation of a chromosomal change are likely to operate much more fervently. The extent of the neocentromere phenomenon in lower organisms remains unknown because of the fact that these organisms are infrequently karyotyped. In addition to humans, neocentromere formation has been reported in Drosophila (26, 35, 36) and a number of plant species (43). The most extensively studied neocentromeres in plants are “maize knobs,” which have been shown to act as facultative centromeres at meiosis and provide meiotic drive advantage (43, 44).
Studies in primates have indicated that the evolutionary repositioning of a centromere within a particular chromosome might have occurred via neocentromere formation (5). Evidence for this comes from the observation that the centromere of the X chromosome occupies different locations in three different primate species without any change to the order of DNA markers, suggesting that centromere repositioning has occurred by the emergence of a neocentromere rather than by the relocation of normal centromeric DNA from a different site. According to this hypothesis, a neocentromere, such as that described in this study, might over evolutionary time acquire α-satellite DNA from other chromosomes, thereby evolving into a conventional centromere and eliminating evidence of its neocentromeric origins. A possible intermediate stage in this progression has been described recently in rice, in which the centromere of chromosome 8 is composed predominantly of unique-sequence DNA but contains a rudimentary amount of rice centromeric satellite DNA (CentO) (45). The PD-NC4 chromosome may be the earliest stage in the progression from neocentromere to mature centromere, representing a living human example of karyotype evolution “in progress.”
Supplementary Material
Acknowledgments
We thank Tim Yen for anti-CENP-I antibody. This work was supported by the National Health and Medical Research Council and National Institute of General Medical Sciences/National Institutes of Health.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BAC, bacterial artificial chromosome; FISH, fluorescence in situ hybridization; PD-NC, pseudodicentric-neocentric chromosome; PFGE, pulse-field gel electrophoresis; TSA, trichostatin A; Mb, megabase(s).
References
- 1.Warburton, P. E., Haaf, T., Gosden, J., Lawson, D. & Willard, H. F. (1996) Genomics 33, 220-228. [DOI] [PubMed] [Google Scholar]
- 2.Haaf, T. & Willard, H. F. (1997) Chromosoma 106, 226-232. [DOI] [PubMed] [Google Scholar]
- 3.Charlesworth, B., Sniegowski, P. & Stephan, W. (1994) Nature 371, 215-220. [DOI] [PubMed] [Google Scholar]
- 4.Montefalcone, G., Tempesta, S., Rocchi, M. & Archidiacono, N. (1999) Genome Res. 9, 1184-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ventura, M., Archidiacono, N. & Rocchi, M. (2001) Genome Res. 11, 595-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.du Sart, D., Cancilla, M. R., Earle, E., Mao, J. I., Saffery, R., Tainton, K. M. A., Kalitsis, P., Martyn, J., Barry, A. E. & Choo, K. H. A. (1997) Nat. Genet. 16, 144-153. [DOI] [PubMed] [Google Scholar]
- 7.Amor, D. J. & Choo, K. H. A. (2002) Am. J. Hum. Genet. 71, 695-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan, B. A. & Warburton, P. E. (1999) in Chromosome Structural Analysis: A Practical Approach, ed. Bickmore, W. A. (Oxford Univ. Press, Oxford), pp. 81-101.
- 9.Craig, J. M., Earle, E., Canham, P., Wong, L. H., Anderson, M. & Choo, K. H. A. (2003) Hum. Mol. Genet. 12, 3109-3121. [DOI] [PubMed] [Google Scholar]
- 10.Liu, S. T., Hittle, J. C., Jablonski, S. A., Campbell, M. S., Yoda, K. & Yen, T. J. (2003) Nat. Cell Biol. 5, 341-345. [DOI] [PubMed] [Google Scholar]
- 11.Saffery, R., Irvine, D. V., Griffiths, B., Kalitsis, P., Wordeman, L. & Choo, K. H. A. (2000) Hum. Mol. Genet. 9, 175-185. [DOI] [PubMed] [Google Scholar]
- 12.Archidiacono, N., Antonacci, R., Marzella, R., Finelli, P., Lonoce, A. & Rocchi, M. (1995) Genomics 25, 477-484. [DOI] [PubMed] [Google Scholar]
- 13.Earnshaw, W. C., Ratrie, H., 3rd, & Stetten, G. (1989) Chromosoma 98, 1-12. [DOI] [PubMed] [Google Scholar]
- 14.Vagnarelli, P. B. & Earnshaw, W. C. (2001) Chromosoma 110, 393-401. [DOI] [PubMed] [Google Scholar]
- 15.Alonso, A., Mahmood, R., Li, S., Cheung, F., Yoda, K. & Warburton, P. E. (2003) Hum. Mol. Genet. 12, 2711-2721. [DOI] [PubMed] [Google Scholar]
- 16.Lo, A. W., Craig, J. M., Saffery, R., Kalitsis, P., Irvine, D. V., Earle, E., Magliano, D. J. & Choo, K. H. A. (2001) EMBO J. 20, 2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo, A. W., Magliano, D. J., Sibson, M. C., Kalitsis, P., Craig, J. M. & Choo, K. H. A. (2001) Genome Res. 11, 448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vissel, B. & Choo, K. H. (1991) Nucleic Acids Res. 19, 271-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo, A. W., Liao, G. C., Rocchi, M. & Choo, K. H. A. (1999) Genome Res. 9, 895-908. [DOI] [PubMed] [Google Scholar]
- 20.Oakey, R. & Tyler-Smith, C. (1990) Genomics 7, 325-330. [DOI] [PubMed] [Google Scholar]
- 21.Nakano, M., Okamoto, Y., Ohzeki, J. I. & Masumoto, H. (2003) J. Cell Sci. 116, 4021-4034. [DOI] [PubMed] [Google Scholar]
- 22.Saffery, R., Sumer, H., Hassan, S., Wong, L. H., Craig, J. M., Todokoro, K., Anderson, M., Stafford, A. & Choo, K. H. A. (2003) Mol. Cell 12, 509-516. [DOI] [PubMed] [Google Scholar]
- 23.Rivera, H., Vasquez, A. I., Ayala-Madrigal, M. L., Ramirez-Duenas, M. L. & Davalos, M. P. (1996) Ann. Genet. 39, 236-239. [PubMed] [Google Scholar]
- 24.Bukvic, N., Susca, F., Gentile, M., Tangari, E., Ianniruberto, A. & Guanti, G. (1996) Hum. Genet. 97, 453-456. [DOI] [PubMed] [Google Scholar]
- 25.Tyler-Smith, C., Gimelli, G., Giglio, S., Floridia, G., Pandya, A., Terzoli, G., Warburton, P. E., Earnshaw, W. C. & Zuffardi, O. (1999) Am. J. Hum. Genet. 64, 1440-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Platero, J. S., Ahmad, K. & Henikoff, S. (1999) Mol. Cell 4, 995-1004. [DOI] [PubMed] [Google Scholar]
- 27.Choo, K. H. A. (2001) Dev. Cell 1, 165-177. [DOI] [PubMed] [Google Scholar]
- 28.Harrington, J. J., Van Bokkelen, G., Mays, R. W., Gustashaw, K. & Willard, H. F. (1997) Nat. Genet. 15, 345-355. [DOI] [PubMed] [Google Scholar]
- 29.Ikeno, M., Grimes, B., Okazaki, T., Nakano, M., Saitoh, K., Hoshino, H., McGill, N. I., Cooke, H. & Masumoto, H. (1998) Nat. Biotechnol. 16, 431-439. [DOI] [PubMed] [Google Scholar]
- 30.Floridia, G., Zatterale, A., Zuffardi, O. & Tyler-Smith, C. (2000) EMBO Rep. 1, 489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schueler, M. G., Higgins, A. W., Rudd, M. K., Gustashaw, K. & Willard, H. F. (2001) Science 294, 109-115. [DOI] [PubMed] [Google Scholar]
- 32.Spence, J. M., Critcher, R., Ebersole, T. A., Valdivia, M. M., Earnshaw, W. C., Fukagawa, T. & Farr, C. J. (2002) EMBO J. 21, 5269-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohzeki, J., Nakano, M., Okada, T. & Masumoto, H. (2002) J. Cell Biol. 159, 765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmad, K. & Henikoff, S. (2002) Proc. Natl. Acad. Sci. USA 99, Suppl. 4, 16477-16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams, B. C., Murphy, T. D., Goldberg, M. L. & Karpen, G. H. (1998) Nat. Genet. 18, 30-37. [DOI] [PubMed] [Google Scholar]
- 36.Maggert, K. A. & Karpen, G. H. (2001) Genetics 158, 1615-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White, M. J. D. (1978) in Modes of Speciation (Freeman, San Francisco).
- 38.Hedrick, P. W. (1981) Evolution (Lawrence, Kans.) 35, 322-332. [Google Scholar]
- 39.Sandler, L. & Novitski, E. (1957) Am. Nat. 91, 105-110. [Google Scholar]
- 40.Pardo-Manuel de Villena, F. & Sapienza, C. (2001) Genetics 159, 1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pardo-Manuel de Villena, F. & Sapienza, C. (2001) Hum. Genet. 108, 31-36. [DOI] [PubMed] [Google Scholar]
- 42.Henikoff, S., Ahmad, K. & Malik, H. S. (2001) Science 293, 1098-1102. [DOI] [PubMed] [Google Scholar]
- 43.Hiatt, E. N., Kentner, E. K. & Dawe, R. K. (2002) Plant Cell 14, 407-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhoades, M. M. & Vilkomerson, H. (1942) Proc. Natl. Acad. Sci. USA 28, 433-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P. B., Kim, M., Jones, K. M., Henikoff, S., Buell, C. R. & Jiang, J. (2004) Nat. Genet. 36, 138-145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.