Abstract
UV light induces the expression of a wide variety of genes. At present, it is unclear how cells sense the extent of DNA damage and alter the expression of UV-induced genes appropriately. UV light induces DNA damage that blocks transcription, and the probability that a gene sustains transcription-blocking DNA damage is proportional to locus size and dose of UV light. Using colon carcinoma cells that express a temperature-sensitive variant of p53 and undergo p53-dependent apoptosis after UV irradiation, we found that the number of p53-induced genes identified by oligonucleotide microarray analysis decreased in a UV dose-dependent manner. This was associated with a statistically significant shift in the spectrum of p53-induced genes toward compact genes with fewer and smaller introns. Genes encoding proapoptotic proteins involved in the initiation of the mitochondrial apoptotic cascade were prominent among the compact p53 target genes, whereas genes encoding negative regulators of p53 and the mitochondrial apoptotic pathway were significantly larger. We propose that the shift in spectrum of UV-responsive gene expression caused by passive effects of UV lesions on transcription acts as a molecular dosimeter, ensuring the elimination of cells sustaining irreparable transcription-blocking DNA damage.
Keywords: DNA damage, microarray, p53, transcription, stress responses
The human genome contains between 26,000 and 39,000 genes distributed throughout ≈3 billion base pairs of DNA (1). The coordinated regulation of this many genes presents a daunting task. Superimposed upon the complexity of transcriptional regulation is the fact that many DNA damaging agents can induce damage that inhibits elongation by RNA polymerase II (2). These transcription-blocking DNA lesions pose an obvious difficulty to the coordinated regulation of gene expression in those cells that have sustained sufficient damage. It is not fortuitous that highly conserved DNA repair pathways coupling transcription to DNA repair have evolved and that mechanisms exist to eliminate cells with excessive transcription-blocking lesions.
Transcription-coupled nucleotide excision repair (TC-NER) preferentially removes transcription-blocking lesions from the transcribed strand of active genes, facilitating the recovery from DNA damage-induced transcriptional arrest (3-5). Much of what is known about TC-NER and DNA damage-induced transcriptional stress stems from studies using the model DNA damaging agent, UV light. Cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4PPs), the most common lesions induced by short-wavelength UV light, block transcription and are substrates for TC-NER (6, 7). After exposure to cytotoxic doses of UV light, cells fail to recover transcription and undergo apoptosis (4, 8), and this is thought to occur primarily through the mitochondrial pathway involving cytochrome c, Apaf1, and caspase 9 (9-11). There is a very tight correlation between the sustained inhibition of transcription and UV-induced apoptosis, indicating that cells sense sustained transcriptional stress as an indicator of irreparable DNA damage (4, 8, 12, 13).
UV light activates signaling cascades that ultimately result in the activation of transcription-factors such as AP-1 and the p53 tumor suppressor (14, 15). However, UV-induced DNA damage also inhibits transcription by posing a physical impediment to elongation by RNA polymerases (2). The likelihood that a gene sustains DNA damage after UV irradiation is proportional to gene size because the induction of UV photoproducts is a stochastic event (16). A moderate dose of 10 J/m2 of UV light that induces apoptosis in <20% of UV-irradiated primary human fibroblasts or most other DNA repair-proficient human cell lines (4, 13, 17-20) yields ≈2.4 lesions per strand of an average gene (Fig. 4, which is published as supporting information on the PNAS web site) (1, 7, 21-25). This high lesion frequency led us to hypothesize that transcription-blocking UV lesions could dramatically effect the expression of UV-induced genes. Here we report that the spectrum of genes induced by UV light or by p53 in the presence of UV-induced DNA damage is significantly altered in a dose-dependent manner. The genes induced at cytotoxic doses of UV light were compact, with fewer and smaller introns, and were enriched for initiators of apoptosis acting through the mitochondrial pathway. We propose that cells use this passive mechanism of gene regulation to ensure that cells sustaining irreparable DNA damage are eliminated by apoptosis.
Materials and Methods
Cell Culture and UV Treatment. The HT29-tsp53 colorectal carcinoma cells expressing a temperature-sensitive variant of p53 and HT29-neo vector control cells (26) were obtained from Mats Ljungman and Jon Maybaum (University of Michigan, Ann Arbor). Cells were grown at the restrictive temperature (38°C) in 10-cm tissue culture dishes in DMEM supplemented with 10% FCS (GIBCO/BRL) and 5 μg/ml gentamicin (Sigma). Growth medium was removed before UV iradiation, and fresh medium was immediately replaced after treatment. Fluence was determined with a UV radiometer (Ultraviolet Products) to deliver ≈1 J/m2/s from a germicidal bulb (Philips) emitting predominately at 254 nm.
RNA Isolation. Cells were seeded 48 h before treatment. At the time of treatment, cells were irradiated with the indicated doses of UV light and were either returned immediately to the restrictive temperature (38°C) or switched to the permissive temperature (32°C). Cells were harvested 6 h after treatment. Total RNA was isolated by using the RNeasy RNA isolation kit (Qiagen, Valencia, CA) according to manufacturer's specifications. The yield of total cellular RNA was not dramatically affected by UV dose by this time.
Quantitative RT-PCR. Five micrograms of total RNA was reverse-transcribed by using first-strand cDNA synthesis kit (MBI Fermentas). Quantitative RT-PCR was performed by using the Sybr green fluorescent DNA stain (Molecular Probes), a LightCycler 2 quantitative PCR machine (Roche Diagnostics) and light-cycler software version 3 (Roche Diagnostics). The primers used were CRYAB (AGCCGCCTCTTTGACCAGTTCTT and GCGGTGACAGCAGGCTTCTCTTC), p21WAF-1/CDK-N1A (CCTCAAATCGTCCAGCGACCTT and CATTGTGGGAGGAGCTGTGAAA), FAS (CTCATCTTAATGGCCTAATGCA and GCTTCAGTTTATAACTATCTTCAC), DDB2 (CCACCTTCATCAAAGGGATTGG and CTCGGATCTCGCTCTTCTGGTC), serpinB5/maspin (CTTTTCTGTGGATGCCGATT and CCTGCCAGGGCTTAACATAA), XP-C (AAGTTCACTCGCCTCGGTTGC and TTCTTTCCTGATTTTAGCCTTTTT), SMAD-3 (AGGCGTGCGGCTCTACTACATC and GGGCGT T TCTGGT TGGACTG), EFNB1 (GGAGGCAGACAACACTGTCA and GAACAATGCCACCTTGGAGT), LOC254531 (CCGAATGTGAGTT TGTAGG and T T TGGTGTCTGGGAGGTG), MAX (CTCTCGTGGTATGTATGGG and AGTAGGA A AGGAAGTGGGATG), MYB (AACTCCTACACCATTCAAAC and CTGCTCCTCCATCTTTCC), PMAIP1/NOXA (TAAAGCAAGAATGGAAGAC and GACCGAAGAAATCAACAC), and ACTIN (GGGCATGGGTCAGAAGGAT and GTGGCCATCTCTTGCTCGA).
Microarray. Twenty micrograms of each RNA sample was labeled by using the Super Script II (Invitrogen) and the Enzo BioArray HighYield RNA transcript labeling kit (Affymetrix). Labeled complementary RNA was hybridized to the Affymetrix U95A microarrays containing 12558 probe sets according to the specifications of the manufacturer (Affymetrix) at the Affymetrix gene expression facility at the Ottawa Hospital Research Institute (Ottawa). Affymetrix microarray suite 5.0 software was used to compare each sample to mock-irradiated controls maintained at the restrictive temperature. A nonparametric Wilcoxon signed rank test was used to determine whether statistically meaningful differences in probe cell intensities were detected between samples (change calls were determined by using γ1H and γ1L values of 0.0025). We considered genes to be differentially expressed under a given condition if and only if they were statistically (P ≤ 0.0025) increased or decreased 2-fold in all experiments compared to samples collected from cells maintained at the restrictive temperature. The NetAffx database was used to determine the identity of differentially expressed genes (27).
The National Centre for Biotechnology Information Locuslink database (www.ncbi.nlm.nih.gov/LocusLink) was used to obtain detailed information regarding the physical makeup of all differentially expressed genes. The size of genes, mRNAs, and introns did not fit a Gaussian distribution, so statistical analysis was performed on log-transformed data by using the PRISM software package (GraphPad, San Diego). However, similar results were obtained with nontransformed data by using a nonparametric Mann-Whitney U test.
Chromatin Immunoprecipitation. The chromatin immunoprecipitation assay was performed as described by Kallesen and Rosen (http://public.bcm.tmc.edu/rosenlab/protocols/ChIP.pdf), which represents a minor modification of the method of Boyd and Farnham (28). Briefly, proteins and DNA were crosslinked with 1% formaldehyde in growth medium. Chromatin was isolated and sonicated to an average length of 700 bp with a microtip sonifier (Branson Sonifier) in 20-sec bursts. Debris was cleared by centrifugation, and the resulting supernatants were frozen in liquid nitrogen and stored at -80°C.
Upon analysis, the chromatin was diluted 5-fold and precleared with protein A-agarose (Roche Diagnostics) for 50 min at 4°C. Twenty percent of the precleared chromatin was retained as a positive control for PCR amplification. The remainder of the solution was divided into equal aliquots and incubated overnight at 4°C, either without antibody or with 5 μg of p53 antibody (Ab-1 antibody, Oncogene Science, or phospho-Ser-15 antibody, Cell Signaling Technologies). Immune complexes were collected overnight at 4°C with the addition of 60 μl of protein A-agarose (Roche Diagnostics).
Crosslinks were reversed by addition of NaCl to a final concentration of 0.3 M with RNase A and incubated at 65°C for 4-5 h. DNA was precipitated in 70% ethanol, collected by centrifugation, and proteinase K treated for 2 h at 45°C. DNA was purified by using QiaQuick spin columns (Qiagen) and was eluted in 10 mM Tris, pH 8.0. Semiquantitative PCRs were performed by using a GenAmp PCR System 9700 (Applied Biosystems). The binding of p53 to the serbinB5 (maspin), CDKN1A (p21WAF1), and FAS p53 response elements was determined by using the following primer pairs: CTTTTCTGTGGATGCCGATT and CCTGCCAGGGCTTAACATAA, GTGGCTCTGATTGGCTTTCTG and CTGAAAACAGGCAGCCCAAG (29), and GGGACCCCGGTTGGAGAG and CTGCTTCGGTGCTGACTTATTTC, respectively. Amplification of upstream sequences of the GAPDH (GTATTCCCCCAGGTTTACAT and TTCTGTCTTCCACTCACTCC; ref. 29), XAB2 (AACGAGCTGGGACCCTCAGT and TATCAGTTTTTGGGGGCCGAGT) and KiSS-1 (CCTGGGGCCCGCACTTAGC and CCCCCGCACCTTCTCCATTTG) promoters and enhancers served as negative controls.
Results and Discussion
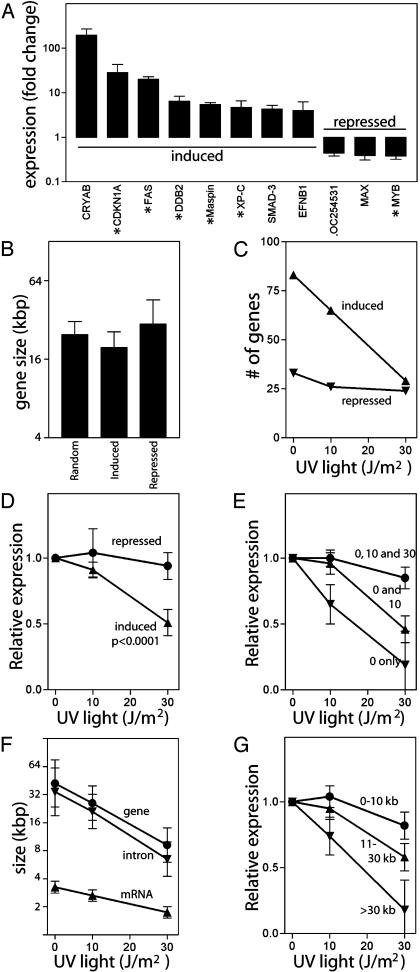
To evaluate the inhibitory affect of UV photoproducts on DNA damage-induced gene expression in a well controlled model system, we examined gene expression profiles in a human colorectal carcinoma cell line harboring a temperature sensitive allele of p53 (26). At the restrictive temperature (38°C), this variant of p53 is in a mutant conformation and cytoplasmic, but becomes nuclear and functional at the permissive temperature (32°C) (30). This variant is ideal for these studies because conditional expression of active p53 does not require de novo transcription of the transgene nor does it require a DNA damage signal. Statistically significant changes in gene expression were determined for each individual experiment by using the MICROARRAY SUITE 5.0 software (Affymetrix). We found 83 genes were induced at least 2-fold at the permissive temperature in each of four independent experiments. Thirty-three of these genes have previously been reported to be p53-induced (31-38). By similar criteria, 33 genes were repressed at the permissive temperature. The differential expression of several gene products was confirmed by using real-time RT-PCR (Fig. 1A). In all instances, RT-PCR data confirmed the differential expression detected by microarray analysis.
Fig. 1.
The effect of UV light on expression of genes induced at the permissive temperature. (A) Fold increase in gene expression at the permissive temperature was determined by real-time RT-PCR. Known p53-regulated genes are denoted with asterisks. (B) The mean size of genes induced or repressed at the permissive temperature compared to the size of randomly selected genes. The effect of the UV light on the number (C) and expression (D) of genes induced or repressed at the permissive temperature. Expression in D for each gene was determined and normalized to the induced level of expression of that gene at the permissive temperature alone and is expressed as the mean level of expression determined from all genes in the group. (E) Genes were grouped by behavior to UV light: induced at 0 (inverted triangle), 0 and 10 (triangle), or 0, 10, and 30 (circle) J/m2. Relative expression was determined as indicated in D. (F) The mean size of loci (circles), mean size of mRNA (triangles), and the total intron size (inverted triangles) are indicated for each group in E. (G) Genes were sorted into three groups by gene size (≤10 kb, 11-30 kb, and >30 kb, n = 28, 26, and 27, respectively) and the mean effect of UV light on gene expression in each group was determined. Error bars in A and D-G indicate the 95% confidence interval of the indicated mean. The P value in D was determined by one-way ANOVA.
We obtained information regarding the physical makeup of all differentially expressed genes (Tables 3-5, which are published as supporting information on the PNAS web site). For statistical purposes, detailed information regarding the physical makeup of 132 randomly selected genes was also obtained (Table 6, which is published as supporting information on the PNAS web site). As expected, the average size of the randomly selected genes closely approximated that reported by the human genome project (1). Overall, the mean size of the genes induced at the permissive temperature was not significantly different from the size of randomly selected genes (Fig. 1B).
We determined the effect of UV light on the spectrum and expression of these p53 target genes. Of the 83 genes induced at the permissive temperature, only 29 were still induced after exposure to 30 J/m2 (Fig. 1C). In contrast, the number of repressed genes was not dramatically altered by UV light (Fig. 1C). Whereas the expression of p53-induced genes was also strongly inhibited by UV light (P < 0.0001), the expression of the p53-repressed genes remained largely unaffected (Fig. 1D). Thus, UV irradiation dramatically altered the spectrum and expression of p53-induced but not p53-repressed genes.
Genes induced at the permissive temperature were grouped by behavior after exposure to UV light (i.e., induced at “0 J/m2 only,” “0 and 10 J/m2,” or “0, 10, and 30 J/m2”) (Fig. 1E). There was a significant UV dose-dependent decrease in the size of genes induced but not repressed at the permissive temperature (Fig. 1F, P < 0.0001, ANOVA). This was associated with a decrease in the number and average size of introns, but mRNA size was not significantly different from that of randomly selected genes (P = 0.005, 0.004, and 0.17, respectively). The p53-induced genes were sorted into groups based on gene size, and the effect of UV light on the expression of each group of genes was determined. The expression of large genes was strongly inhibited, whereas the expression of small genes was not (Fig. 1G). Because many of these putative p53-regulated genes identified through our microarray analysis have not been independently confirmed to be p53-regulated, we performed a similar analysis on the 43 previously reported target genes (31-40) (Table 5). Again, genes induced at higher doses of UV light were significantly smaller (Table 1), and this was associated with a significant decrease in the average number and size of introns (P < 0.005, t test). Thus, UV light inhibits the p53 response in a dose-dependent manner, resulting in the preferential induction of compact p53 target genes with fewer and smaller introns.
Table 1. Effect of UV light on the expression of known p53-responsive genes.
| Expression | UV, J/m2 | n | Median gene size, bp | mRNA size, log2 bp | Gene size, log2 bp | Gene size, bp† | Total intron size, log2 bp |
|---|---|---|---|---|---|---|---|
| Random‡ | NA | 132 | 29,104 | 11.1 ± 0.1 | 14.6 ± 0.2 | 24,800 | 14.4 ± 0.2 |
| p53 induced | All | 43 | 13,751 | 11.1 ± 0.1 | 14.0 ± 0.2 | 16,500 | 13.7 ± 0.3 |
| 0 | 33 | 23,036 | 11.1 ± 0.1 | 14.1 ± 0.3 | 17,600 | 13.8 ± 0.3 | |
| 10 | 32 | 11,934 | 11.1 ± 0.1 | 13.7 ± 0.3 * | 13,300 | 13.5 ± 0.3 * | |
| 30 | 18 | 8,176 | 10.8 ± 0.2 | 13.2 ± 0.3 ** | 9,400 | 12.8 ± 0.3 ** |
Asterisks indicate that the indicated value is significantly different from randomly selected genes using a Student's test at *, P ≤ 0.05 or **, P ≤ 0.005. NA, not applicable.
For clarity, mean gene size is presented in a linear form but was determined from the mean of transformed values by using the following formula: (mean gene size = 2meanlog2 gene size).
A random number generator was used to generate a random series of Locuslink accession numbers.
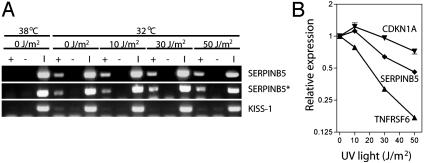
To ensure that p53 was associated with consensus response elements in vivo at high doses of UV light, we performed chromatin immunoprecipitation experiments. An interaction between p53 and the SERPINB5 (maspin, Fig. 2A), CDKN1A (p21WAF1, data not shown), and TNFRSF6 (Fas, data not shown) p53-response elements in vivo by chromatin immunoprecipitation was detected even though UV light inhibited the expression of SERPINB5 and TNFRSF6 (Fig. 2B). The interactions were apparent for all three promoters using two different p53 antibodies. We interpret these results to indicate that dose-dependent variation in the spectrum of p53-induced genes cannot be explained by a decrease in the binding of p53 to its response elements.
Fig. 2.
The effect of UV dose on p53 binding and p53 target gene expression. (A) Representative chromatin immunoprecipitation experiments demonstrate an interaction of p53 and ser15 phosphorylated p53 (asterisk) with the SERPINB5 p53-response element at all doses of UV light. KiSS-1 served as a negative control. +, antibody; -, the no-antibody control; I, input DNA. (B) The effect of UV light on representative p53 target genes was determined by real-time RT-PCR. Values are expressed relative to the corresponding mean determined for p53 target genes induced at the permissive temperature alone. Values represent the mean ± standard error.
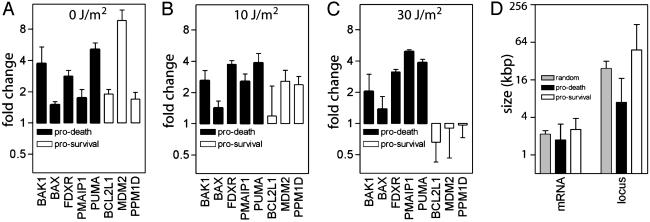
We have previously reported that the HT29-tsp53 cells used here undergo extensive apoptosis at the permissive temperature after exposure to 30 but not after exposure to 0 or 10 J/m2 of UV light (20). Of the 43 known p53 target genes differentially expressed in our microarray experiments, 17 have been reported to regulate the induction of apoptosis. Of these, five are inducers of apoptosis through the mitochondrial apoptotic pathway (Fig. 3) important for both p53- and UV-induced apoptosis (9-11, 32, 33, 41-45). All five genes implicated in the p53-mediated mitochondrial pathway were highly resistant to inhibition by UV light (Fig. 3 A-C), whereas the expression of genes known to inhibit p53-mediated apoptosis was strongly blocked by UV light (Fig. 3 A-C). The proapoptotic genes were significantly smaller than the prosurvival genes (Fig. 3D). The large size of MDM2 and PPM1D genes ensures that feedback inhibition is blocked at higher doses, permitting cells to be eliminated by apoptosis. Taken together, UV irradiation resulted in the expression of a restricted set of small p53-responsive genes at the expense of larger p53 targets, and this coincides with the induction of apoptosis in a dose-dependent manner.
Fig. 3.
p53 target genes involved in mitochondrial apoptotic pathways are resistant to inhibition of gene expression by UV light. (A-C) The fold change in the expression of genes encoding antiapoptotic (open symbols) and proapoptotic (filled symbols) proteins after exposure to 0 (A), 10 (B), or 30 (C)J/m2. (D) The mean size of the mRNA and locus of genes encoding p53-regulated proapoptotic proteins or survival promoting proteins (in A-C).
To determine whether gene size had an effect on the p53-independent induction of UV-responsive genes, we assessed the effect of UV light on gene expression at the restrictive temperature, as well. We found that the size of genes induced after exposure to high doses of UV light at the restrictive temperature were again smaller (P = 0.005 and P < 0.0001, respectively, Table 2). Furthermore, we determined the average size of genes reported to be induced by UVC, UVB, and γ radiation in a variety of cell types by several other groups (38, 46-49). In striking support of our results, we found that these previously reported UVB- and UVC-induced genes were also significantly smaller than randomly selected genes (Table 7, which is published as supporting information on the PNAS web site). The compact UV-induced genes included p53-responsive genes, AP-1 responsive immediate early genes, cytokines, chemokines, and histones (38, 46-49). In contrast, exposure of tumor cells to γ radiation did not result in the striking induction of compact genes (Table 7). In fact, UV-induced genes were significantly smaller than genes induced by ionizing radiation (0.05 < P < 0.0001, depending on the studies compared). This is consistent with the fact that γ radiation does not strongly inhibit transcription (8). Therefore, gene size is an important determinant of UV-induced but not γ-radiation-induced gene expression.
Table 2. Size of genes induced by UV light at the restrictive temperature.
| UV | n | Median | Gene size* | Mean† | P‡ |
|---|---|---|---|---|---|
| Random§ | 132 | 29,104 | 14.6 ± 0.2 | 24,800 | |
| 10 J/m2 | 14 | 10,340 | 13.1 ± 0.4 | 8,800 | 0.005 |
| 30 J/m2 | 17 | 5,822 | 12.6 ± 0.3 | 6,200 | <0.0001 |
The size of individual genes in bp were log2 transformed, and the mean (±SEM) of these log2 transformed values is presented.
For clarity, mean gene size is presented in a linear form, but was determined from the mean of transformed values by using the formula: mean gene size = 2mean gene size (log2).
Each mean was compared to the mean determined from randomly selected genes by using a t test.
Randomly selected genes.
In summary, we found that the induction of UV-induced gene expression is subject to a strong gene size constraint. The gamut of UV-induced genes was enriched for compact genes with fewer and smaller introns. We propose that this gene size constraint has played a significant role in the evolution of UV-response pathways and that the dose-dependent change in the spectrum of p53-induced and UV-induced genes acts as an elegant molecular dosimeter. Selective gene expression based on gene size represents a gene regulatory mechanism that acts at the level of transcript elongation because of blocked RNA polymerases. We propose that differences in gene expression caused by gene size are used to interpret the extent of irreparable transcription-blocking DNA damage sustained by the cell. This dose-dependent pattern of expression explains in large part the tight association between the UV-induced inhibition of transcription and the induction of apoptosis (4, 8, 12, 13, 20).
Supplementary Material
Acknowledgments
We thank Drs. Mats Ljungman and Jon Maybaum (University of Michigan) for providing the HT29-tsp53 cell line used in these studies and Dr. Douglas Gray for critical reading of the manuscript. We appreciate the technical assistance of the Ottawa Health Research Institute Gene Expression Facility. This work was supported by the National Cancer Institute of Canada with funds from the Terry Fox Run and the Ontario Premier's Research Excellence Award. B.C.M. is a Research Scientist of the Canadian Cancer Society.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 2.Tornaletti, S. & Hanawalt, P. (1999) Biochimie 81, 139-146. [DOI] [PubMed] [Google Scholar]
- 3.Mayne, L. & Lehmann, A. (1982) Cancer Res. 42, 1473-1478. [PubMed] [Google Scholar]
- 4.Ljungman, M. & Zhang, F. (1996) Oncogene 13, 823-831. [PubMed] [Google Scholar]
- 5.McKay, B. C., Chen, F., Clarke, S. T., Wiggin, H. E., Harvey, L. M. & Ljungman, M. (2001) Mutat. Res. 485, 93-105. [DOI] [PubMed] [Google Scholar]
- 6.Mellon, I., Spivak, G. & Hanawalt, P. C. (1987) Cell 51, 241-249. [DOI] [PubMed] [Google Scholar]
- 7.van Hoffen, A., Venema, J., Meschini, R., Vanzeeland, A. A. & Mullenders, L. H. F. (1995) EMBO J. 14, 360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ljungman, M., Zhang, F., Chen, F., Rainbow, A. J. & McKay, B. C. (1999) Oncogene 18, 583-592. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida, H., Kong, Y. Y., Yoshida, R., Elia, A. J., Hakem, A., Hakem, R., Penninger, J. M. & Mak, T. W. (1998) Cell 94, 739-750. [DOI] [PubMed] [Google Scholar]
- 10.Hakem, R., Hakem, A., Duncan, G. S., Henderson, J. T., Woo, M., Soengas, M. S., Elia, A., de la Pompa, J. L., Kagi, D., Khoo, W., et al. (1998) Cell 94, 339-352. [DOI] [PubMed] [Google Scholar]
- 11.Sitailo, L. A., Tibudan, S. S. & Denning, M. F. (2002) J. Biol. Chem. 277, 19346-19352. [DOI] [PubMed] [Google Scholar]
- 12.McKay, B. C., Becerril, C. & Ljungman, M. (2001) Oncogene 20, 6805-6808. [DOI] [PubMed] [Google Scholar]
- 13.McKay, B. C. & Ljungman, M. (1999) Neoplasia 1, 276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrlich, P., Blattner, C., Knebel, A., Bender, K. & Rahmsdorf, H. J. (1997) Biol. Chem. 378, 1217-1229. [DOI] [PubMed] [Google Scholar]
- 15.Ljungman, M. (2000) Neoplasia 2, 208-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauerbier, W. & Hercules, K. (1978) Annu. Rev. Genet. 12, 329-363. [DOI] [PubMed] [Google Scholar]
- 17.McKay, B. C., Ljungman, M. & Rainbow, A. J. (1998) Oncogene 17, 545-555. [DOI] [PubMed] [Google Scholar]
- 18.Queille, S., Drougard, C., Sarasin, A. & Daya-Grosjean, L. (2001) J. Invest. Dermatol. 117, 1162-1170. [DOI] [PubMed] [Google Scholar]
- 19.McKay, B. C., Becerril, C., Spronck, J. C. & Ljungman, M. (2002) DNA Rep. 1, 811-820. [DOI] [PubMed] [Google Scholar]
- 20.McKay, B. C., Chen, F., Perumalswami, C. R., Zhang, F. & Ljungman, M. (2000) Mol. Biol. Cell 11, 2543-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venema, J., Mullenders, L. H. F., Natarajan, A. T., van Zeeland, A. A. & Mayne, L. V. (1990) Proc. Natl. Acad. Sci. USA 87, 4707-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venema, J., van Hoffen, A., Natarajan, A. T., van Zeeland, A. A. & Mullenders, L. H. F. (1990) Nucleic Acids. Res. 18, 443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans, M. K., Robbins, J. H., Ganges, M. B., Tarone, R. E., Nairn, R. S. & Bohr, V. A. (1993) J. Biol. Chem. 268, 4839-4847. [PubMed] [Google Scholar]
- 24.Tolbert, D. M. & Kantor, G. J. (1996) Cancer Res. 56, 3324-3330. [PubMed] [Google Scholar]
- 25.Ford, J. M. & Hanawalt, P. C. (1997) J. Biol. Chem. 272, 28073-28080. [DOI] [PubMed] [Google Scholar]
- 26.Merchant, A. K., Loney, T. L. & Maybaum, J. (1996) Oncogene 13, 2631-2637. [PubMed] [Google Scholar]
- 27.Liu, G., Loraine, A. E., Shigeta, R., Cline, M., Cheng, J., Valmeekam, V., Sun, S., Kulp, D. & Siani-Rose, M. A. (2003) Nucleic Acids Res. 31, 82-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyd, K. E. & Farnham, P. J. (1999) Mol. Biol. Cell 19, 8393-8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaeser, M. D. & Iggo, R. D. (2002) Proc. Natl. Acad. Sci. USA 99, 95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gannon, J. & Lane, D. (1991) Nature 349, 802-806. [DOI] [PubMed] [Google Scholar]
- 31.Vousden, K. H. & Lu, X. (2002) Nat. Rev. Cancer 2, 594-604. [DOI] [PubMed] [Google Scholar]
- 32.Nakano, K. & Vousden, K. H. (2001) Mol. Cell 7, 683-694. [DOI] [PubMed] [Google Scholar]
- 33.Oda, E., Ohki, R., Murasawa, H., Nemoto, J., Shibue, T., Yamashita, T., Tokino, T., Taniguchi, T. & Tanaka, N. (2000) Science 288, 1053-1058. [DOI] [PubMed] [Google Scholar]
- 34.Zou, Z., Gao, C., Nagaich, A. K., Connell, T., Saito, S., Moul, J. W., Seth, P., Appella, E. & Srivastava, S. (2000) J. Biol. Chem. 275, 6051-6054. [DOI] [PubMed] [Google Scholar]
- 35.Kannan, K., Amariglio, N., Rechavi, G., Jakob-Hirsch, J., Kela, I., Kaminski, N., Getz, G., Domany, E. & Givol, D. (2001) Oncogene 20, 2225-2234. [DOI] [PubMed] [Google Scholar]
- 36.Kannan, K., Amariglio, N., Rechavi, G. & Givol, D. (2000) FEBS Lett. 470, 77-82. [DOI] [PubMed] [Google Scholar]
- 37.Yu, J., Hwang, P. M., Rago, C., Kinzler, K. W. & Vogelstein, B. (1999) Proc. Natl. Acad. Sci. USA 96, 14517-14522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, R., Gish, K., Murphy, M., Yin, Y., Notterman, D., Hoffman, W. H., Tom, E., Mack, D. H. & Levine, A. J. (2000) Genes Dev. 14, 981-993. [PMC free article] [PubMed] [Google Scholar]
- 39.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307-310. [DOI] [PubMed] [Google Scholar]
- 40.Polyak, K., Xia, Y., Zweier, J. L., Kinzler, K. W. & Vogelstein, B. (1997) Nature 389, 300-305. [DOI] [PubMed] [Google Scholar]
- 41.Soengas, M. S., Alarcon, R. M., Yoshida, H., Giaccia, A. J., Hakem, R., Mak, T. W. & Lowe, S. W. (1999) Science 284, 156-159. [DOI] [PubMed] [Google Scholar]
- 42.Bates, S. & Vousden, K. H. (1999) Cell Mol. Life Sci. 55, 28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, J., Wang, Z., Kinzler, K. W., Vogelstein, B. & Zhang, L. (2003) Proc. Natl. Acad. Sci. USA 100, 1931-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyashita, T., Krajewsky, S., Krajewska, M., Wang, H., Lin, H., Hoffman, B., Lieberman, D. & Reed, J. (1994) Oncogene 9, 1799-1805. [PubMed] [Google Scholar]
- 45.Pohl, U., Wagenknecht, B., Naumann, U. & Weller, M. (1999) Cell Physiol. Biochem. 9, 29-37. [DOI] [PubMed] [Google Scholar]
- 46.Dazard, J.-E., Gal, H., Amariglio, N., Rechavi, G., Domany, E. & Givol, D. (2003) Oncogene 22, 2993-3006. [DOI] [PubMed] [Google Scholar]
- 47.Gentile, M., Latonen, L. & Laiho, M. (2003) Nucleic Acids Res. 31, 4779-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amundson, S. A., Bittner, M., Chen, Y., Trent, J., Meltzer, P. & Fornace, A. J. J. (1999) Oncogene 18, 3666-3672. [DOI] [PubMed] [Google Scholar]
- 49.Guo, Y., Chang, H., Tsai, J., Huang, J., Li, C., Young, K., Wu, L., Lai, M., Liu, H. & Huang, W. (2002) Environ. Mol. Mutagen 40, 122-128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.