Abstract
The p38 mitogen-activated protein kinase pathway regulates innate immune responses in evolutionarily diverse species. We have previously shown that the Caenorhabditis elegans p38 mitogen-activated protein kinase, PMK-1, functions in an innate immune response pathway that mediates resistance to a variety of microbial pathogens. Here, we show that tir-1, a gene encoding a highly conserved Toll/IL-1 resistance (TIR) domain protein, is also required for C. elegans resistance to microbial pathogens. RNA interference inactivation of tir-1 resulted in enhanced susceptibility to killing by pathogens and correspondingly diminished PMK-1 phosphorylation. Unlike all known TIR-domain adapter proteins, overexpression of the human TIR-1 homologue, SARM, in mammalian cells was not sufficient to induce expression of NF-κB or IRF3-dependent reporter genes that are activated by Toll-like receptor signaling. These data reveal the involvement of a previously uncharacterized, evolutionarily conserved TIR domain protein in innate immunity that is functionally distinct from other known TIR domain signaling adapters.
During the past decade, a variety of studies have demonstrated a high degree of evolutionary conservation between the mammalian innate immune system and that of Drosophila melanogaster (1-3). In contrast, relatively little is known about innate immunity in the nematode Caenorhabditis elegans. Recent work has shown that C. elegans is killed by a variety of bacterial and fungal pathogens, including Pseudomonas aeruginosa (4-12). When this experimental host-pathogen system was used, a forward genetic screen for C. elegans mutants exhibiting enhanced susceptibility to P. aeruginosa-mediated killing (Esp phenotype) identified two components of a mitogen-activated protein kinase (MAPK) signaling cascade, the MAPK kinase kinase, NSY-1, and the MAPK kinase, SEK-1, that are required for pathogen resistance (13). RNA interference (RNAi) inactivation of pmk-1, the gene encoding the p38 MAPK that functions downstream of NSY-1 and SEK-1, also resulted in enhanced susceptibility to killing by bacterial pathogens (11, 13), and immunoblot analysis showed diminished activation of PMK-1 in nsy-1 and sek-1 mutants. Furthermore, pathogen-elicited apoptosis in C. elegans depended on PMK-1 signaling (14). The central role that PMK-1 plays in mediating C. elegans pathogen resistance parallels the critical function that its mammalian homolog, p38 MAPK, plays in the mammalian immune response to bacterial lipopolysaccharide and to proinflammatory cytokines (15-18). These results show that C. elegans shares at least one key conserved feature of the innate immune response with mammals.
The requirement of the PMK-1 MAPK pathway for immunity to diverse pathogens [Gram-negative (13, 14) and Gram-positive (11) bacteria] suggests that various upstream signaling inputs may converge on PMK-1. To further characterize the C. elegans innate immune response, we sought to identify factors that act upstream of PMK-1. In Drosophila and mammals, transmembrane Toll and Toll-like receptors (TLRs) as well as their corresponding intracellular “adapter” proteins, all of which contain Toll/IL-1 resistance (TIR) domains (for review, see refs. 19-24), function upstream of a conserved p38 MAPK cascade (for review see refs. 22 and 25). TIR domains allow these receptors and adapter proteins to homo- and heterooligomerize, forming multiprotein complexes that initiate signaling.
Three types of TIR domain-containing proteins have been described (19). The Ig subgroup consists of transmembrane receptors such as the IL-1 receptor (IL-1R) that have extracellular immunoglobulin domains. Members of the leucine-rich repeat (LRR) subgroup are also transmembrane receptors, but unlike Ig proteins, contain extracellular LRRs. Most LRR-TIR proteins, which include the mammalian TLRs, function as receptors for microbial pathogen-associated molecular patterns such as lipopolysaccharide or peptidoglycan. The final subgroup consists of the intracellular adapter proteins, MyD88, Mal (also known as TIRAP), TRIF (also known as TICAM-1), and TRAM (also known as TIRP and TICAM-2), which transduce signals from the transmembrane Ig and LRR receptors to the downstream components of the pathway (for review see refs. 19-23). Studies performed with gene-targeted mice and small interfering RNA silencing experiments have revealed that MyD88 is an essential component of signaling by all IL-1R/TLR superfamily members with the exception of TLR3 (26-28). In contrast to MyD88, the generation of Mal/TIRAP-, TRIF-, and TRAM-deficient mice revealed that these adapters mediate signaling by some, but not all TLRs. Mal/TIRAP functions in TLR2 and TLR4 signaling (29, 30), TRIF plays a critical role in TLR3 and TLR4 signaling (31-33), whereas the function of TRAM appears to be restricted to the TLR4 pathway (34, 35). Complex formation between IL-1R/TLR family members and their respective adapters activates a signaling pathway that that initiates both a MAPK cascade, resulting in the activation of c-Jun N-terminal kinase and p38 MAPKs, and a series of signaling reactions that ultimately lead to the activation of the transcription factor NF-κB. A subset of TLRs that includes TLR3 and TLR4, also activates another transcription factor, IRF3 (for review see refs. 19-23 and 25).
The TIR domains present in these transmembrane receptors and their adapter proteins play a critical role in mammalian innate immunity. For example, the lipopolysaccharide response of transgenic mice is abrogated by a single point mutation in the DNA sequence encoding the TIR domain of TLR4 that blocks the interaction of TLR4 with downstream TIR domain-containing adapter proteins (36). In C. elegans, only two genes encode TIR domain proteins (37). tol-1 encodes a Toll receptor homologue, and tir-1 (F13B10.1) encodes a protein that is homologous to the mammalian gene known as SARM. The absence of transmembrane motifs in the primary amino acid sequence of TIR-1 and SARM suggests that these proteins may function as adapters in a manner similar to MyD88. Whether SARM mediates TLR signaling like other members of this protein subgroup, however, has not been established.
Previous studies have shown that mutations in C. elegans tol-1 have no effect on pathogen susceptibility (10). In contrast, we report here that RNAi of C. elegans tir-1 enhances susceptibility of nematodes to killing by bacterial pathogens and that activated PMK-1 levels in tir-1 RNAi-inactivated worms are diminished, suggesting that tir-1 acts upstream of pmk-1. We also show that overexpression of SARM fails to activate either NF-κΒ- or IRF-3-dependent reporter gene expression, two transcription factors that are sensitive to TLR signaling, suggesting that SARM is functionally distinct from other mammalian TIR domain-containing proteins.
Materials and Methods
RNAi. Escherichia coli HT115 carrying the RNAi vector L4440 or L4440-derived plasmids engineered to express double-stranded RNA (dsRNA) that targets the C. elegans T07H6.5, Y51H4A.17, tir-1 (F13B10.1), or pmk-1 (B0218.3) genes [Ahringer RNAi library (38), L4440:189F9, L4440:119H12, L4440:69G10, and pDK177 (13), respectively] were cultured in LB containing 100 μg/ml ampicillin for 12-15 h at 37°C, seeded onto RNAi agar plates (NGM agar, 25 μg/ml carbenicillin, and 5 mM isopropylthiogalactosidase) to induce dsRNA expression, and incubated overnight at room temperature. Six to eight worms at the L4 larval stage were added to each plate, incubated at 20°C, and allowed to lay eggs. Progeny of these worms at the L4 larval stage were used in subsequent experiments. Worms grown under the same conditions on E. coli HT115 expressing dsRNA targeting unc-22 were included as a positive control to confirm the efficacy of RNAi (39).
Killing Assays. P. aeruginosa (slow) killing assays were performed as described (8, 12). Briefly, PA14 was cultured in LB containing 100 μg/ml rifampicin, seeded on modified nematode growth medium (0.35% instead of 0.25% peptone), and incubated first for 24 h at 37°C and then for at least 8 h at room temperature before adding worms fed with RNAi-expressing bacteria. Enterococcus faecalis strain OG1RF was cultured in brain heart infusion medium for 4 h at 37°C and seeded onto blood heart infusion agar plates containing 50 μg/ml gentamicin as described (6). Plates were incubated overnight at 37°C and cooled to room temperature before beginning the experiment. A total of 25-30 L4 stage worms grown on dsRNA-expressing bacterial strains were transferred to pathogen plates. All killing assays were conducted at 25°C. Worms were scored as dead when they no longer responded to touch.
Lifespan Analysis. Lifespan assays were performed as described (40). C. elegans were grown on either RNAi bacteria as described above or the laboratory E. coli feeder strain OP50 to the L4 developmental stage and then transferred to RNAi plates containing ≈80 μg/ml 5-fluorodeoxyuridine (FUDR) to prevent progeny growth and the appropriate dsRNA-producing bacteria. FUDR was added to these plates just before the addition of worms by adding a concentrated stock onto the outside edges of preseeded RNAi plates so as not to disturb the bacterial lawn. The plates were allowed to dry before seeding with worms. Plates were scored approximately every 24 h thereafter. Worms were transferred to fresh RNAi/FUDR plates every 3-5 days as needed to provide an ample amount of food over the course of the experiment.
PMK-1 Immunoblot Analysis. Synchronized L1 C. elegans (41) were placed on RNAi plates on which a lawn of RNAi-expressing bacteria had been grown as described above, and allowed to develop into young adults at 20°C. Approximately 1,500-3,000 animals were collected in M9, pelleted by gentle centrifugation, lysed in Laemmli sample buffer (Bio-Rad) containing 5% 2-mercaptoethanol, frozen at -20°C, and thawed in a boiling water bath for 5 min. Samples were separated by SDS/PAGE on a 10% Tris·HCl polyacrylamide gel (Bio-Rad) and transferred to nitrocellulose. Total PMK-1, phosphorylated PMK-1, and β-tubulin were detected by immunoblotting described (13, 42). Anti-PMK-1 antiserum was kindly provided by K. Matsumoto (Department of Molecular Biology, Graduate School of Science, Nagoya University, Nagoya, Japan).
Cloning hSARM and Transfection Assays. The presence of several point mutations, including a nonsense mutation in the 5′ end of a full-length human SARM cDNA clone (IMAGE clone 5240465, GenBank accession no. BI918439, Open Biosystems) was revealed by DNA sequence analysis and comparison to the complete SARM cDNA sequence (GenBank accession no NP_055892) (43). These mutations were corrected by PCR with two primers that spanned the mutated sequences (5′-CCGGAATTCATGGGGGCGGTGGCACGGGCCCATGGTGGGCTGCGGGTGG-3′ and 5′-GGCCCACGGCCGCAGCAGC-3′). The corrected DNA fragment was then ligated to the rest of the SARM cDNA, already cloned in frame with a C-terminal FLAG tag in the pEF-Bos expression vector (44), by using an EagI site inside the coding sequence. The entire cDNA was sequenced to ensure the absence of point mutations introduced by PCR. The expression of full-length hSARM-FLAG was confirmed by anti-FLAG immunoblotting (see Fig. 5C).
Fig. 5.
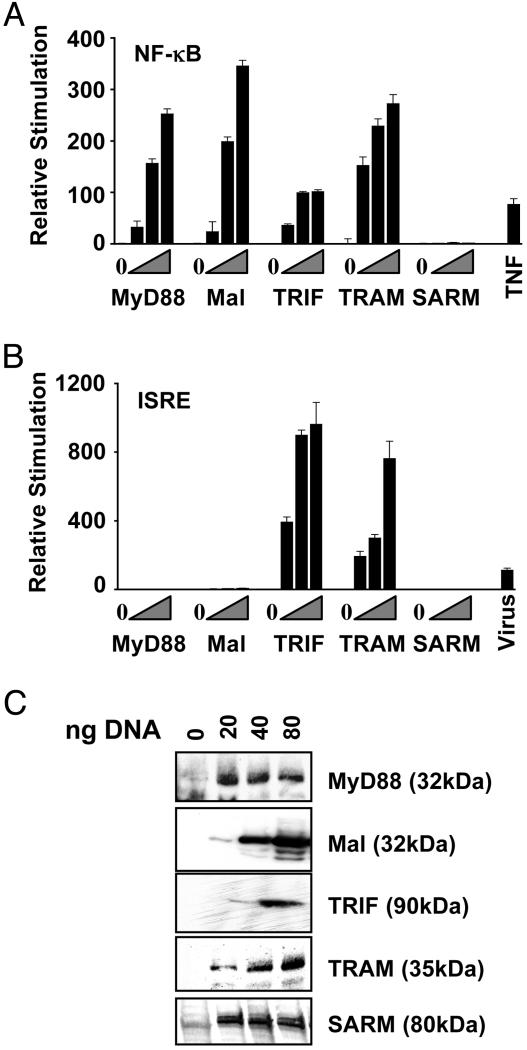
TLR reporter gene expression is not induced by human SARM over-expression. (A) HEK 293 cells were cotransfected with an NF-κB reporter construct and 0, 20, 40, or 80 ng of expression plasmids encoding Flag-tagged MyD88, Mal/TIRAP, TRIF/TICAM-1, TRAM, or SARM cDNAs. Triangles indicate increasing amounts of each expression plasmid added to the transfections. Cells treated with TNFα were included as a positive control. (B) HEK 293 cells were cotransfected with an ISRE reporter construct and 0, 20, 40, or 80 ng of expression plasmids encoding Flag-tagged MyD88, Mal/TIRAP, TRIF, TRAM, or SARM cDNAs. Triangles indicate increasing amounts of each expression plasmid added to the transfections. Cells exposed to Sendai virus were included as a positive control. (C) Flag or AU1 immunoblots of transfected cell lysates used in reporter studies in A and B. The amount of plasmid DNA transfected is noted. Data are expressed as mean relative stimulation ± SD for a representative experiment from a minimum of three separate experiments, each performed in triplicate.
The ISG54-ISRE, NF-κB-luc, pEF-Bos Mal/TIRAP-flag, pCDNA3-MyD88-AU1, pEF-Bos TRIF-flag, and pEF-Bos TRAM-flag constructs were as described (34). The NF-κB-dependent reporter, HIV LTR-κB-luc, contains several NF-κB DNA-binding sites upstream of the firefly luciferase reporter gene. Similarly, the IRF3-dependent reporter, ISG54-ISRE, contains the IFN-stimulated regulatory element (ISRE) of the IFIT2 gene encoding ISG54 situated upstream of luciferase. HEK293 cells (5 × 104 cells per well, 96-well plate) were transfected with 40 ng of either the NF-κB or the ISG-54 reporter construct and cotransfected with plasmids encoding MyD88, Mal/TIRAP, TRIF, TRAM, or SARM cDNAs. As positive controls, cells transfected with the NF-κB reporter construct were exposed to 100 ng/ml tumor necrosis factor α (TNFα) for 16 h, and cells transfected with the ISG54 reporter construct were exposed to Sendai Virus (Charles River Breeding Laboratories) for 24 h before harvesting. In all cases, cells were also transfected with 40 ng of a reporter construct containing the thymidine kinase promoter upstream of the Renilla reniforms (sea pansy) luciferase gene to normalize transfection efficiencies. Approximately 24 h later, cell lysates were prepared and luciferase and Renilla reporter gene activity was measured by using the Dual Glo Luciferase Assay system (Promega). Cell lysates were also subjected to immunoblot analysis for epitope-tagged (Flag or AU1) adapter proteins as indicated.
Results
C. elegans tir-1 Is Required for Resistance to Bacterial Pathogens. We used a reverse genetics approach to identify signaling components involved in the C. elegans innate immune response. Several C. elegans genes encoding proteins that contain motifs commonly found in proteins with immune function in mammals were selected. This set of C. elegans genes included T07H6.5, which encodes a protein that contains several complement control protein (CCP) or Sushi domains, commonly found in complement proteins; Y51H4A.17, a gene that encodes a STAT DNA binding and STAT protein interaction domains; and F13B10.1, also known as tir-1 (37), a gene that, as described above, encodes a TIR domain, a motif found in Toll/TLR and adapter signaling proteins.
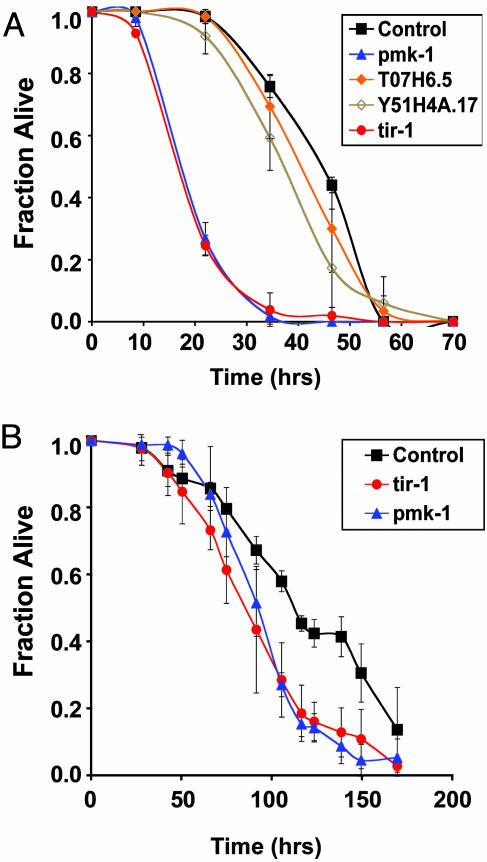
The contribution of these genes to pathogen resistance in C. elegans was evaluated by RNAi. C. elegans were fed E. coli expressing dsRNA that targeted the endogenous mRNA transcripts of each of the selected genes for degradation. The susceptibility of these RNAi-inactivated nematodes to bacterial pathogens was then assessed. As previously observed, RNAi inactivation of pmk-1 reduced the LT50 (time when half the worms were scored dead) of wild-type N2 worms by ≈40% compared with control worms fed the same E. coli strain carrying the plasmid used for dsRNA production with no gene sequence cloned in between the two T7 promoter sites (Fig. 1A) (13). Survival of T07H6.5 and Y51H4A.17 RNAi-inactivated worms on P. aeruginosa was similar to control worms. In contrast, as shown in Fig. 1 A, nematodes fed tir-1 dsRNA died significantly faster than control worms, at a similar rate to pmk-1 RNAi-inactivated worms. RNAi inactivation of tir-1 also resulted in enhanced susceptibility to killing by the Gram-positive bacterial pathogen Enterococcus faecalis (Fig. 1B), suggesting that TIR-1 is required for conferring resistance to a broad range of bacterial pathogens.
Fig. 1.
Survival of RNAi-inactivated C. elegans exposed to bacterial pathogens. (A) Survival of vector control (black square), pmk-1 (blue triangle), T07H6.5 (orange diamond), Y51H4A.17 (brown open diamond), or tir-1 (red circle) RNAi-inactivated C. elegans exposed to Pseudomonas aeruginosa strain PA14. Experiments were performed in duplicate. Error bars represent standard deviations. (B) Survival of vector control (black square), pmk-1 (blue triangle), or tir-1 (red circle) RNAi-inactivated C. elegans exposed to Enterococcus faecalis strain OG1RF. Experiments were performed in triplicate. Results are representative of two independent experiments. Error bars represent standard deviations.
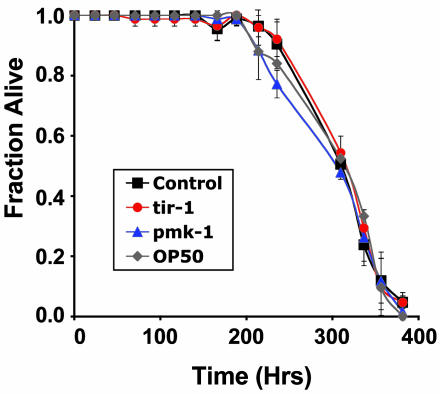
tir-1 Inactivation Does Not Adversely Affect C. elegans Longevity. Previous reports indicated that RNAi-inactivation of T07H6.5, Y51H4A.17, or tir-1 had no effect on nematode development (45, 46). In accordance with these observations, we found that N2 worms exposed to T07H6.5, Y51H4A.17, or tir-1 RNAi developed at a normal rate, reaching adulthood concomitantly with worms exposed to control RNAi. As a measure of relative fitness, the lifespan of tir-1 RNAi-inactivated worms was measured. As shown in Fig. 2, the longevity of tir-1 RNAi-treated worms was equivalent to that of worms exposed to control RNAi and to worms fed the normal E. coli feeding strain OP50 lacking any RNAi expression construct. These results indicated that the enhanced pathogen susceptibility of the nematodes exposed to tir-1 RNAi was not a consequence of a nonspecific decrease in worm fitness.
Fig. 2.
Lifespan of tir-1 RNAi-inactivated C. elegans. Lifespan assays of C. elegans maintained on E. coli feeding strain OP50 (gray diamond) or HT115 expressing vector control (black square), pmk-1 (blue triangle), or tir-1 (red circle) dsRNA. Experiments were performed in duplicate. Standard deviations are shown.
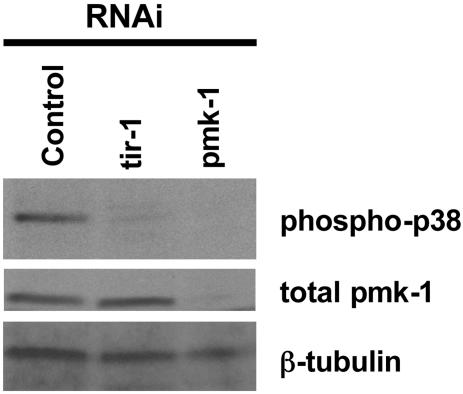
tir-1 Is Required for PMK-1 p38 MAPK Activation in C. elegans. Previously, we reported that esp-2/sek-1(ag1) and esp-8/nsy-1(ag3), two mutants with strong Esp phenotypes, have diminished levels of phosphorylated PMK-1 (13). To determine whether TIR-1 also acts upstream of PMK-1, we used a similar immunoblotting procedure to measure activated PMK-1 levels in worms in which tir-1 was inactivated by RNAi. As shown in Fig. 3, activated PMK-1 in tir-1 RNAi-targeted worms was significantly reduced. Under the same growth conditions, pmk-1 RNAi resulted in a reduction in both the activated and total amounts of PMK-1 detected, confirming that RNAi treatment reduced target protein levels. Consistent with the conclusion that TIR-1 functions in the same pathway as PMK-1, tir-1 RNAi inactivation did not further enhance the susceptibility of esp-2/sek-1(ag1) nematodes to P. aeruginosa (data not shown).
Fig. 3.
Immunoblot analysis of vector, pmk-1, or tir-1 RNAi-inactivated young adult stage N2 lysates. (Top) Immunoblot with an antibody that recognizes the doubly phosphorylated activated form of p38 MAP kinase (Promega V121A). (Middle) Total PMK-1 immunoblot. (Bottom) Anti-β-tubulin immunoblot analysis (loading control). Immunoblots are representative of at least two experiments.
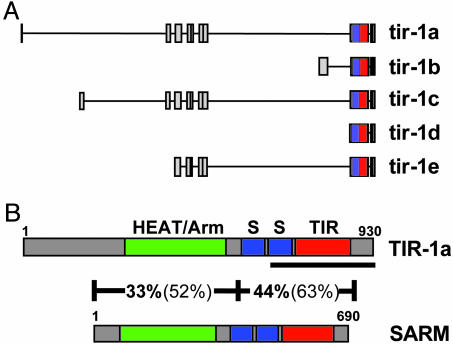
tir-1 Is Highly Conserved in Flies and Mammals. As described above, tir-1 is the only C. elegans gene identified by bioinformatic analysis that encodes an intracellular TIR domain-containing protein that could potentially function as an adapter in TLR signaling (37). However, the tir-1 gene has five known isoforms confirmed by partial cDNAs, tir-1a, tir-1b, tir-1c, tir-1d, and tir-1e (Fig. 4A) (37). In addition to the TIR domain, the proteins encoded by all five TIR-1 isoforms contain two sterile α motifs (SAM), which have previously been shown to mediate protein-protein interactions in large protein complexes (47). Regions in TIR-1A, TIR-1C, and TIR-1E also share moderate homology with HEAT/armadillo-like repeats, motifs that work together to provide a surface for protein docking, facilitating the assembly of protein complexes (43, 48).
Fig. 4.
(A) Genomic structure of five known isoforms of tir-1 (see the WormBase web site, www.wormbase.org, Release WS115). Each box represents an individual exon. Each line represents intronic DNA. Note that each isoform includes exon 8, which encodes both the SAM domains (blue) and the TIR domain (red). (B) Domain architecture of the C. elegans TIR-1A and H. sapiens SARM proteins. SAM domains (S, blue), TIR domains (red), and HEAT/Armadillo repeats (green) are denoted in the figure. The amino acid length of each protein is noted. Results of blastp analysis of two different sections of the TIR-1 and SARM protein sequences are shown. Percentage amino acid identity is shown in bold, and percentage amino acid homology is shown in parentheses. Additional HEAT/Armadillo repeats overlap the SAM and TIR domains but were omitted from the figure for the sake of clarity. The TIR-1 protein encoded by the gene sequence targeted by the RNAi clone used in this study is underlined. Note that this RNAi-targeted gene segment includes the TIR domain. Because the region of the tir-1 gene targeted by RNA interference is shared among all TIR-1 isoforms, the expression of each isoform was inhibited by this treatment.
TIR-1 is highly conserved between nematodes, flies, and mammals. TIR-1A has been described as a homologue of the D. melanogaster protein, Ect4 (also known as CG7915) and the mouse and human SARM (for sterile α and HEAT/Armadillo motifs) proteins (43, 49). CLUSTALW alignment of the C. elegans, D. melanogaster, Mus musculus, and Homo sapiens proteins reveals the extent of conservation of TIR-1 (see Fig. 6, which is published as supporting information on the PNAS web site). Indeed, the high level of conservation between TIR-1and human SARM is striking (illustrated in Fig. 4B). BLASTP analysis of TIR-1A against the human SARM amino acid sequence reveals 38% amino acid identity and 57% amino acid homology over 81% of the SARM sequence that includes the SAM and TIR domains.
TLR Signaling Reporter Gene Expression Is Not Induced by SARM Over-Expression. In light of the well established role of TIR domain-containing adapter proteins in TLR signaling in mammals, we assessed whether the human homologue of TIR-1, SARM, functions in classical TLR-mediated responses. Upon activation, different TLRs recruit specific TLR adapter proteins to the receptor complex to initiate the TLR signaling pathway. In addition to the activation of MAPK cascades, TLR signaling induces gene expression from NF-κB-regulated promoters (50). As previously observed, overexpression of all known TLR adapter proteins, MyD88, Mal, TRIF, and TRAM, induced the expression of a reporter gene construct containing several NF-κB DNA binding sites upstream of the firefly luciferase gene (Fig. 5A). Under these same conditions, however, SARM overexpression did not stimulate NF-κB-mediated luciferase expression (Fig. 5A).
In addition to NF-κB activation, TRIF and TRAM activate the transcription factor IRF-3 independently of MyD88 to mediate type I IFN and chemokine production in TLR3/4 signaling (34, 51). As reported previously, overexpression of each of these adapters activated expression of a luciferase reporter gene linked to an IRF3-dependent promoter element, the ISRE (Fig. 5B). SARM overexpression, however, failed to initiate IRF-3-dependent reporter gene expression (Fig. 5B). Immunoblot analysis confirmed the expression of full-length SARM and the TLR adapter proteins in this experiment (Fig. 5C).
Discussion
Here, we have used the study of immunity in C. elegans to gain insight into evolutionarily conserved mechanisms of innate immunity. We show that the C. elegans tir-1 gene is required for pathogen resistance to both Gram-positive and Gram-negative bacterial pathogens, suggesting a central role for tir-1 in a broad C. elegans response to microbial invasion. These data have been corroborated by recent independent work by Couillault et al., who have shown that tir-1 is also required for anti-fungal immunity in C. elegans (55). We also show that TIR-1 functions upstream of PMK-1, a homologue of the mammalian p38 MAPK. Although it is unclear at what point TIR-1 intersects the NSY-1/SEK-1/PMK-1 pathway, the regulation of PMK-1 activation by TIR-1 appears to be a critical component of TIR-1-mediated immunity. Consistent with a role for TIR-1 in the NSY-1-SEK-1-PMK-1 pathway, no further enhancement of the Esp phenotype was displayed by sek-1 mutant worms upon tir-1 RNAi inactivation.
Although the precise function of TIR-1 in innate immune signaling is not known, the presence of conserved motifs in addition to the TIR domain may provide some insight into the mechanism of TIR-1 action. SAM domains allow SAM-containing proteins to multimerize, forming large signaling complexes through homo- and hetero-oligomerization. Moderate homology to HEAT/Armadillo repeats throughout the conserved region of the TIR-1 primary amino acid sequence further indicates a role for TIR-1 in protein-protein interactions. Together, the known participation of TIR, SAM, and HEAT/Armadillo motifs in protein complexes is consistent with a pivotal role for TIR-1 in signaling.
In mammalian cells, the functions of four TIR-domain intracellular adapter proteins in immune signaling have been verified by the construction of transgenic loss of function mice. Mutations in MyD88, Mal/TIRAP, TRIF/TICAM-1, and TRAM all result in impaired cytokine/IFN induction in response to ligand engagement of particular IL-1R/TLR family members (26-29, 31-35, 52). Furthermore, the activation of NF-κB or IRF3 has also been shown to be abrogated or impaired when individual adapter molecules are absent (25, 26, 31, 32, 35). Because of its TIR domain, it has been inferred that SARM functions like the other four TLR adapter proteins to activate NF-κB- or IRF3-dependent reporter gene expression (53). Although our results show that SARM activated neither NF-κB- nor IRF3-dependent reporters, they do not rule out the possibility that SARM functions in a mammalian immune response pathway. SARM may not function in the embryonic kidney cells used in this experiment because of tissue or cell-type specificity or SARM may require activation by an upstream signaling component before it can signal effectively. It is also possible that SARM may function in an NF-κB- or IRF3-independent TLR signaling pathway, or alternatively, SARM may mediate TLR-independent immune responses.
In addition to our experimental results, there are several reasons to postulate that SARM may function independently of TLR signaling. Phylogenetic analysis suggests that, among the family of TIR domain adapter proteins defined by MyD88, Mal/TIRAP, TRIF/TICAM-1, TRAM, and SARM, the TIR domain of SARM is distinct. As noted by O'Neill and colleagues, although Box 1 of the consensus TIR motif is maintained in the SARM sequence, Box 2 and Box 3 of the SARM TIR amino acid sequence, which partially encode sections of the hydrophobic core of the TIR domain, diverge from the TIR domain sequences in other known TIR proteins (53, 54). In addition, the presence of SAM domains and HEAT/Armadillo repeats in the SARM protein sequence is unique among the known TLR signaling adapters. Thus, SARM may interact with proteins not yet defined as TIR domain-containing protein interactors.
A second reason to suggest that SARM functions differently than the other TIR domain adapter proteins is that, in C. elegans, the single Toll receptor homologue, TOL-1, does not appear to be required for pathogen resistance, indicating that TIR-1 and TOL-1 do not function in the same pathway in immune defense in the worm (10). In addition, even though several other components of the Toll signaling pathway have C. elegans homologues (trf-1, pik-1, and ikb-1 are C. elegans homologues of dTRAF, pelle, and cactus, respectively), C. elegans mutants containing deletions in each of these genes do not display enhanced susceptibility to a variety of bacterial and yeast pathogens (10). Finally, no NF-κB homologue has been identified in the C. elegans genome.
Because among the family of mammalian TIR-domain adapter proteins, only SARM is conserved in C. elegans, we speculate that SARM/TIR-1 may be the most ancient member of this class of signaling proteins and that the evolution of innate immunity involving TLR signaling through NF-κB in insects and mammals required the further evolution of the TIR domain adapter family. Our data in C. elegans suggest that the ancestral member, SARM/TIR-1, may be involved in a correspondingly ancient TLR/TOL-1-independent signaling pathway that activates p38 MAPK/PMK-1 and predates the involvement of TLRs in innate immune responses.
In summary, the demonstrated role of TIR-1 in C. elegans immunity, the structural differences between TIR-1 and known TIR domain adapter proteins, and the absence of data demonstrating a role for TLR signaling in C. elegans, suggest that TIR-1-mediated p38 activation may represent an ancient feature of innate immune signaling that predates the evolution of TLR signaling. The extent of the conservation of the TIR-1 amino acid sequence through evolution as well as the inability of SARM, the human homologue of TIR-1, to activate TLR signaling reporter gene expression suggests that SARM may be involved in mammalian immunity, functioning upstream of p38, perhaps independent of TLR, NF-κB, or IRF3 signaling.
Acknowledgments
We thank J. Xu for technical assistance and J. Ahringer, A. Fire, and K. Matsumoto for materials. We thank C. Sifri and J. Ewbank for sharing results before publication. N.T.L. is supported by a National Science Foundation postdoctoral research fellowship in Microbial Biology. K.A.F. is supported by the Wellcome Trust. D.H.K. is supported by a postdoctoral fellowship from the Howard Hughes Medical Institute, by a National Institutes of Health K08 Career Development Award, and by a Burroughs Wellcome Fund career award in the biomedical sciences. This work was supported by National Institutes of Health Grants GM48707 (to F.M.A.) and AI52455 (to D.T.G.).
Abbreviations: MAPK, mitogen-activated protein kinase; RNAi, RNA interference; dsRNA, double-stranded RNA; TLR, Toll-like receptors; IL-1R, IL-1 receptor; ISRE, IFN-stimulated regulatory element; TIR, Toll/IL-1 resistance; SAM, sterile α motifs.
References
- 1.Hoffmann, J. A. & Reichhart, J. M. (2002) Nat. Immunol. 3, 121-126. [DOI] [PubMed] [Google Scholar]
- 2.Jasper, H. & Bohmann, D. (2002) Mol. Cell 10, 967-969. [DOI] [PubMed] [Google Scholar]
- 3.Kimbrell, D. A. & Beutler, B. (2001) Nat. Rev. Genet. 2, 256-267. [DOI] [PubMed] [Google Scholar]
- 4.Aballay, A., Yorgey, P. & Ausubel, F. M. (2000) Curr. Biol. 10, 1539-1542. [DOI] [PubMed] [Google Scholar]
- 5.Couillault, C. & Ewbank, J. J. (2002) Infect. Immun. 70, 4705-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garsin, D. A., Sifri, C. D., Mylonakis, E., Qin, X., Singh, K. V., Murray, B. E., Calderwood, S. B. & Ausubel, F. M. (2001) Proc. Natl. Acad. Sci. USA 98, 10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labrousse, A., Chauvet, S., Couillault, C., Kurz, C. L. & Ewbank, J. J. (2000) Curr. Biol. 10, 1543-1545. [DOI] [PubMed] [Google Scholar]
- 8.Mahajan-Miklos, S., Tan, M. W., Rahme, L. G. & Ausubel, F. M. (1999) Cell 96, 47-56. [DOI] [PubMed] [Google Scholar]
- 9.O'Quinn, A. L., Wiegand, E. M. & Jeddeloh, J. A. (2001) Cell Microbiol. 3, 381-393. [DOI] [PubMed] [Google Scholar]
- 10.Pujol, N., Link, E. M., Liu, L. X., Kurz, C. L., Alloing, G., Tan, M. W., Ray, K. P., Solari, R., Johnson, C. D. & Ewbank, J. J. (2001) Curr. Biol. 11, 809-821. [DOI] [PubMed] [Google Scholar]
- 11.Sifri, C. D., Begun, J., Ausubel, F. M. & Calderwood, S. B. (2003) Infect. Immun. 71, 2208-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan, M. W., Mahajan-Miklos, S. & Ausubel, F. M. (1999) Proc. Natl. Acad. Sci. USA 96, 715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, D. H., Feinbaum, R., Alloing, G., Emerson, F. E., Garsin, D. A., Inoue, H., Tanaka-Hino, M., Hisamoto, N., Matsumoto, K., Tan, M. W. & Ausubel, F. M. (2002) Science 297, 623-626. [DOI] [PubMed] [Google Scholar]
- 14.Aballay, A., Drenkard, E., Hilbun, L. R. & Ausubel, F. M. (2003) Curr. Biol. 13, 47-52. [DOI] [PubMed] [Google Scholar]
- 15.Wajant, H., Grell, M. & Scheurich, P. (1999) Cytokine Growth Factor Rev. 10, 15-26. [DOI] [PubMed] [Google Scholar]
- 16.Kyriakis, J. M. & Avruch, J. (2001) Physiol. Rev. 81, 807-869. [DOI] [PubMed] [Google Scholar]
- 17.Guha, M. & Mackman, N. (2001) Cell Signalling 13, 85-94. [DOI] [PubMed] [Google Scholar]
- 18.Dong, C., Davis, R. J. & Flavell, R. A. (2002) Annu. Rev. Immunol. 20, 55-72. [DOI] [PubMed] [Google Scholar]
- 19.Dunne, A. & O'Neill, L. A. (2003) Sci STKE, re3. [DOI] [PubMed]
- 20.Qureshi, S. T. & Medzhitov, R. (2003) Genes Immun. 4, 87-94. [DOI] [PubMed] [Google Scholar]
- 21.Beutler, B. & Rehli, M. (2002) Curr. Top. Microbiol. Immunol. 270, 1-21. [DOI] [PubMed] [Google Scholar]
- 22.Barton, G. M. & Medzhitov, R. (2003) Science 300, 1524-1525. [DOI] [PubMed] [Google Scholar]
- 23.Takeda, K. & Akira, S. (2003) Cell Microbiol. 5, 143-153. [DOI] [PubMed] [Google Scholar]
- 24.Janssens, S. & Beyaert, R. (2002) Trends Biochem. Sci. 27, 474-482. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, G. & Ghosh, S. (2001) J. Clin. Invest. 107, 13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawai, T., Adachi, O., Ogawa, T., Takeda, K. & Akira, S. (1999) Immunity 11, 115-122. [DOI] [PubMed] [Google Scholar]
- 27.Adachi, O., Kawai, T., Takeda, K., Matsumoto, M., Tsutsui, H., Sakagami, M., Nakanishi, K. & Akira, S. (1998) Immunity 9, 143-150. [DOI] [PubMed] [Google Scholar]
- 28.Schnare, M., Holt, A. C., Takeda, K., Akira, S. & Medzhitov, R. (2000) Curr. Biol. 10, 1139-1142. [DOI] [PubMed] [Google Scholar]
- 29.Horng, T., Barton, G. M., Flavell, R. A. & Medzhitov, R. (2002) Nature 420, 329-333. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto, M., Sato, S., Hemmi, H., Sanjo, H., Uematsu, S., Kaisho, T., Hoshino, K., Takeuchi, O., Kobayashi, M., Fujita, T., et al. (2002) Nature 420, 324-329. [DOI] [PubMed] [Google Scholar]
- 31.Oshiumi, H., Matsumoto, M., Funami, K., Akazawa, T. & Seya, T. (2003) Nat. Immunol. 4, 161-167. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto, M., Sato, S., Hemmi, H., Hoshino, K., Kaisho, T., Sanjo, H., Takeuchi, O., Sugiyama, M., Okabe, M., Takeda, K. & Akira, S. (2003) Science 301, 640-643. [DOI] [PubMed] [Google Scholar]
- 33.Hoebe, K., Du, X., Georgel, P., Janssen, E., Tabeta, K., Kim, S. O., Goode, J., Lin, P., Mann, N., Mudd, S., et al. (2003) Nature 424, 743-748. [DOI] [PubMed] [Google Scholar]
- 34.Fitzgerald, K. A., Rowe, D. C., Barnes, B. J., Caffrey, D. R., Visintin, A., Latz, E., Monks, B., Pitha, P. M. & Golenbock, D. T. (2003) J. Exp. Med. 198, 1043-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto, M., Sato, S., Hemmi, H., Uematsu, S., Hoshino, K., Kaisho, T., Takeuchi, O., Takeda, K. & Akira, S. (2003) Nat. Immunol. 4, 1144-1150. [DOI] [PubMed] [Google Scholar]
- 36.Poltorak, A., He, X., Smirnova, I., Liu, M. Y., Van Huffel, C., Du, X., Birdwell, D., Alejos, E., Silva, M., Galanos, C., et al. (1998) Science 282, 2085-2088. [DOI] [PubMed] [Google Scholar]
- 37.Harris, T. W., Lee, R., Schwarz, E., Bradnam, K., Lawson, D., Chen, W., Blasier, D., Kenny, E., Cunningham, F., Kishore, R., et al. (2003) Nucleic Acids Res. 31, 133-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamath, R. S. & Ahringer, J. (2003) Methods 30, 313-321. [DOI] [PubMed] [Google Scholar]
- 39.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- 40.Lee, S. S., Lee, R. Y., Fraser, A. G., Kamath, R. S., Ahringer, J. & Ruvkun, G. (2003) Nat. Genet. 33, 40-48. [DOI] [PubMed] [Google Scholar]
- 41.Epstein, H. F. & Shakes, D. C. (1995) Caenorhabditis elegans: A Modern Biological Analysis of an Organism (Academic, San Diego), Vol. 48.
- 42.Reinhart, B. J. & Ruvkun, G. (2001) Genetics 157, 199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mink, M., Fogelgren, B., Olszewski, K., Maroy, P. & Csiszar, K. (2001) Genomics 74, 234-244. [DOI] [PubMed] [Google Scholar]
- 44.Mizushima, S. & Nagata, S. (1990) Nucleic Acids Res. 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S. J., Copley, R. R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. (2000) Nature 408, 331-336. [DOI] [PubMed] [Google Scholar]
- 46.Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003) Nature 421, 231-237. [DOI] [PubMed] [Google Scholar]
- 47.Kim, C. A. & Bowie, J. U. (2003) Trends Biochem. Sci. 28, 625-628. [DOI] [PubMed] [Google Scholar]
- 48.Andrade, M. A., Petosa, C., O'Donoghue, S. I., Muller, C. W. & Bork, P. (2001) J. Mol. Biol. 309, 1-18. [DOI] [PubMed] [Google Scholar]
- 49.Stathopoulos, A., Van Drenth, M., Erives, A., Markstein, M. & Levine, M. (2002) Cell 111, 687-701. [DOI] [PubMed] [Google Scholar]
- 50.Karin, M. & Ben-Neriah, Y. (2000) Annu. Rev. Immunol. 18, 621-663. [DOI] [PubMed] [Google Scholar]
- 51.Doyle, S., Vaidya, S., O'Connell, R., Dadgostar, H., Dempsey, P., Wu, T., Rao, G., Sun, R., Haberland, M., Modlin, R. & Cheng, G. (2002) Immunity 17, 251-263. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto, M., Sato, S., Mori, K., Hoshino, K., Takeuchi, O., Takeda, K. & Akira, S. (2002) J. Immunol. 169, 6668-6672. [DOI] [PubMed] [Google Scholar]
- 53.O'Neill, L. A., Fitzgerald, K. A. & Bowie, A. G. (2003) Trends Immunol. 24, 286-290. [DOI] [PubMed] [Google Scholar]
- 54.Xu, Y., Tao, X., Shen, B., Horng, T., Medzhitov, R., Manley, J. L. & Tong, L. (2000) Nature 408, 111-115. [DOI] [PubMed] [Google Scholar]
- 55.Couillault, C., Pujol, N., Sabatier, L., Guichou, J.-F., Kohara, Y. & Ewbank, J. J. (2004) Nat. Immunol., in press. [DOI] [PubMed]