Abstract
Fas ligand- (FasL) mediated apoptosis is an important element of tissue-specific organ damage. We have developed biologically active small exocyclic peptide mimetics that disable apoptotic functions of Fas. The most effective mimetic binds to both its receptor and FasL with comparable affinity. In vitro, the most effective antagonist blocked FasL-induced cytotoxicity completely and specifically. In vivo, the antagonistic mimetic also prevented Concanavilin A (Con A) induced hepatitis, a CD4+ T cell-mediated animal model of liver injury. Although current approaches prevent Fas receptor signaling by excluding FasL binding to Fas, the small molecule mimetics reported here disable Fas by promoting a defective Fas-FasL receptor complex. This event desensitizes FasL-mediated apoptosis by inhibiting extracellular signal regulated kinase activity and up-regulating NF-κB.
Keywords: inhibitor, rational drug design
Fas (CD95/APO-1) and its specific ligand [Fas ligand (FasL)/CD95L] are members of the tumor necrosis factor receptor (TNFR) and TNF families of proteins, respectively (1). The interaction between Fas and FasL triggers a cascade of subcellular events that results in a definable cell death process in Fas-expressing targets. Fas is a 45-kDa type I membrane protein expressed constitutively in various tissues, including spleen, lymph nodes, liver, lung, kidney, and ovary (2). FasL is a 40-kDa type II membrane protein, and its expression is predominantly restricted to lymphoid organs and perhaps certain immune-privileged tissues (3). In humans, FasL can induce cytolysis of Fas-expressing cells, as either a membrane-bound or a 17-kDa soluble form, which is released through metalloproteinase-mediated proteolytic shedding (4, 5). The FasL/Fas system has been implicated in the control of immune response and inflammation and response to infection, neoplasia, and death of parenchymal cells in several organs (1, 6, 7). Defects of the FasL/Fas system can limit lymphocyte apoptosis and lead to lymphoproliferation and autoimmunity (8).
Concanavilin A (Con A) induced hepatitis is an experimental murine model of human autoimmune hepatitis (9). T cell activation plays a crucial role in the process of Con A-induced hepatitis (9, 10). Hepatic injury seems to be induced by several different mechanisms involving Fas-FasL (11-13), the perforin-granzyme system (14), IFN-γ (12), and TNF-α-mediated cytotoxicity (10, 15-18). Hepatic damage depends primarily on the Fas-FasL system, because FasL-defective gld/gld mice or Fas-defective lpr/lpr mice are resistant to liver injury induced by Con A treatment (12, 13).
Macromolecules such as monoclonal anti-FasL antibody and recombinant soluble Fas protein are potential candidate antagonists for clinical studies (19, 20).
Peptidomimetics that are constructed to resemble secondary structural features of the targeted protein represent an approach to overcome some of the limitations of macromolecules and can mimic inhibitory features of large molecules such as antibody (21) and soluble receptors (21, 22).
We found that a potent small molecular species we have developed is specific to Fas and may lead to the formation of defective receptor ensembles. The molecular and biological features suggest that the previously undescribed mimetics may have therapeutic applications in disease states mediated by Fas.
Experimental Protocol
Materials. Human recombinant TNF-α was obtained from Roche Diagnostics. Flag-tagged soluble human Fas ligand (FasL-Flag) and human Fas extra cellular domain-IgGFc fusion protein (Fas-Fc) were purchased from Kamiya Biomedical (Seattle). Human recombinant TNFR (type 1) extracellular domain-IgGFc fusion protein was obtained from R & D Systems. Anti-Flag-horseradish peroxidase antibody, hydrogen peroxide solution, 3, 3′, 5,5′-tetramethylbenzidine, and Con A were from Sigma.
Cell Line. American Type Culture Collection Jurkat cells were grown in RPMI medium 1640 supplemented with 10% heat inactivated FCS/L-glutamine (2 mM)/penicillin (100 units/ml)/streptomycin (100 μg/ml) at 37°C in a humidified 5% CO2 atmosphere.
Mice. Eight-week-old C57BL/6 (B6) mice were purchased from CLEA Japan (Tokyo). All mice used were maintained under specific pathogen-free conditions in our animal facility.
Molecular Modeling. Computer modeling and structural analysis were performed by using both QUANTA and INSIGHT (Molecular Simulations, San Diego). The molecular model of the human Fas-FasL complex was built by using the crystal structure of the TNFR and the molecular model of Fas (23) as described (24).
Peptide Synthesis and Cyclization. Peptides were synthesized and purified by the Chemistry Laboratory of the University of Pennsylvania. The peptides containing internal cysteine residues were refolded, oxidized, and purified as described (22).
Solid-Phase Ligand-Binding Assay. Binding of Flag-tagged soluble FasL and peptide to Fas was determined by using the standard solid-phase binding assay by using ELISA (25). Briefly, the Fas-Fc fusion protein (250 ng/ml) was immobilized onto a 96-well ELISA plate (Costar). After blocking and subsequent washing, Flag-tagged soluble FasL (100 ng/ml) peptide was added to the Fas-Fc for 2 h. The plate was washed and incubated with anti-FLAG(M2) for 1-h washes, and horseradish peroxidase antibody was added for 1 h. Finally, the plate was washed, and the enzyme reaction was started. Absorbance at 450 nm was measured with an ELISA reader.
Biosensor Analysis. All experiments were carried out on a BIAcore 3000 instrument (Biacore, Uppsala, Sweden) as described (26). Approximately 1,500 resonance units of FasL-Flag, Fas-Fc, TNF-α or TNFRI-Fc were immobilized on research-grade CM5 sensor chips (Biacore) by using standard N-ethyl-N-dimethylaminopropyl carbodiimide/N-hydroxysuccinimide coupling. Surface plasmon resonance measurements were carried out at a flow rate of 20 μl·min-1. Data were analyzed with BIA EVALUATION 3.0 software (Biacore).
Cytotoxicity Assay. Twenty microliters of Jurkat cells at 1 × 105 cells/ml was plated in 96-well U-bottom plates. FasL-Flag (120 ng/ml in culture medium) was preincubated with an equal volume of peptide sample in PBS for 1 h at 37°C,and20 μl of mixture was added to each well. After an incubation period of 24 h, each culture was pulsed with 1 μCi (1 Ci = 37 GBq) of thymidine for 24 h before harvesting on glass fiber filters. Incorporation of the radioactive label was measured by liquid scintillation counting (Wallac, Turku, Finland) and expressed as the arithmetic mean cpm of triplicate cultures.
Flow Cytometry Assay for Apoptosis. Apoptotic cells were detected by annexin V-FITC binding to phosphatidylserine expressed on the cell membrane in the early phase of apoptosis by using a commercial kit purchased from Roche as described by Vermes et al. (27). Briefly, 1 × 105 Jurkat cells were cultured with FasL-Flag (250 ng/ml) in the presence or absence of the peptide sample for 3 h. The cells were then washed and resuspended for 10 min in buffer containing calcium, FITC-conjugated annexin V, and propidium iodide (PI). Cells were analyzed by FACScan (Becton Dickinson). Early apoptotic cells were expressed as percentage of cells positive for annexin V and negative for PI.
Western Blotting. Downstream molecules involved in Fas signaling during apoptosis were examined. Jurkat cells (1 × 106/well) were cultured in six-well plates for 12 h, treated with or without 1 mM Kp7-6 for 2 h, and then treated with FasL at 100 ng/ml for indicated periods. Cells were then washed with chilled PBS and treated with lysis buffer. Cell lysates (15-30 μg) were separated by 12% SDS/PAGE, electroblotted onto nitrocellulose membranes (Osmonics, Westborough, MA), and probed with antiphospho-IκBα, anti-IκBα, antiphospho extracellular regulated kinase (ERK)1/2, anti-ERK2, and anti-β-actin Abs (Cell Signaling Technology, Beverly, MA). The membranes were then developed by using the enhanced chemiluminescence system(Amersham Pharmacia Biosciences).
Administration of Con A and Measurement of Serum Transaminase Activity. Hepatic damage was induced by injection of a single dose of 0.5 mg of Con A dissolved in pyrogen-free saline and administered to mice via the tail vein. Anti-FasL monoclonal antibody [MFL-4 (28)] or Fas mimetic peptide (Kp7-6 or Kp1-1) was diluted with pyrogen-free saline and injected in a single dose i.p. 30 min before Con A.
Blood samples were collected from mice at 12 h after Con A injection, and the serum was taken by centrifugation. Serum activities of alanine aminotransferase and aspartate aminotransferase were measured by Lippi-Guidi's method (Iatrozyme TA-LQ, Dia-Iatron, Tokyo) (29).
Statistical Analysis. Results are expressed as mean ± SE and analyzed by Student's t test or ANOVA where appropriate. Post hoc comparisons were performed by using the Scheffé test. A 95% confidence interval was used to define statistical significance.
Results
Molecular Model of Fas Receptor Complex. Fas, a member of the TNF superfamily, shares significant structural homology with the TNFR. The structure of the TNFR contains distinct “cystine-knot” repeating subdomains (30). Loop structures in the first three domains as well as β turns in proteins are considered to mediate roles in molecular recognition and binding (31). To develop a cystine-knot peptide mimetic, we identified critical sites of protein-protein interaction that might be disrupted or influenced by small molecules.
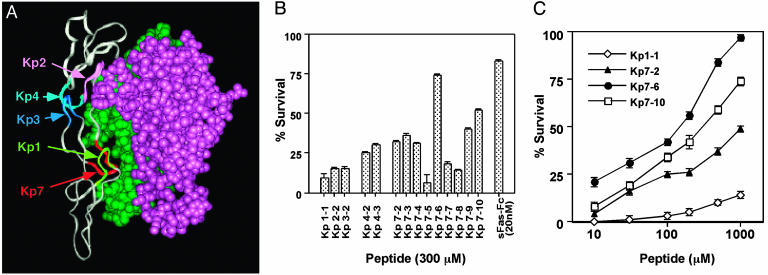
We developed a molecular model of the Fas and FasL complex (Fig. 1A) by using the crystal structure of the TNFR complex as well as other published models (23, 32). The overall features of the receptor-ligand interaction were noted to be very similar to that of the TNFR-ligand complex (22). Fas-FasL contact sites predicted by the molecular model are consistent with other mutation analysis data (33). Our Fas-FasL structural model suggested five critical surfaces by which FasL can bind to its receptor compared to three sites identified in the TNFR (22). The amino acids in the loops Kp1-7 adopt well defined conformations (i.e., statistically allowed conformations) as judged by Ramachandran plots (34) and profile analyses (35).
Fig. 1.
Identification of FasL inhibitor. (A) Molecular model and 3D structure of the critical binding site in the FasL-Fas complex. Molecular interaction analyses between FasL and Fas identified the major sites of interaction that are shown; monomeric Fas (gray), dimeric FasL (green and violet), Kp1 (yellowish green), Kp2 (pink), Kp3 (blue), Kp4 (turquoise), and Kp7 (red). (B) Inhibition of FasL binding to the Fas receptor by exocyclic peptidemimetics in a binding assay. Inhibition activities of several mimetics from different regions were compared at 300 nM each peptide and 20 nM soluble Fas receptor. Inhibition (%) by several doses of peptides was calculated and plotted. The results represent the means and standard deviations derived from three independent experiments. (C) Inhibition of FasL-induced cytolysis in Jurkat cells by the antagonistic peptides. Incorporation of [3H]thymidine obtained with culture medium alone and with 30 ng/ml of FasL was used as reference for 100% and 0% survival, respectively. Survival (%) obtained over several doses of peptides is plotted. The results represent the arithmetic mean cpm of triplicate cultures.
Several peptide analogs were designed from the identified relevant loop structures. Each mimetic was optimized for its ability to mimic the binding conformation of the loop and for its ring size, which we have determined to be critical to reduce the inherent flexibility of mimetics. A set of mimetics that were selected for biological assay is shown in Table 1.
Table 1. Sequences of exocyclic peptidomimetics derived from Fas.
| Fas receptor | Exocyclic peptidomimetic | |
|---|---|---|
| Kp | ||
| 119 CNSTVC 124 | Kp1-1 | YCNSTVCY |
| Kp2 | ||
| 77 DKAHFSSKC 85 | Kp2-2 | YCDKAEHFCY |
| Kp3 | ||
| 103 CTRTQ 107 | Kp3-2 | YCNTRTQNTCY |
| Kp4 | ||
| 69 CQEGKEY 75 | Kp4-2 | YCQEKEYCY |
| Kp4-3 | YCQERKEYCY | |
| Kp7 | ||
| 91 CDEGHGL 97 | Kp7-2 | YCDEGHLCY |
| Kp7-3 | YCDEGLCY | |
| Kp7-4 | YCDEGYFCY | |
| Kp7-5 | YCDEGEYCY | |
| Kp7-6 | YCDEHFCY | |
| Kp7-7 | YCDEHGLCY | |
| Kp7-8 | YCDEHGQCY | |
| Kp7-9 | YCDEKFCY | |
| Kp7-10 | YCDEQFCY | |
Peptidomimetics were cyclized and constrained with cysteine disulfide bridges.
Inhibition of FasL Binding to the Receptor by Mimetics. The inhibition of FasL-Flag binding (100 ng/ml) to the Fas-Fc fusion protein immobilized onto plastic plates was then used to evaluate one aspect of mimetic activity. First-generation mimetics (Kp1-1, -2-2, -3-2, -4-2, and -7-2) were designed from different deduced binding sites of Fas to FasL and screened by using a binding inhibition assay (Fig. 1B). The results indicated that the Kp7 loop is a suitable surface for the design of mimetics as a template. We have reengineered and designed second and later generations of exocyclic peptides derived from the Kp7 loop surface. By analysis of the interaction site between FasL and Fas, the acidic residues (Asp and Glu) in the Kp7 loop appear to represent the most relevant residues involved in the interaction. Modification of other residues of Kp7 led to some improvement of inhibitory activities as seen with the Kp7 series (Fig. 1B).
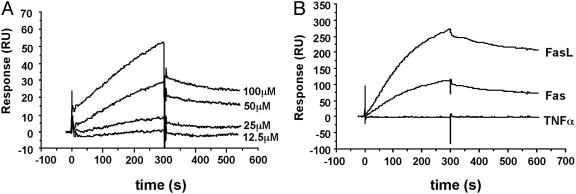
Binding Affinity and Specificity of Mimetics. The Kp7 peptide analogs were expected to bind FasL and inhibit the interaction of FasL and Fas. To investigate the kinetics of binding of Kp7-6, the exocyclic species that mediated the best inhibitory activity to FasL, we performed surface plasmon resonance (BIAcore) analysis. FasL-Flag was immobilized onto a sensor chip and various concentrations of Kp7-6 were passed over the surface. The kon and koff rate constants were estimated to be 68.5 M-1·s-1 and 7.65 × 10-4·s-1, respectively, and a Kd value of 11.2 μM was obtained from the ratio of the dissociation/association rate constants (Fig. 2A). Interestingly, Kp7-6 also bound to immobilized Fas (Fig. 2B). The kon and koff rate constants were estimated to be 24.1 M-1·s-1 and 3.18 × 10-4·s-1, respectively, and the Kd of value 13.2 μM. Kp7-6 bound neither to the TNFR nor to TNF-α, suggesting that the mimetic is specific to FasL and interacts with the Fas receptor assembly.
Fig. 2.
Biosensor analysis of mimetic binding to immobilized FasL. The sensorgram shows the relative response in resonance units after background subtraction vs. time in seconds. The association phase injection time was 300 s followed by dissociation in buffer. (A) Sensorgrams showing the binding of Kp7-6 to FasL at Kp7-6 concentrations indicated in the graph. (B) Sensorgrams showing the interaction of Kp7-6 with Fas, FasL, and TNF-α.
Inhibition of FasL-Induced Cytotoxicity by Mimetics. To evaluate the effect of mimetics on Fas-mediated cytotoxicity, FasL-sensitive Jurkat cells were stimulated with the soluble FasL-Flag fusion protein in the presence or absence of various concentrations of mimetics. Generally, the inhibitory effects of mimetics on Fas-mediated cytotoxicity were consistent with the results of the FasL-binding inhibition (Fig. 1B). As shown in Fig. 3A, Kp7-6 showed a dose-dependent inhibitory activity, and at high doses, Kp7-6 protected >90% cells from Fas-mediated cytotoxicity. Kp7-10 also showed a dose-dependent inhibition (Fig. 1C). The cyclic peptides did not mediate any cytotoxicity for Jurkat cells in the range of concentration tested (data not shown). Neither Kp7-6 nor Kp7-10 used in combination with members of the Kp4 mimetics showed any significant synergy, suggesting that inhibition of the Kp7-binding site by itself is sufficient to antagonize FasL activity (data not shown).
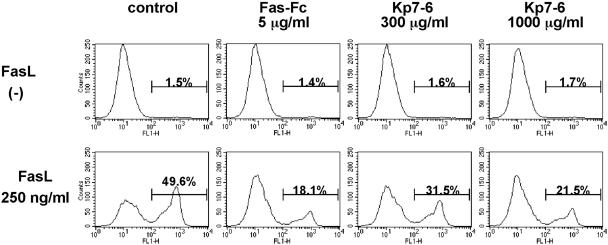
Fig. 3.
Inhibition of FasL-induced apoptosis in Jurkat cells by the antagonistic peptide. Jurkat cells were treated with 250 ng/ml of FasL in the presence or absence of Kp7-6 for 3 h and stained with FITC-conjugated annexin V and propidium iodide. Ten thousand cells were analyzed in each condition. The number above each bar indicates the percentage of apoptotic (annexin-V+) cells.
Inhibition of FasL-Induced Apoptosis by Mimetics. To evaluate whether the designed mimetics can inhibit FasL-induced apoptosis, we analyzed phosphatidylserine (PS) externalization on the cell membrane by using annexin V. During the early apoptotic process, the cell membrane remains intact (as opposed to necrosis), and translocation of PS to the outer leaflet of the plasma membrane is a common feature of apoptosis that can be quantitatively measured using annexin V-FITC binding (27). Jurkat cells treated with FasL-Flag (200 ng/ml) for 3 h showed a marked increase in PS exposure compared with untreated cells (Fig. 3). The increase in Jurkat cell cytolysis was prevented by Kp7-6 in a dose-dependent manner (Fig. 1C). These results show that Kp7-6 specifically protected FasL-induced cell lysis.
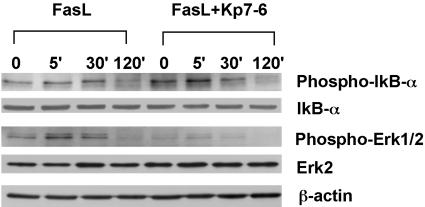
Modulation of NF-κB and ERK Activities by Mimetics. To evaluate how the designed mimetics alter FasL-mediated apoptosis, we analyzed IκBα, ERK, and c-jun N-terminal kinase (JNK) activation in Jurkat cells during the treatment of FasL. As shown in Fig. 4, Kp7-6 induced the phosphorylation of IκBα by 5 min in the presence of FasL. FasL alone did not activate IκBα significantly. These results indicate that Kp7-6 can cause activation of NF-κB, a constituent in a process that leads to inhibition of FasL-induced apoptosis. However, the IκBα effect was transient. We also noted that FasL leads to phosphorylation of ERK (by 5-30 min). ERK activation is thought to correlate with cell sensitivity to killing signals (36). We noted that ERK phosphorylation was significantly inhibited by Kp7-6, suggesting that the mimetic “desensitized” cells from death signals. These combined activities, namely phosphorylation of IκBα and reduction of ERK phosphorylation, are unexpected and represent a mechanistic approach to modulate apoptosis that follows receptor modulation.
Fig. 4.
Activation of the downstream NF-κB pathway and inhibition of mitogen-activated protein kinase kinase P42/44 in Jurkat cells by Kp7-6. Jurkat cells were pretreated with Kp7-6 for 2 h and then treated with 100 ng/ml of FasL at the indicated time course. The cells were lysed and analyzed by Western blotting. The expression of phospho-IκBα,IκBα, phospho-P42/44, and ERK-2 was analyzed. The expression of β-actin was analyzed as loading control. Kp7-6 at 1 mM clearly enhanced phosphorylation of IκBα in the presence of FasL at 5 min and significantly inhibited phosphorylation of Erk1/2 at 5-30 min, compared with the treatment of FasL alone.
The unexpected combination of enhanced survival signals (IκBα) and decreased apoptotic signals (inhibition of ERK phosphorylation) indicates that creating disabled receptor ensembles is a previously undescribed therapeutic approach. The JNK pathway was also examined, and no alteration in the activation signal either with or without pretreatment of Kp7-6 (data not shown) was seen, indicating that JNK activation was not relevant for desensitization of cells.
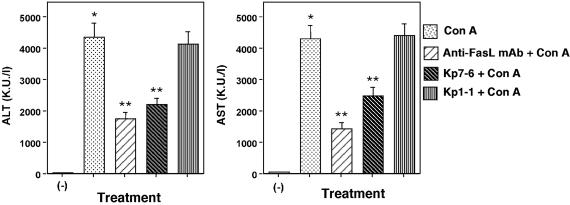
Protection of Mice Against Con A-Induced Liver Injury by the Antagonistic Fas Mimetic Peptide. Next we investigated the in vivo effects of the mimetic, Kp7-6. FasL-induced apoptosis is one of the primary and dominant pathways by which liver cells undergo apoptosis under various conditions such as viral infection, drug toxicity, and other lesions (8, 15, 37). Several studies have found that blocking Fas signaling by either RNA interference or oligonucleotides limits the extent of liver damage (38, 39). We chose to investigate the antagonistic effect of Kp7-6 in vivo using the Con A-induced hepatitis model. C57BL/6 mice were pretreated i.p. with anti-FasL monoclonal antibody, Fas mimetic peptides, or saline. Thirty minutes later, animals were challenged i.v. with Con A or saline. The induction of liver damage and inflammatory hepatitis was evaluated by measuring the activities of two transaminases (alanine aminotransferase and aspartate aminotransferase) in the serum 12 h after Con A treatment. As shown in Fig. 5, the activities of both transaminase were reduced in Kp7-6-pretreated mice, indicating Kp7-6 blocked Fas-mediated hepatic injury in vivo. On the other hand, Kp1-1, a control mimetic, did not block hepatic injury
Fig. 5.
Protection of mice against Con A-induced liver injury by the antagonistic Fas mimetic peptide. C57BL/6 mice were injected with 0.5 mg of Con A in saline or saline only as negative controls i.v. Anti-FasL monoclonal antibody (MFL-4, 0.5 mg/mouse) or Fas mimetic peptide (Kp7-6 or Kp1-1, 3 mg per mouse) in saline was injected i.p. 30 min before Con A. Blood samples were obtained 12 h after Con A injection, and serum alanine aminotransferase and aspartate aminotransferase levels were measured. *, P < 0.0001 vs. control; **, P < 0.01 vs. Con A-treated mice.
Discussion
Dysregulation of Fas has been implicated in several pathological conditions including cancer, autoimmunity, angiogenesis, and organ damage (19, 40). Fas has been shown to possess dual functions, namely modulation of apoptosis and proliferation (41). Thus, altering Fas regulation in a unilateral manner either by using genetic ablation or preventing FasL signals (by antibody or soluble receptors that exclude ligand binding to the receptor) may result in different outcomes than achieved by creating a dysfunctional Fas receptor complex. Here we have shown that rationally designed small molecules that form disabled receptor ensembles regulate Fas/FasL-mediated apoptotic function in a more novel manner than simply preventing ligand-receptor interactions.
A molecular model of Fas revealed that overall folding and interaction patterns with FasL are similar to those seen with the TNFR-TNF-β complex (32). In our earlier work, we found that the third domain of TNFR was critical for TNF-α to bind to its cognate receptor (22). Peptide mimetics derived from different regions of Fas indicated that the loop (Kp7) in the second domain was most critical (Fig. 1 A and B). Thus our results suggest that membrane proximal domains of TNF super family receptors may be critical for receptor-ligand stabilization and essential for signaling. This is also in agreement with our structural observations of the erbB receptor family (which are also type II receptors) where the membrane proximal domain, which shares structural homology to the TNFR, is critical for signaling (42). Thus, we propose that an antibody or small molecule that binds to membrane proximal domains will have more pronounced effects on signaling aspects of the receptor system.
The Kd of the Kp7-6-FasL interaction was of less affinity than that noted for Fas-FasL interactions (43). Although Kp7-6 is a weak binding species, it is able to alter Fas functions in vitro and in vivo. The dissociation rate (koff) of Kp7-6 was similar to an antigen-antibody interaction, which suggests that Kp7-6 forms a stable receptor complex (26). The koff value is considered a critical indicator in the development of therapeutics with biological activity (44, 45) and generally correlates with potent biological effects (46). These observations help explain the observed biological activity of Kp7-6.
Kp7-6 is selective to the Fas receptor complex. Selectivity of small molecules binding to large receptor complexes is difficult to achieve due to the limited surface area available for the small molecule to interact with its receptor. Selectivity is notable for Kp7-6, because several members of the superfamily share significant homology. To assess the specificity of this interaction, Fas, FasL, and TNF-α were immobilized onto a sensor chip. Kp7-6 bound to FasL but not to TNF-α, which indicates Kp7-6 binds to FasL specifically (Fig. 2B). On the other hand, Kp7-6 bound not only to FasL but also to Fas (Fig. 2B), yet Kp7-6 did not bind to the closely related TNFRs. These observations are reminiscent of features of the soluble TNFRI (p55), which has been shown to form antiparallel homodimeric complexes in the absence of ligand (30). Our results in this study suggest that soluble Fas may also form such antiparallel dimers, and the Kp7 loop may be involved in dimer formation. Thus these results suggest that Kp7-6 is selective to Fas and may function like a minisoluble Fas receptor.
Small molecules such as Kp7-6 may disable Fas by a different mechanism from the macromolecular anti-Fas antibodies. Unlike anti-Fas macromolecular species that have been shown to limit Con A-induced hepatitis, Kp7-6 is a small molecule, not large enough to preclude the interaction of FasL with Fas. This raises the possibility that Kp7-6 acts by promoting a defective signaling complex. The Kp7-6 mimetic, described above, is derived from the Fas receptor and has the propensity to bind to both the receptor and its ligand with comparable affinity. This suggests that Kp7-6 could either bind to Fas and prevent FasL from forming a stable complex with the receptor or bind to both FasL and Fas and form a defective signaling complex. A proposed scheme is shown in Fig. 6.
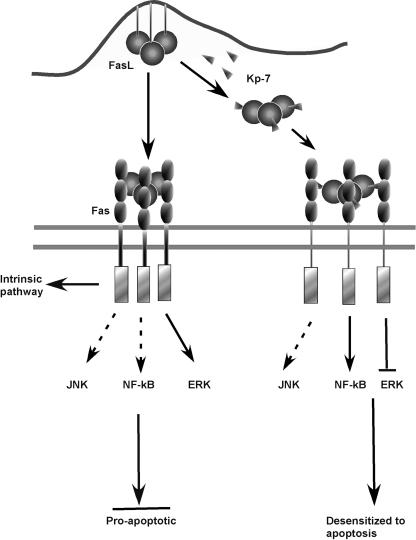
Fig. 6.
Proposed mechanism by which Kp7-6 desensitize Fas/FasL proapoptotis signals. Fas-FasL-mediated apoptosis follows two major pathways: (i) intrinsic and (ii) extrinsic. Key apoptotic signal regulators (NF-κB, JNK, and ERK) in the pathways (Left) and the signal modification obtained by Kp7-6 (Right) are shown. Kp7-6 binding to either FasL or Fas or both creates a defective receptor complex ensemble. As a consequence, NF-κB is activated, but ERK activation is inhibited. The combined modulation of these two key regulator leads to “desensitization” of cells to apoptosis. The JNK pathway remains unaltered.
Furthermore, Fas/FasL-induced apoptosis follows two major pathways: (i) an intrinsic pathway, often referred to as a caspase-dependent mitochondrial pathway (47, 48); and (ii) an extrinsic pathway that is caspase independent and involves NF-κB and JNK (40). A secondary signaling process, the ERK pathway, has been shown to be proapoptotic or to act as an attenuator of apoptosis (36, 49). Our studies revealed that the FasL-Kp7-6 complex weakly stimulated NF-κB but suppressed ERK activation. Kp7-6 had no effect on the JNK activation state. Activation of NF-κB is closely linked to regulation of antiapoptotic molecules (41, 50, 51). Thus, weak activation of NF-κB is consistent with the antiapoptotic events observed. In several studies, activation of JNK has been shown to be not relevant for apoptotic signals (49, 52), and indeed JNK activation was unaltered by the FasL-Kp7-6 complex, indicating that JNK may play an insignificant role in Fas-mediated signals in this system.
The FasL-Kp7-6 complex inhibited ERK activation. In general, activation of ERK is considered an indicator of cell proliferation (53-55). However, under some circumstances, depending on the cell type and the level of Fas expression, activation of ERK plays a secondary role: to sensitize cells to FasL-induced apoptosis (36, 49, 56). Thus, in the context of Fas/FasL receptor signaling, disabling ERK by the FasL-Kp7-6 complex may be considered a “FasL desensitization” stimulus. Kp7-6 thus desensitizes or attenuates FasL-mediated apoptosis. By creating a defective complex, Kp7-6 protects cells from FasL-induced apoptosis by a complex pattern of signals, the concurrence of which has not been greatly appreciated.
The specific mechanism by which apoptotic receptors such as Fas and TNFR1 (p55) mediate Con A-induced hepatitis is not fully understood. Genetic studies using Fas-deficient lpr/lpr mice indicate mechanisms other than that mediated by the Fas-FasL system also involved in this process (12, 16). Nevertheless, inhibition of Fas has some beneficial effect in limiting liver damage. Thus, Fas is a primary drug target against liver injury, and several approaches have been reported including gene therapy (57), interference RNA (39), and monoclonal antibody (11). Kp7-6 represents a previously unreported small molecule that can effectively disable Fas receptor function in vitro and that limits liver injury in vivo comparable to the effects of the anti-Fas monoclonal antibody.
Kp7-6 is a rationally fashioned peptide mimetic that demonstrates specific anti-FasL activity in vitro and in vivo. Signaling studies show that Kp7-6 is able to protect cells from Fas-mediated apoptosis by promoting NF-κB activity and by inhibiting ERK activity. Our work suggests that small molecular drugs can be developed to modulate cell surface receptors using a novel approach that in essence creates spacers between functional receptor proteins and creates dysfunctional receptor complexes. We believe the development of small molecules that are specific for other members of the TNFR family may represent a general way of studying functional aspects of receptor subdomains that will facilitate the rational development of therapeutics.
Acknowledgments
We thank the Biosensor/Interaction Analysis and Structural Biology Core Group, Department of Medicine, University of Pennsylvania, for assistance with Biacore binding studies and the Protein Chemistry Laboratory, University of Pennsylvania, for peptide synthesis and purification. This work was supported by the Abramson Family Cancer Research Institute and by grants from the National Institutes of Health, the National Cancer Institute, the National Institute of Neurological Disorders and Stroke, and the Cancer Center, University of Pennsylvania.
Abbreviations: FasL, Fas ligand; ERK, extracellular regulated kinase; JNK, c-jun N-terminal kinase; TNF, tumor necrosis factor; TNFR, TNF receptor.
References
- 1.Nagata, S. & Golstein, P. (1995) Science 267, 1449-1456. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe-Fukunaga, R., Brannan, C. I., Itoh, N., Yonehara, S., Copeland, N. G., Jenkins, N. A. & Nagata, S. (1992) J. Immunol. 148, 1274-1279. [PubMed] [Google Scholar]
- 3.Suda, T., Takahashi, T., Golstein, P. & Nagata, S. (1993) Cell 75, 1169-1178. [DOI] [PubMed] [Google Scholar]
- 4.Kayagaki, N., Kawasaki, A., Ebata, T., Ohmoto, H., Ikeda, S., Inoue, S., Yoshino, K., Okumura, K. & Yagita, H. (1995) J. Exp. Med. 182, 1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mariani, S. M., Matiba, B., Baumler, C. & Krammer, P. H. (1995) Eur. J. Immunol. 25, 2303-2307. [DOI] [PubMed] [Google Scholar]
- 6.Biancone, L., Martino, A. D., Orlandi, V., Conaldi, P. G., Toniolo, A. & Camussi, G. (1997) J. Exp. Med. 186, 147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krammer, P. H. (1999) Adv. Immunol. 71, 163-210. [DOI] [PubMed] [Google Scholar]
- 8.Famularo, G., Nucera, E., Marcellini, S. & De Simone, C. (1999) Med. Hypotheses 53, 50-62. [DOI] [PubMed] [Google Scholar]
- 9.Tiegs, G., Hentschel, J. & Wendel, A. (1992) J. Clin. Invest. 90, 196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuhara, H., O'Neill, E., Seki, N., Ogawa, T., Kusunoki, C., Otsuka, K., Satoh, S., Niwa, M., Senoh, H. & Fujiwara, H. (1994) J. Exp. Med. 179, 1529-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seino, K., Kayagaki, N., Takeda, K., Fukao, K., Okumura, K. & Yagita, H. (1997) Gastroenterology 113, 1315-1322. [DOI] [PubMed] [Google Scholar]
- 12.Tagawa, Y., Sekikawa, K. & Iwakura, Y. (1997) J. Immunol. 159, 1418-1428. [PubMed] [Google Scholar]
- 13.Tagawa, Y., Kakuta, S. & Iwakura, Y. (1998) Eur. J. Immunol. 28, 4105-4113. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe, Y., Morita, M. & Akaike, T. (1996) Hepatology 24, 702-710. [DOI] [PubMed] [Google Scholar]
- 15.Gantner, F., Leist, M., Lohse, A. W., Germann, P. G. & Tiegs, G. (1995) Hepatology 21, 190-198. [DOI] [PubMed] [Google Scholar]
- 16.Toyabe, S., Seki, S., Iiai, T., Takeda, K., Shirai, K., Watanabe, H., Hiraide, H., Uchiyama, M. & Abo, T. (1997) J. Immunol. 159, 1537-1542. [PubMed] [Google Scholar]
- 17.Kusters, S., Tiegs, G., Alexopoulou, L., Pasparakis, M., Douni, E., Kunstle, G., Bluethmann, H., Wendel, A., Pfizenmaier, K., Kollias, G., et al. (1997) Eur. J. Immunol. 27, 2870-2875. [DOI] [PubMed] [Google Scholar]
- 18.Ksontini, R., Colagiovanni, D. B., Josephs, M. D., Edwards, C. K., III, Tannahill, C. L., Solorzano, C. C., Norman, J., Denham, W., Clare-Salzler, M., MacKay, S. L., et al. (1998) J. Immunol. 160, 4082-4089. [PubMed] [Google Scholar]
- 19.Maggi, C. A. (1998) Pharmacol. Res. 38, 1-34. [DOI] [PubMed] [Google Scholar]
- 20.Okuda, Y., Sakoda, S., Fujimura, H., Nagata, S., Yanagihara, T. & Bernard, C. C. (2000) Biochem. Biophys. Res. Commun. 275, 164-168. [DOI] [PubMed] [Google Scholar]
- 21.Park, B. W., Zhang, H. T., Wu, C., Berezov, A., Zhang, X., Dua, R., Wang, Q., Kao, G., O'Rourke, D. M., Greene, M. I. & Murali, R. (2000) Nat. Biotechnol. 18, 194-198. [DOI] [PubMed] [Google Scholar]
- 22.Takasaki, W., Kajino, Y., Kajino, K., Murali, R. & Greene, M. I. (1997) Nat. Biotechnol. 15, 1266-1270. [DOI] [PubMed] [Google Scholar]
- 23.Bajorath, J. (1999) J. Comput. Aided Mol. Des. 13, 409-418. [DOI] [PubMed] [Google Scholar]
- 24.Cheng, X., Kinosaki, M., Takami, M., Choi, Y., Zhang, H. & Murali, R. (2004) J. Biol. Chem. 279, 8269-8277. [DOI] [PubMed] [Google Scholar]
- 25.Schneider, P., Bodmer, J. L., Holler, N., Mattmann, C., Scuderi, P., Terskikh, A., Peitsch, M. C. & Tschopp, J. (1997) J. Biol. Chem. 272, 18827-18833. [DOI] [PubMed] [Google Scholar]
- 26.Berezov, A., Zhang, H. T., Greene, M. I. & Murali, R. (2001) J. Med. Chem. 44, 2565-2574. [DOI] [PubMed] [Google Scholar]
- 27.Vermes, I., Haanen, C., Steffens-Nakken, H. & Reutelingsperger, C. (1995) J. Immunol. Methods 184, 39-51. [DOI] [PubMed] [Google Scholar]
- 28.Kayagaki, N., Yamaguchi, N., Nagao, F., Matsuo, S., Maeda, H., Okumura, K. & Yagita, H. (1997) Proc. Natl. Acad. Sci. USA 94, 3914-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lippi, U. & Guidi, G. (1970) Clin. Chim. Acta 28, 431-437. [DOI] [PubMed] [Google Scholar]
- 30.Naismith, J. H., Devine, T. Q., Kohno, T. & Sprang, S. R. (1996) Structure (Cambridge, U.K.) 4, 1251-1262. [DOI] [PubMed] [Google Scholar]
- 31.Leszczynski, J. F. & Rose, G. D. (1986) Science 234, 849-855. [DOI] [PubMed] [Google Scholar]
- 32.Banner, D. W., D'Arcy, A., Janes, W., Gentz, R., Schoenfeld, H. J., Broger, C., Loetscher, H. & Lesslauer, W. (1993) Cell 73, 431-445. [DOI] [PubMed] [Google Scholar]
- 33.Beltinger, C., Bohler, T., Karawajew, L., Ludwig, W. D., Schrappe, M. & Debatin, K. M. (1998) Br. J. Haematol. 102, 722-728. [DOI] [PubMed] [Google Scholar]
- 34.Ramachandran, G. N. & Venkatachalam, C. M. (1968) Biopolymers 6, 1255-1262. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, K. Y. & Eisenberg, D. (1994) Protein Sci. 3, 687-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran, S. E., Holmstrom, T. H., Ahonen, M., Kahari, V. M. & Eriksson, J. E. (2001) J. Biol. Chem. 276, 16484-16490. [DOI] [PubMed] [Google Scholar]
- 37.Kakinuma, C., Takagaki, K., Yatomi, T., Nakamura, N., Nagata, S., Uemura, A. & Shibutani, Y. (1999) Toxicol. Pathol. 27, 412-420. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, H., Cook, J., Nickel, J., Yu, R., Stecker, K., Myers, K. & Dean, N. M. (2000) Nat. Biotechnol. 18, 862-867. [DOI] [PubMed] [Google Scholar]
- 39.Song, E., Lee, S. K., Wang, J., Ince, N., Ouyang, N., Min, J., Chen, J., Shankar, P. & Lieberman, J. (2003) Nat. Med. 9, 347-351. [DOI] [PubMed] [Google Scholar]
- 40.Sharma, K., Wang, R. X., Zhang, L. Y., Yin, D. L., Luo, X. Y., Solomon, J. C., Jiang, R. F., Markos, K., Davidson, W., Scott, D. W. & Shi, Y. F. (2000) Pharmacol. Ther. 88, 333-347. [DOI] [PubMed] [Google Scholar]
- 41.Wajant, H., Pfizenmaier, K. & Scheurich, P. (2003) Cytokine Growth Factor Rev. 14, 53-66. [DOI] [PubMed] [Google Scholar]
- 42.Berezov, A., Chen, J., Liu, Q., Zhang, H. T., Greene, M. I. & Murali, R. (2002) J. Biol. Chem. 277, 28330-28339. [DOI] [PubMed] [Google Scholar]
- 43.Starling, G. C., Bajorath, J., Emswiler, J., Ledbetter, J. A., Aruffo, A. & Kiener, P. A. (1997) J. Exp. Med. 185, 1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benveniste, M. & Mayer, M. L. (1991) Br. J. Pharmacol. 104, 207-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yiallouros, I., Vassiliou, S., Yiotakis, A., Zwilling, R., Stocker, W. & Dive, V. (1998) Biochem. J. 331, 375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moosmayer, D., Wajant, H., Gerlach, E., Schmidt, M., Brocks, B. & Pfizenmaier, K. (1996) J. Interferon Cytokine Res. 16, 471-477. [DOI] [PubMed] [Google Scholar]
- 47.Waring, P. & Mullbacher, A. (1999) Immunol. Cell Biol. 77, 312-317. [DOI] [PubMed] [Google Scholar]
- 48.Yin, X. M. & Ding, W. X. (2003) Curr. Mol. Med. 3, 491-508. [DOI] [PubMed] [Google Scholar]
- 49.Wilson, D. J., Alessandrini, A. & Budd, R. C. (1999) Cell Immunol. 194, 67-77. [DOI] [PubMed] [Google Scholar]
- 50.Mandal, M., Maggirwar, S. B., Sharma, N., Kaufmann, S. H., Sun, S. C. & Kumar, R. (1996) J. Biol. Chem. 271, 30354-30359. [DOI] [PubMed] [Google Scholar]
- 51.Wajant, H., Haas, E., Schwenzer, R., Muhlenbeck, F., Kreuz, S., Schubert, G., Grell, M., Smith, C. & Scheurich, P. (2000) J. Biol. Chem. 275, 24357-24366. [DOI] [PubMed] [Google Scholar]
- 52.Low, W., Smith, A., Ashworth, A. & Collins, M. (1999) Oncogene 18, 3737-3741. [DOI] [PubMed] [Google Scholar]
- 53.Cobb, M. H., Hepler, J. E., Cheng, M. & Robbins, D. (1994) Semin. Cancer Biol. 5, 261-268. [PubMed] [Google Scholar]
- 54.Marais, R. & Marshall, C. J. (1996) Cancer Surv. 27, 101-125. [PubMed] [Google Scholar]
- 55.Fan, M. & Chambers, T. C. (2001) Drug Resist. Updat. 4, 253-267. [DOI] [PubMed] [Google Scholar]
- 56.Engedal, N. & Blomhoff, H. K. (2003) J. Biol. Chem. 278, 10934-10941. [DOI] [PubMed] [Google Scholar]
- 57.Fujino, M., Kawasaki, M., Funeshima, N., Kitazawa, Y., Kosuga, M., Okabe, K., Hashimoto, M., Yaginuma, H., Mikoshiba, K., Okuyama, T., et al. (2003) Gene Ther. 10, 1781-1790. [DOI] [PubMed] [Google Scholar]