Abstract
Genetic and physical mapping of the RP17 locus on 17q identified a 3.6-megabase candidate region that includes the gene encoding carbonic anhydrase IV (CA4), a glycosylphosphatidylinositol-anchored protein that is highly expressed in the choriocapillaris of the human eye. By sequencing candidate genes in this region, we identified a mutation that causes replacement of an arginine with a tryptophan (R14W) in the signal sequence of the CA4 gene at position -5 relative to the signal sequence cleavage site. This mutation was found to cosegregate with the disease phenotype in two large families and was not found in 36 unaffected family members or 100 controls. Expression of the mutant cDNA in COS-7 cells produced several findings, suggesting a mechanism by which the mutation can explain the autosomal dominant disease. In transfected COS-7 cells, the R14W mutation (i) reduced the steady-state level of carbonic anhydrase IV activity expressed by 28% due to a combination of decreased synthesis and accelerated turnover; (ii) led to up-regulation of immunoglobulin-binding protein, double-stranded RNA-regulated protein kinase-like ER kinase, and CCAAT/enhancer-binding protein homologous protein, markers of the unfolded protein response and endoplasmic reticulum stress; and (iii) induced apoptosis, as evidenced by annexin V binding and terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling staining, in most cells expressing the mutant, but not the WT, protein. We suggest that a high level of expression of the mutant allele in the endothelial cells of the choriocapillaris leads to apoptosis, leading in turn to ischemia in the overlying retina and producing autosomal dominant retinitis pigmentosa.
Keywords: unfolded protein response, choriocapillaris, annexin V, terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling staining, endoplasmic reticulum stress
Retinitis pigmentosa (RP) is the term used to describe a group of heterogeneous progressive retinal degenerative disorders characterized by restriction in visual fields and night blindness, which may progress to complete blindness in later life. Of the 13 loci so far associated with dominant RP, only one, namely RP17, has remained uncloned. This locus was first mapped to chromosome 17 by Bardien et al. in 1995 (1). Four years later, the RP17 locus was refined to map in an ≈1-centimorgan region between the markers D17S1604 and D17S948, based on haplotype analysis of two apparently unrelated South African families (2). This locus was subsequently linked to autosomal dominant RP in a Dutch kindred (3).
Construction of a sequence assembly across the candidate region was undertaken, and bioinformatic analysis and annotation of the region led to construction of an improved map. Further bioinformatic analysis revealed a list of nine candidate genes in the region. The gene, carbonic anhydrase 4 (CA4), was ranked the most attractive positional candidate. The carbonic anhydrase IV (CA IV) protein is a glycosylphosphatidylinositol membrane-anchored zinc metalloenzyme expressed on the luminal surface of microcapillaries and highly expressed in the choriocapillaris of the eye (4, 5).
We report here that analysis of the CA4 gene from affected members of the South African families identified a mutation in the signal sequence, substituting tryptophan for arginine at residue -5 relative to the cleavage site of the signal peptidase. The previously undescribed C to T change at base 40 of the cDNA sequence was not detected in 36 unaffected relatives and 100 unrelated control individuals. The infrequent coding of tryptophan at the -5 position of signal peptides (6) raised the possibility that this mutation may affect signal peptide cleavage after translocation of the nascent polypeptide into the endoplasmic reticulum (ER) lumen. Mutations in either the hydrophobic domain or the signal peptidase recognition domain of signal peptides can delay or block removal of the signal sequence (7-9). As a consequence, impaired protein folding, impaired disulfide bond formation, incomplete glycosylation, and diminished translocation from ER to Golgi (10) can result in accumulation and rapid degradation of some of the gene product in the ER (11, 12).
There are at least two examples where a mutation in the signal sequence in one allele causes a dominantly inherited hormone deficiency disease, presumably by leading to apoptosis of the vasopressin and the parathyroid hormone-producing cells, respectively (13-15). More commonly, protein conformational disorders result from mutations in the sequence of the mature protein that impair normal folding, resulting in ER accumulation of aggregated proteins and rapid turnover. Several forms of autosomal dominant RP (16) and autosomal dominant diabetes in the Akita mouse (17) are examples.
Because CA IV is highly expressed in the choriocapillaris, but not in the retina, we hypothesized that the R14W mutation in the CA4 gene leads to accumulation of unfolded forms of CA IV in the ER of endothelial cells of the choriocapillaris, inducing the unfolded protein response (UPR), and that the chronic ER stress results in apoptosis of these cells, leading in turn to ischemia of the overlying retina and RP. To test the hypothesis that this is an apoptosis-inducing mutation, we studied the effects of the R14W mutation on CA IV enzyme activity and steady-state protein level and on the rates of biosynthesis, conversion of unfolded to mature enzyme, and turnover of CA IV in the presence and absence of a proteasomal inhibitor in transfected COS-7 cells. We also studied the effects of the mutant protein on the expression of Ig-binding protein (BiP) (an ER stress chaperone), double-stranded RNA-regulated protein kinase-like ER kinase (PERK) (an ER stress-inducible kinase), CCAAT/enhancer-binding protein homologous protein (CHOP) (a PERK-inducible proapoptotic transcription factor), and on binding of annexin V at the cell membrane and terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining, two markers of apoptosis. The results presented here show that the R14W mutation in the CA4 gene is an apoptosis-inducing mutation, providing a mechanism whereby this mutation could be the disease-causing mutation in RP17.
Materials and Methods
Patient bloods and DNA samples, genotyping and sequencing, restriction analysis and bioinformatics, mapping and annotation, as well as methods for metabolic labeling and immunoprecipitation, are all described in Supporting Methods, which is published as supporting information on the PNAS web site.
Antibodies. Rabbit anti-human CA IV antiserum was produced by using affinity purified recombinant secretory CA IV enzyme from Chinese hamster ovary secretion media as described (5). Goat anti-rabbit IgG-peroxidase, rabbit anti-goat IgG-FITC, and goat anti-rabbit IgG-rhodamine conjugates were from Sigma. Goat anti-BiP, goat anti-PERK, and mouse anti-CHOP were from Santa Cruz Biotechnology. The TUNEL assay kit was purchased from Roche Applied Science (Mannheim, Germany). Annexin V-FITC conjugate was from Sigma.
Construction of Mutant CA4 cDNA and Vector for Expression in COS-7 Cells. The amino acid arginine (R) at position 14 in the signal sequence of human CA4 was changed to tryptophan (W) by mutating nucleotide C at position 40 to a T, changing the codon CGG (for R) to TGG (for W), using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The presence of the mutation was confirmed by sequencing and restriction analysis. The C to T change creates a new restriction site for Msc I (New England Biolabs) in the mutant. The WT and R14W mutant human CA4 cDNAs were digested with EcoRI and the R1 inserts subcloned into mammalian expression vector pCXN at the EcoRI site (18).
Transfection of COS-7 Cells. COS-7 cells on p-60 plates or chamber slides were transfected with 5 or 1 μg of cDNA, respectively, using the DEAE-dextran procedure (19), followed by 1 h of incubation with 100 mM chloroquine (20). After chloroquine treatment, cells were fed normal DMEM containing 10% FBS. For control experiments, COS-7 cells were transfected with pCXN vector alone. After 72 h, lysates were produced by sonication of COS-7 cells in 25 mM Tris, pH 7.5, plus 1% Nonidet P-40, containing the protease inhibitors PMSF (1 mM) and orthophenortholine (1 mM).
CA Assay. The CA IV activity of the cell lysates was determined by using the end-point titration method of Maren (21). The protein concentration of the cell lysate was determined by microLowry, using BSA as a standard (22).
SDS/PAGE and Western Blot Analysis. SDS/PAGE was performed under reducing and nonreducing conditions, according to Laemmli's procedure (23). The polypeptides were electrophoretically transferred to Immobilon membranes and characterized by immunostaining by using anti-human CA IV antibodies followed by incubation with peroxidase-conjugated goat anti-rabbit IgG. The peroxidase activity was visualized by chemiluminescent substrate. For CHOP analysis, 100 μg of total cell protein was used from COS-7 cells transfected with vector only with or without 10 or 20 μg/ml tunicamycin for 3 h at 37°C before harvesting, and COS-7 cells transfected with WT or R14W CA4 cDNA. The proteins were separated by gel electrophoresis and transferred to Immobilon membranes, which were visualized with monoclonal anti-CHOP antibody (Santa Cruz Biotechnology) at a 1:500 dilution.
Immunocytochemical Staining. The COS-7 cells transfected on chamber slides with WT or R14W mutant CA4 cDNAs and vector alone were treated with warm 3% paraformaldehyde in PBS for 40 min at room temperature. Fixed cells were rinsed with PBS and treated with 50 mM NH4Cl in PBS for 10 min. For permeabilized cells, the COS-7 cells were treated with 0.3% Triton X-100 two times for 5 min. The cells were rinsed with PBS and incubated with 0.2% gelatin in PBS for 15 min. The first antibody was used at a concentration of 10 μg/ml in 0.2% gelatin in PBS and was incubated with the cells at room temperature for 1.5 h. The cells were rinsed with 0.2% gelatin in PBS and incubated with secondary antibodies conjugated with FITC or rhodamine at a 1:200 dilution in 0.2% gelatin in PBS at room temperature for 1 h. The cells were rinsed with 0.2% gelatin in PBS followed by PBS and water. The slides were embedded in antiquencher solution containing 4′,6-diamidino-2-phenylindole (DAPI) and coverslip edges were sealed with nail polish. The slides were viewed and the images recorded by Olympus microscope.
Results
Genetic and Physical Mapping of the RP17 Interval and Identification of the R14W Mutation. The refined map of the RP17 region published in 1999 (3) was used as a starting point, and the genetic markers described (see Table 2, which is published as supporting information on the PNAS web site) were reexamined in the extended families to confirm the recombination events defining the candidate region. The primers used in this analysis are also listed in Tables 3 and 4, which are published as supporting information on the PNAS web site. Reanalysis of the genotyping data for marker D17S1604 revealed that a de novo mutation had occurred in one affected individual, resulting in misinterpretation of the haplotyping results. This observation led to a reevaluation of the candidate region to include D17S1606 as a centromeric marker (24). The distal marker, originally D17S948, was confirmed, and examination of more proximal markers showed that the recombination event at D17S948 could be refined to a new marker D17S1838, thereby reducing the candidate region by ≈3 megabases (Mb). Bioinformatic analysis and sequence assembly produced a region of 6 Mb that contained no gaps between D17S1606 and D17S1838. Examination of the sequence assembly added new microsatellite markers in the proximal region that were examined for recombination events, and the centromeric marker was redefined as RP17AC12, which is located 240 kb distal to D17S1606. Pedigrees showing the haplotypes across the region and the two recombination events are shown in Fig. 6, which is published as supporting information on the PNAS web site. Genotyping of a number of newly identified markers (RP17AC1, RP17AC3, RP17AC7, and RP17AC8) did not reduce the candidate region.
Bioinformatic examination of the published RP17 candidate region yielded nine candidate genes between D17S1604 and D17S948: RPS6KB1, LOC51136, CA4, NY-REN-60, PRO2492, TBX2, APPB2, PPMID, and BRIP1 (nomenclature according to the Human Genome Organisation Gene Nomenclature Committee, except for PRO2492, which is a predicted protein). Bioinformatic investigation by “upstream sequence analysis” flagged two genes in the region for closer examination: RPS6KB1 and CA4. Both genes were analyzed by sequencing of the exons and intron-exon junctions by using direct sequencing of PCR products.
No sequence variants were observed in the RPS6KB1 gene. Analysis of the CA4 gene identified a previously undescribed sequence change at base 40 of the cDNA sequence in 24 affected individuals; this change was not detected in 36 unaffected relatives and 100 unrelated individuals from the same population. The C to T transition mutation (Fig. 7, which is published as supporting information on the PNAS web site) leads to a change from an arginine to a tryptophan in the signal sequence at position -5 relative to the signal peptidase cleavage site. This signal sequence variant is predicted not to alter the sequence of the mature CA IV (Fig. 8, which is published as supporting information on the PNAS web site).
The R14W mutation creates an Msc I restriction endonuclease site in the PCR fragment for Exon 1a. A restriction fragment length polymorphism assay was used to demonstrate cosegregation of the Msc I recognition site with the disease phenotype in 101 individuals from two large families with autosomal dominant RP (Fig. 7B). Restriction analysis of 101 individuals in the extended family showed consistent segregation of the mutation with the RP phenotype. The change was not observed in 100 control subjects matched for age and population group.
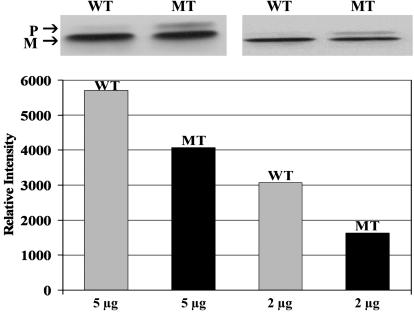
R14W Mutation Affects Steady-State Levels of CA IV. Transfected COS-7 cells were compared for CA IV activity in lysates from cells expressing either WT or R14W CA4 cDNAs. In 10 independent transfections, the R14W mutation reduced the amount of CA IV activity expressed by ≈28% on average (Table 1). In four independent experiments, a cotransfecting plasmid expressing Escherichia coli β-galactosidase was included to control for efficiency of transfection, and the results were comparable (data not shown). The amount of immunoreactive CA IV protein detected by Western blot after nonreducing SDS/PAGE was correspondingly decreased (Fig. 1). Note that in the nonreducing gel, one can observe both the unfolded precursor form of CA IV, which migrates more slowly, and the mature form. The mature form migrates faster due to formation of its two disulfide bridges, which give it a more compact structure (5). Although both forms are evident in lysates expressing WT and R14W mutant CA IV (Fig. 1), the R14W CA IV expressing cells have a larger fractional amount of the slower-migrating precursor. This observation suggests that folding and disulfide bond formation of the precursor to the mature form of CA IV is slower as a consequence of the R14W mutation.
Table 1. Comparison of steady-state CA IV activity in lysates from transfected COS-7 cells expressing WT or R14W CA IV.
| cDNA | CA activity, units/mg protein* | CA IV activity |
|---|---|---|
| WT | 1.85 ± 0.56 (n = 10) | 100 |
| MT | 1.33 ± 0.36 (n = 10) | 72 ± 2 |
Average of two independent measurements per transfection. n, number of transfections.
Fig. 1.
Effect of the R14W mutation on the steady-state level of CA IV protein. Aliquots of lysates from COS-7 cells transfected with WT or R14W mutant (MT) CA4 cDNAs containing 2 or 5 μg of cell protein were analyzed by SDS/PAGE under nonreducing conditions. The forms of CA IV were characterized by Western blot by using CA IV specific antibodies (Upper). The 37-kDa unfolded precursor (P) migrates slower than the mature 35-kDa protein (M). The polypeptide band intensities were analyzed by an image analyzer and the relative intensity for the bands plotted (Lower).
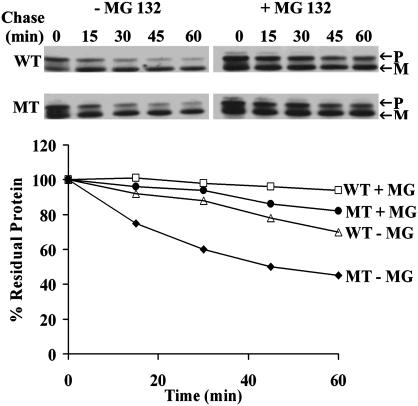
R14W Mutation Affects the Rates of Biosynthesis, Maturation, and Turnover of CA IV. Biosynthesis was studied in COS cells transfected with WT and mutant CA4 cDNAs by prelabeling with 35S-translabel for 15-120 min and examining the radiolabeled newly synthesized CA IV after immunoprecipitation and SDS/PAGE. The results in Fig. 9, which is published as supporting information on the PNAS web site, show that both the precursor and mature forms of the mutant CA IV accumulate at a rate 15-20% slower than those of the WT CA IV. Next, we compared their rates of conversion of precursor to mature enzyme by pulse-labeling for 15 min, chasing for 2-15 min with nonradioactive media, and analyzing the radioactivity in immunoprecipitated precursor and mature polypeptide bonds identified by SDS/PAGE and fluorography. Fig. 10, which is published as supporting information on the PNAS web site, shows that the rate of conversion from radioactive precursor to radioactive mature CA IV is 15-30% slower for R14W than for WT CA IV. We suspected that delayed maturation of mutant CA IV might expose the unfolded nondisulfide bonded precursor to accelerated degradation in the ER and/or proteosome (11-14). To test this hypothesis, we carried out pulse-labeling of WT and mutant enzyme expressed in COS-7 cells in the presence of 10 μg/ml MG 132, an inhibitor of proteosomal degradation (25). The Western blot shown in the Fig. 2 Upper shows both the precursor (P) and mature (M) forms of WT and R14W mutant CA IV in the presence and absence of MG 132. The results presented in Fig. 2 Lower show that pulse-labeled total R14W CA IV (precursor plus mature) is degraded much more rapidly than the WT CA IV. In both cases, the degradation was markedly slower in the presence of the inhibitor. These results suggest that at least some of the reduction in activity resulting from the R14W signal sequence mutation in the CA4 gene is due to accelerated degradation.
Fig. 2.
Effect of the R14W mutation on the stability of CA IV protein in the absence and presence of MG 132. COS-7 cells transfected with WT or R14W mutant (MT) CA4 cDNAs were pulse-labeled for 30 min and chased for 15-60 min in the absence (-) and presence (+) of MG 132 (10 μg/ml). CA IV immunoprecipitates were analyzed by SDS/PAGE under nonreducing conditions followed by fluorography. Precursor (P) and mature (M) polypeptides are marked (Upper). The percent residual protein remaining was calculated from the incorporation of radioactive amino acids in the total polypeptides (precursor + mature) (Lower). The mutant CA IV was degraded faster than the WT and was protected by the proteasomal inhibitor, MG 132.
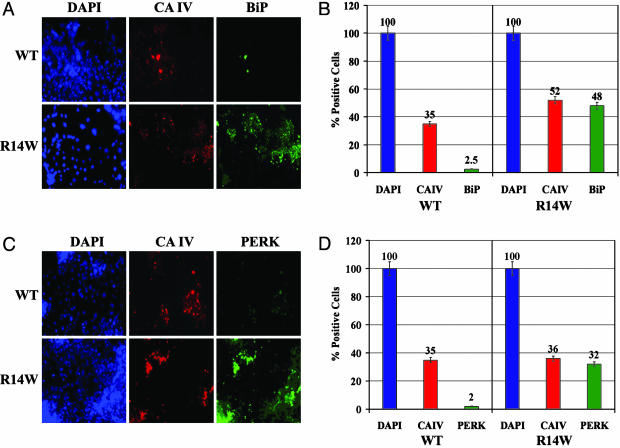
R14W Expression Up-Regulates Production of Proteins Associated with the UPR. Immunofluorescent staining of unpermeabilized, transfected COS-7 cells showed no difference between WT and R14W CA IV (Fig. 11, which is published as supporting information on the PNAS web site). However, permeabilized cells showed greater staining in the perinuclear region, suggesting some ER retention (Fig. 11). When exposed to an excess of unfolded proteins in the ER, mammalian cells respond by up-regulation of ER chaperones, including BiP, GRP78, and GRP94, up-regulation and activation of the ER kinase, PERK, and induction of CHOP (GADD153). To look for evidence that R14W CA IV expression induces the UPR, we compared the levels of BiP and PERK (by immunohistochemical staining) and CHOP (by Western blot) in transfected COS cells expressing either WT or mutant CA IV. Fig. 3A shows one field examined for total cell nuclei (DAPI), CA IV (rhodamine stain), and BiP (FITC). Fig. 3B summarizes data from many fields showing that, of 193 COS-7 cells expressing R14W CA IV, 92% expressed BiP. By contrast, only 7% of 181 cells expressing WT CA IV expressed BiP. Fig. 3C shows one field examined for activated PERK. Fig. 3D summarizes data from many fields in which 89% of the 337 COS-7 cells expressing R14W CA IV stained for PERK, whereas only 6% of the 389 cells expressing WT CA IV stained for PERK.
Fig. 3.
Effect of R14W mutation on induction of the ER proteins, BiP, and PERK, in transfected COS-7 cells. (A) One of many fields of COS-7 cells transfected with WT or R14W mutant (R14W) CA4 cDNA that were fixed, permeabilized, and treated with a mixture of anti-CA IV and anti-BiP antibodies, followed by secondary antibodies conjugated with FITC or rhodamine, respectively. The slides were visualized for DAPI (blue; for cell nucleus), CA IV (red), and BiP (green). (B) Data summarizing counts from 500 WT and 350 R14W cells total in many fields, showing the percent of total cells that were positive for CA IV and BiP expression. (C) COS-7 cells transfected with WT or R14W mutant (R14W) CA4 cDNA were fixed, permeabilized, and treated with a mixture of anti-CA IV and anti-PERK antibodies, followed by secondary antibodies conjugated with FITC or rhodamine, respectively. The cells were visualized for DAPI (blue), CA IV (red), and PERK (green). (D) Data summarizing analysis of many fields containing 580 WT and 377 R14W cells total, showing percent of total cells positive for CA IV and PERK expression.
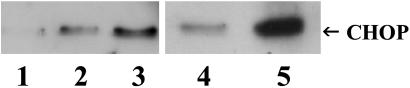
Expression of CHOP in COS-7 cells was examined by Western blot (Fig. 4). A low level of CHOP expression was detected in vector-only transfected cells (lane 1). However, exposure of vector-only transfected cells to 10 or 20 μg/ml tunicamycin for 3 h (lanes 2 and 3, respectively), which blocks N-linked glycosylation and disrupts folding of many secretory proteins, markedly up-regulated CHOP expression. Lanes 4 and 5 compare CHOP expression in cells expressing WT or R14W CA IV. Although CHOP is detectable in cells expressing WT CA IV (lane 4), it is dramatically increased in cells expressing R14W CA IV (lane 5). Thus, the signal sequence mutation in R14W CA IV clearly leads to up-regulation of all three markers of UPR and ER stress (BiP, PERK, and CHOP) in transfected COS-7 cells.
Fig. 4.
Effect of R14W mutation on induction of CHOP in transfected COS-7 cells. Western blot analysis of control COS-7 cells expressing vector-only (lane 1, negative control) or vector-only exposed to 10 μg/ml (lane 2, positive control) or 20 μg/ml (lane 3, positive control) tunicamycin for 3 h, which is known to cause ER stress and up-regulation of CHOP. Lanes 4 and 5 are lysates from COS-7 cells transfected with WT and R14W mutant CA4 cDNAs, respectively. The arrow indicates CHOP (28 kDa).
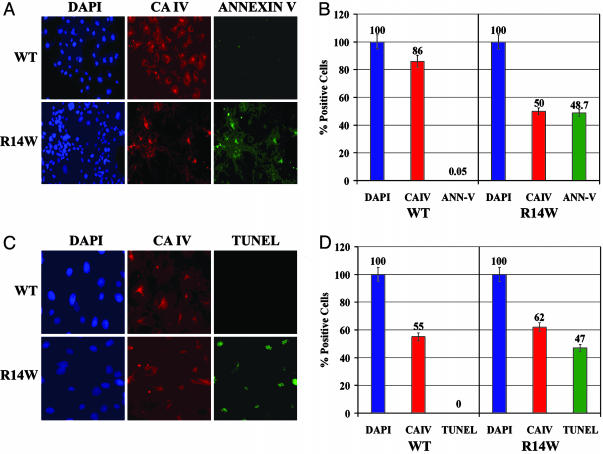
R14W CA IV Expression Induces Apoptosis. When the cells' defenses against excessive ER stress are overwhelmed, cells undergo apoptosis (26). One marker indicating cells have entered the apoptotic pathway is annexin V binding (cells undergoing apoptosis can no longer maintain the asymmetry of phospholipids in the plasma membrane). When that occurs, the phosphatidylserine and phosphatidylcholine, which are exteriorized, allow annexin V to bind to the cell surface. Another marker for apoptosis is TUNEL staining, indicating that nuclear DNA is undergoing fragmentation en route to cell death. Fig. 5A shows one of many fields examined for annexin V binding. Fig. 5B summarizes data from many fields in which, of 113 COS-7 cells expressing R14W CA IV, >97% showed annexin V binding. In contrast, of 106 COS-7 cells expressing WT CA IV, none showed annexin V binding. Fig. 5C shows one of the many fields examined for TUNEL staining. Fig. 5D summarizes data from many fields in which, of 46 COS-7 cells expressing R14W CA IV, 76% showed a positive signal for TUNEL staining. By contrast, TUNEL-positive cells were rare among cells expressing the WT CA IV (0 of 32 COS-7 cells). These data show that, when expressed in COS-7 cells, R14W is an apoptosis-inducing mutation.
Fig. 5.
Effect of R14W mutation on induction of apoptosis in transfected COS-7 cells. (A) One of many fields of COS-7 cells transfected with WT or R14W (R14W) mutant CA4 cDNAs, which were fixed and incubated with anti-CA IV followed by secondary antibody conjugated with rhodamine or FITC-annexin V. The slides were visualized for DAPI (blue), CA IV (red), and annexin V (green). (B) Data summarizing counts from many fields of 121 WT and 330 R14W cells total, showing the percent of total cells stained for CA IV and annexin V. (C) One of many fields of COS-7 cells transfected with WT or R14W (R14W) mutant CA4 cDNAs that were fixed, permeabilized, and incubated with anti-CA IV antibody followed by secondary antibody conjugated with rhodamine. The slides were visualized for DAPI (blue), CA IV expression (red), and TUNEL reaction (green). (D) Data summarizing counts from many fields of 54 WT and 63 R14W cells total, showing the percent of total cells positive for CA IV expression and TUNEL reaction.
Discussion
The initial concept that elements of the phototransduction cascade are primarily involved in retinal degenerative disorders has proven to be mostly correct. However, several loci have been implicated where no obvious retina-specific genes, or indeed phototransduction-specific proteins, were identified. For example, we previously mapped RP13 to 17p, within which interval the culprit PRPC8 gene was shown to be causative of disease (27). The product is a component of the spliceosome, which must be the basis for the retinal pathology. Several other reports have implicated “nonretina-specific” genes in retinal degenerative disorders (28).
We suggest that RP17 is another example of retinal pathology caused by the R14W mutation in the “nonretina-specific gene,” CA4. The mutation leads to accumulation of unfolded proteins in the ER, which is known to be a threat to all living cells (29). When faced with ER stress, cells activate the UPR. Prolonged activation of the UPR activates the apoptotic pathway. The results presented here show that the expression of the R14W mutant CA IV in transfected COS-7 cells activates the UPR (evident from increased BiP, PERK, and CHOP) and leads to apoptosis (evident from annexin V binding and TUNEL staining).
A growing number of previously unrelated disorders such as diabetes, prion diseases, cystic fibrosis, and several neurodegenerative disorders share the pathological features of aggregated misfolded protein deposits (16). In most protein folding diseases, a mutation in the primary sequence of the mature proteins, not in the signal sequence, leads to misfolding and prolonged activation of the UPR. Nearly half of the forms of autosomal dominant RP fit in this category, with causative mutations impairing folding of the rhodopsin protein (16, 30). However, there are two clear examples of hormone deficiency diseases where the mutations producing the autosomal dominant diseases are in the signal sequences of hormone precursors, and the diseases almost certainly result from apoptosis of the hormone-producing cells. One disorder results from a signal sequence mutation in preproparathyroid hormone (13, 14), the other from a mutation in the signal sequence of preprovasopressin (15). We propose that the R14W signal sequence mutation in the CA4 gene of RP17 patients provides a third example of an apoptosis-inducing signal sequence mutation, which produces an autosomal dominant disease.
Were CA IV highly expressed in the retina, a disease mechanism similar to forms of RP involving rhodopsin mutations could be proposed. However, CA IV expression is not detectable in retina by histochemistry (4). If present at all, it must be expressed there at a very low level. It is highly expressed in the endothelial cells of the choriocapillaris, the capillary bed that nourishes the retina. It therefore seems more plausible that RP17 is primarily (or initially) a disease of the choriocapillaris. In this scenario, the pathology in the retina is most likely secondary to accumulation over years of apoptotic damage to the endothelial cells of the capillaries, which provide the retina its blood supply. This retinopathy might be secondary to ischemia or to injury by inflammatory mediators produced by the ER-stressed capillary endothelial cells. It is not clear whether R14W CA IV expression has deleterious effects on other capillary beds where CA IV is expressed, or on epithelial cells in kidney and gut where CA IV is also expressed. Different cell types are known to show different levels of sensitivity to UPR (31). If other cell types are not affected, the basis for the sensitivity of the choriocapillaris would be important to explore.
The attractive feature of the proposition that RP17 is a protein folding disease is that it suggests an approach to therapy. Several chemical chaperones are currently being used in other diseases involving impaired protein folding (32). Members of one class, called disease-specific chemical chaperones, bind to the active site of the mutant protein and stabilize it or destabilize the folding transition state to compensate for the mutation. This is the mechanism whereby, in some forms of RP, folding defects in rhodopsin were corrected by using retinal-based ligands (30). This approach was also used by Fan et al. (33) in Fabry disease, a lysosomal storage disease. They showed that competitive inhibitors of α-galactosidase A can rescue the misfolding and mistrafficking associated with certain Fabry disease-causing mutations. For RP17, we suggest screening the wide array of well-characterized carbonic anhydrase inhibitors for such a stabilizing effect on R14W CA IV. The second class is called nonspecific chemical chaperones, which are not disease-specific. In one example, osmolytes are given to reduce the concentration of the misfolded proteins in the ER and enhance translocation of some fraction of protein that would otherwise be trapped in the ER. Perlmutter and colleagues (34) have used 4-phenylbutyric acid as a nonspecific chemical chaperone for treatment of α-1 antitrypsin deficiency. We could hope that application of one of these chemical chaperones might at least delay the onset of the disease in RP17 patients. Clinical trials to evaluate those approaches will be challenging because of the late onset and gradual progression of the disease. For this reason, availability of a good animal model, allowing experimental testing of candidate chemical chaperones for clinical efficacy for RP17, would be highly desirable. Examining the CA4 gene in dogs affected with canine progressive rod-cone degeneration, which maps to the syntenic region of the dog genome (35), is a logical first step in searching for such an animal model.
Supplementary Material
Acknowledgments
We thank Sister L. Bartman, who has been of invaluable assistance in the collection and verification of patient information and the collection of samples for this project, and Prof. Ari Ziskind for performing the necessary electroretinograms on patients for confirming the diagnosis of RP. G.R. has been supported for some of this work by the South African National Research Foundation and by Retina International. Work on the project has also been supported by Retina South Africa and the South African Medical Research Council. This work was also supported by National Institutes of Health Grant DK40163 (to W.S.S.).
Abbreviations: RP, retinitis pigmentosa; UPR, unfolded protein response; ER, endoplasmic reticulum; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling; CA IV, carbonic anhydrase IV; DAPI, 4′,6-diamidino-2-phenylindole; BiP, Ig-binding protein; PERK, double-stranded RNA-regulated protein kinase-like ER kinase; CHOP, CCAAT/enhancer-binding protein homologous protein.
References
- 1.Bardien, S., Ebenezer, N., Greenberg, J., Inglehearn, C. F., Bartmann, L., Goliath, R., Beighton, P., Ramesar, R. & Bhattacharya, S. S. (1995) Hum. Mol. Genet. 4 1459-1462. [DOI] [PubMed] [Google Scholar]
- 2.Bardien-Kruger, S., Greenberg, J., Tubb, B., Bryan, J., Queimado, L., Lovett, M. & Ramesar, R. S. (1999) Eur. J. Hum. Genet. 7 332-338. [DOI] [PubMed] [Google Scholar]
- 3.den Hollander, A. I., van der Velde-Visser, S. D., Pinckers, A. J., Hoyng, C. B., Brunner, H. G. & Cremers, F. P. (1999) Hum. Genet. 104 73-76. [DOI] [PubMed] [Google Scholar]
- 4.Hageman, G. S., Zhu, X. L., Waheed, A. & Sly, W. S. (1991) Proc. Natl. Acad. Sci. USA 88 2716-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waheed, A., Zhu, X. L. & Sly, W. S. (1992) J. Biol. Chem. 267 3308-3311. [PubMed] [Google Scholar]
- 6.von Heijne, G. (1986) Nucleic Acids Res. 14 4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellgaard, L. & Helenius, A. (2001) Curr. Opin. Cell Biol. 13 431-437. [DOI] [PubMed] [Google Scholar]
- 8.von Heijne, G. & Abrahmsen, L. (1989) FEBS Lett. 244 439-446. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X., VanValkenburgh, C., Liang, H., Fang, H. & Green, N. (2001) J. Biol. Chem. 276 2411-2416. [DOI] [PubMed] [Google Scholar]
- 10.Sitia, R. & Braakman, I. (2003) Nature 426 891-894. [DOI] [PubMed] [Google Scholar]
- 11.Kostova, Z. & Wolf, D. H. (2003) EMBO J. 22 2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anjos, S., Nguyen, A., Ounissi-Benkalha, H., Tessier, M. C. & Polychronakos, C. (2002) J. Biol. Chem. 277 46478-46486. [DOI] [PubMed] [Google Scholar]
- 13.Ito, M., Jameson, J. L. & Ito, M. (1997) J. Clin. Invest. 99 1897-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beuret, N., Rutishauser, J., Bider, M. D. & Spiess, M. (1999) J. Biol. Chem. 274 18965-18972. [DOI] [PubMed] [Google Scholar]
- 15.Karaplis, A. C., Lim, S. K., Baba, H., Arnold, A. & Kronenberg, H. M. (1995) J. Biol. Chem. 270 1629-1635. [DOI] [PubMed] [Google Scholar]
- 16.Illing, M. E., Rajan, R. S., Bence, N. F. & Kopito, R. R. (2002) J. Biol. Chem. 277 34150-34160. [DOI] [PubMed] [Google Scholar]
- 17.Izumi, T., Yokota-Hashimoto, H., Zhao, S., Wang, J., Halban, P. A. & Takeuchi, T. (2003) Diabetes 52 409-416. [DOI] [PubMed] [Google Scholar]
- 18.Niwa, H., Yamamura, K. & Miyazaki, J. (1991) Gene 108 193-199. [DOI] [PubMed] [Google Scholar]
- 19.Lopata, M. A., Cleveland, D. W. & Sollner-Webb, B. (1984) Nucleic Acids Res. 12 5707-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luthman, H. & Magnusson, G. (1983) Nucleic Acids Res. 11 1295-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maren, T. H. (1960) J. Pharmacol. Exp. Ther. 130 26-29. [PubMed] [Google Scholar]
- 22.Peterson, G. L. (1979) Anal. Biochem. 100 201-220. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. (1970) Nature 227 680-685. [DOI] [PubMed] [Google Scholar]
- 24.Bardien-Kruger, S., Greenberg, J., Tubb, B., Bryan, J., Queimado, L., Lovett, M. & Ramesar, R. S. (1999) Eur. J. Hum. Genet. 7 332-338, and [DOI] [PubMed] [Google Scholar]; erratum (2004), in press.
- 25.Bence, N. F., Sampat, R. M. & Kopito, R. R. (2001) Science 292 1552-1555. [DOI] [PubMed] [Google Scholar]
- 26.Ron, D. (2002) J. Clin. Invest. 109 443-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKie, A. B., McHale, J. C., Keen, T. J., Tarttelin, E. E., Goliath, R., Lith-Verhoeven, J. J., Greenberg, J., Ramesar, R. S., Hoyng, C. B., Cremers, F. P., et al. (2001) Hum. Mol. Genet. 10 1555-1562. [DOI] [PubMed] [Google Scholar]
- 28.Hims, M. M., Diager, S. P. & Inglehearn, C. F. (2003) Dev. Ophthalmol. 37 109-125. [DOI] [PubMed] [Google Scholar]
- 29.Kaufman, R. J. (2002) J. Clin. Invest. 110 1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noorwez, S. M., Kuksa, V., Imanishi, Y., Zhu, L., Filipek, S., Palczewski, K. & Kaushal, S. (2003) J. Biol. Chem. 278 14442-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaufman, R. J., Scheuner, D., Schroder, M., Shen, X., Lee, K., Liu, C. Y. & Arnold, S. M. (2002) Nat. Rev. Mol. Cell. Biol. 3 411-421. [DOI] [PubMed] [Google Scholar]
- 32.Perlmutter, D. H. (2002) Pediatr. Res. 52 832-836. [DOI] [PubMed] [Google Scholar]
- 33.Fan, J. Q., Ishii, S., Asano, N. & Suzuki, Y. (1999) Nat. Med. 5 112-115. [DOI] [PubMed] [Google Scholar]
- 34.Burrows, J. A., Willis, L. K. & Perlmutter, D. H. (2000) Proc. Natl. Acad. Sci. USA 97 1796-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Acland, G. M., Ray, K., Mellersh, C. S., Gu, W., Langston, A. A., Rine, J., Ostrander, E. A. & Aguirre, G. D. (1998) Proc. Natl. Acad. Sci. USA 95 3048-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.