Abstract
Accumulation of amyloid-β (Aβ) peptides in the brain has been suggested to be the primary event in sequential progression of Alzheimer's disease (AD). Here, we use Drosophila to examine whether expression of either the human Aβ40 or Aβ42 peptide in the Drosophila brain can induce pathological phenotypes resembling AD. The expression of Aβ42 led to the formation of diffused amyloid deposits, age-dependent learning defects, and extensive neurodegeneration. In contrast, expression of Aβ40 caused only age-dependent learning defects but did not lead to the formation of amyloid deposits or neurodegeneration. These results strongly suggest that accumulation of Aβ42 in the brain is sufficient to cause behavioral deficits and neurodegeneration. Moreover, Drosophila may serve as a model for facilitating the understanding of molecular mechanisms underlying Aβ toxicity and the discovery of novel therapeutic targets for AD.
Alzheimer's disease (AD) is a neurodegenerative disorder characterized clinically by progressive decline in memory accompanied by histological changes, including neuronal loss and the formation of neurofibrillary tangles (NFTs) and senile plaques (1). The accumulation of amyloid-β (Aβ)42 peptide, the major component of senile plaques, has been hypothesized to be the primary event in AD pathogenesis (2, 3). The strongest support for the Aβ hypothesis comes from genetic analyses of familial AD (FAD); most FAD mutations identified in Aβ precursor protein (APP), Presenilin1 (PS1) and Presenilin2 (PS2) genes appear to cause excessive accumulation of Aβ42 (4). Secretion of Aβ peptides is a result of sequential cleavage of APP by β-secretase, a type I transmembrane glycosylated aspartyl protease, and γ-secretase, a large protein complex that includes at least four proteins, Presenilins (PS1 or PS2), Nicastrin, Aph-1, and Pen-2 (for review, see ref. 5). The heterogeneity of γ-secretase cleavage gives rise to a series of Aβ peptides, including the major species Aβ40 and a smaller amount of Aβ42.
To study AD pathogenesis in vivo, a number of AD mouse models have been established and have successfully recapitulated AD-like phenotypes, including abundant amyloid deposits, astroglial activation, synaptic loss and dysfunction, behavioral abnormalities, and neurodegeneration (6-15). In addition to these mouse models, the model systems that allow high-throughput genetic screening will facilitate the discovery of genes involved in AD pathogenesis. Furthermore, one of the intriguing issues that have not been elucidated in these transgenic mice is the pathological roles of each specific Aβ species (i.e., Aβ40 and Aβ42), because currently available mouse models mainly rely on overexpression of APP.
We use a Drosophila model (16) to compare the specific pathological roles of Aβ40 and Aβ42. In Drosophila, all components involved in the protein complex responsible for γ-secretase activity are highly conserved (17), whereas β-secretase activity is absent or very low (18). An APP-like protein (APPL) is also present in flies, although the Aβ domain is not conserved. A null mutation of APPL exhibits behavioral deficits, which are rescued by a human APP transgene (19). Drosophila has been used to study the physiological functions of APP and APPL in synaptogenesis (20), axonal transport (21, 22), and apoptosis (22). To determine whether Drosophila can be used as a model to study the molecular basis of AD pathogenesis, we examined the effects of Aβ40 and Aβ42 in the Drosophila brain using the GAL4-UAS system (23). In particular, we were able to separately analyze and determine specific roles for Aβ40 and Aβ42 in progressive learning defects and neurodegeneration.
Methods
Transgenic Flies. All Aβ transgenic Drosophila strains were obtained from Finelli et al. (ref. 16; see also ref. 24). For behavior experiments, the elav-GAL4c155 line was outcrossed with w1118 (isoCJ1), an isogenic line, for five generations.
Western Blot and Mass Spectrometric Analysis. Fly heads were homogenized in RIPA buffer (50 mM Tris·HCl, pH 8.0/0.5% sodium deoxycholate/1% Triton X-100/150 mM NaCl) containing 1% SDS, ultracentrifuged at 100,000 × g for 1 h, and supernatant was collected (SDS-soluble fraction). Protein extracts were immunoprecipitated with the 4G8 antibody (Signet Laboratories, Dedham, MA), separated on 10-20% Tris-Tricine gel (Invitrogen), and blotted with the 6E10 antiserum (Signet), anti-Aβ40 specific (Alpha Diagnostic, San Antonio, TX), or anti-Aβ42 specific (Oncogene Science) antibodies. SDS-insoluble pellets were further homogenized in 70% formic acid (Sigma) followed by ultracentrifugation at 100,000 × g for 1 h, and supernatant was collected [formic acid (FA) fraction]. FA was evaporated by Speed Vac (Savant, SC100), and protein was resuspended in dimethyl sulfoxiside (Sigma). The signal intensity was quantified by using NIH IMAGE 1.6.2. Mass spectrometric analysis was performed as described (25).
Pavlovian Olfactory Associative Learning. Olfactory associative learning was performed as described (26). Briefly, flies were trained by exposure to electroshock paired with one odor {octanol [10-3 (vol/vol)] or methylcyclohexanol [10-3 (vol/vol)]} for 60 s and subsequent exposure to a second odor without electroshock for 60 s. Immediately after training, learning is measured by allowing flies to choose between the two odors for 120 s. The performance index was calculated by subtracting the number of flies making the incorrect choice from those making the correct one, dividing by the total number of flies, and multiplying by 100. Absolute odor avoidance and electric shock reactivity were quantified as described (27).
Climbing Assay. Twenty flies were placed in a plastic vial and gently tapped to the bottom. The number of flies at the top of the vial was counted after 18 s of climbing under red light (Kodak, GBX-2, Safelight Filter). The data shown represent results from a cohort of flies tested serially for 40-60 days. The experiment was repeated more than three times.
Survival Assay. Twenty to 30 flies were placed in a food vial. Each vial was kept on its side at 25°C, 70% humidity, under a 12-h light-dark cycle. Food vials were changed every 2-3 days, and the dead flies were counted at that time. At least 100 flies were prepared for each genotype, and the experiments were carried out more than three times.
Anatomical Study. To detect neurodegeneration, fixed and permeabilized fly brains were stained with NBD C6-ceramide, followed by counterstaining with propidium iodide (Molecular Probes) (28). Samples were cleared by incubation in FocusClear solution (PacGen, Vancouver) and viewed with a Zeiss LSM 510 confocal microscope with a ×40 C-Apochromat water immersion objective lens. To detect amyloid deposits, fixed and permeabilized brains were treated with 10% formic acid (Acros Organics, Fairlawn, NJ), followed by immunostaining with a mouse monoclonal anti-Aβ antibody (Chemicon). ThioflavinS staining was performed following Fay et al. (29). Transmission electron microscope analysis was performed as described (30).
Results
Expression and Accumulation of Aβ40 and Aβ42 in Transgenic Fly Brains. Each Aβ40 or Aβ42 peptide was fused to the rat preproenkephalin signal peptide at the N terminus to ensure secretion of Aβ peptides once expressed (24). The Aβ fusion constructs have been shown to produce secreted Aβ peptides when expressed in human embryonic kidney cells (24) or in Drosophila S2 cells (16). The Aβ peptides were targeted to express in all neurons in Drosophila by using the GAL4-UAS system (23) (driven by the pan-neuronal elav-GAL4c155 driver; see Methods). Several lines of evidence suggested that Aβ40 or Aβ42 was produced appropriately in the fly brain. First, an Aβ signal of the correct size of 4 kDa was readily detected in Western blots of flies that express either Aβ40 or Aβ42 but not in controls (Fig. 1A Left, arrowhead). Putative oligomeric forms of Aβ peptides were also observed, including a 6-kDa band in Aβ40 flies (as better shown in Fig. 1B, asterisk), and 8- and 12-kDa bands in Aβ42 flies (as better shown in Fig. 1C, asterisks). Second, the Aβ peptides were correctly cleaved from the fused signal peptide, as indicated by the precise molecular weight of Aβ40 and Aβ42 measured by mass spectrometry (Fig. 1D; Mr = 4328.9004 for Aβ40 and 4513.2754 Da for Aβ42). Third, the intactness of the C-terminal end of both Aβ40 and Aβ42 was further confirmed by Western blotting with each C-terminal end-specific antibody (Fig. 1B). Finally, the age-dependent accumulation of Aβ peptides showed biophysical features similar to previous characterization (31-33), i.e., Aβ40 was accumulated in the SDS-soluble fraction (Fig. 1C Upper Right), whereas Aβ42 accumulated in the SDS-insoluble/FA-soluble fraction (Fig. 1C Lower Left). From Western blot analysis of a 3-day-old head (Fig. 1 A Left), we estimated that the expression level of peptides (R = signal intensity compared to Aβ42 males, n = 3) was significantly higher for Aβ40 (R = 3.51 ± 0.53 for males and R = 1.42 ± 0.40 for females) than for Aβ42 (R = 1.00 for males and R = 0.19 ± 0.04 for females), even after taking into account the insoluble fraction (Fig. 1 A Right, arrowhead). The higher expression level of Aβ peptides in male than in female flies can be partly explained by the effect of gene dosage compensation, because the GAL4 promoter is located on the X chromosome.
Fig. 1.
Expression and accumulation profiling of Aβ in transgenic fly heads. (A) Expression levels of Aβ peptides in SDS-soluble (Left) and FA fraction (Right) at 3 days old. (B) Confirmation of the intactness of Aβ40 or Aβ42. 6E10 recognizes the common part of Aβ40 and Aβ42, whereas the 40 or 42 antibody is specific to each C terminus. (C) Age-dependent accumulation of Aβ peptides in SDS-soluble (Upper) and FA fraction (Lower). Arrowheads, monomeric Aβ; asterisks, putative oligomeric forms. (D) Mass spectrometric analysis of Aβ peptide from Aβ40 (Upper) or Aβ42 transgenic fly heads (Lower).
Formation of Amyloid Deposits in Aβ42 but Not in Aβ40 Flies. To determine whether expressed and accumulated Aβ peptides form Aβ deposits in the fly brain, we performed whole-mount immunohistochemical staining. In the neuropil region, 48-day-old Aβ42 fly brains showed the presence of abundant amyloid deposits (Fig. 2B, arrowheads), and both the number and size of the deposits were increased during aging (comparing Fig. 2 A and B, arrowheads). In contrast, such clear deposits were not observed in Aβ40 or control brains (Fig. 2 C and D). Importantly, the staining signal observed in Aβ40 brain (Fig. 2C, asterisk) is not an Aβ deposit but the expression of Aβ40 in peduncle structure, which is the axon bundle of mushroom body neurons (Kenyon cells). In the Kenyon cell body region, strong Aβ staining was observed in both Aβ40 and Aβ42 brains (Fig. 2 E-G) but not in control (Fig. 2H), confirming the specificity of the antibody.
Fig. 2.
Detection of Aβ deposits in fly brain. (A-H) Whole-mount Aβ immunostaining (green) and nuclear staining (red) in the neuropil region (A-D) and Kenyon cell layer (E-H). Arrowheads, deposited Aβ42 (A and B); asterisks, the peduncle structure, an axon bundle of Kenyon cells. (I-N) ThioflavinS staining in the Kenyon cell (I-K) and neuropil regions (L-N). ThioflavinS-positive deposits were detected in Aβ42 flies (I, arrows) but not in Aβ40 or control (J and K, arrowheads). The fiber structures seen in I-K are tracheas. Pd, peduncle; Kn, Kenyon cell layer; Ca, calyx; the dendritic structure of Kenyon cells. [Bar (D, H, and N) = 50 μm.]
We also performed thioflavinS staining to label the Aβ deposits containing amyloid fibril structures. The thioflavinS-positive deposits can be observed in the Kenyon cell body region of Aβ42 brains (Fig. 2I, arrows) but not in Aβ40 or control brains (Fig. 2 J and K, arrowheads). In contrast, no thioflavinS staining was detected in the neuropil region (Fig. 2 L-N) even in Aβ42 fly brains, which had abundant immunopositive Aβ deposits as shown above (comparing Fig. 2 B and L). We further analyzed the Aβ42 brains in both the Kenyon cell body and the neuropil regions by transmission electron microscopy; however, there was no evidence of clear amyloid fibril structure in both regions. These results suggest that observed Aβ42 deposits in our fly brain are mainly diffused Aβ deposits without clear amyloid fibril core structures, although some of the deposits were stained by thioflavinS. In summary, both Aβ40 and -42 peptides accumulated during aging (see Fig. 1C); however, only Aβ42 peptides could form diffused amyloid deposits in the fly brains.
Age-Dependent Olfactory Learning Defects Induced by Aβ40 and Aβ42. Learning and memory of these flies were tested by using a Pavlovian olfactory learning assay (26). For 2- to 3-day-old adult flies, no significant defect was observed in either Aβ40 or Aβ42 flies (Fig. 3A). Both Aβ40 and Aβ42 flies began to show a subtle but statistically significant learning defect at 6-7 days old (Fig. 3B, asterisks). This decline became more obvious for 14- to 15-day-old flies (Fig. 3C, asterisks). The defect was greater in male flies than in females, consistent with higher expression of peptides in male flies (see Fig. 1 A and text). Controls did not show the sex difference at any time point (Fig. 3 A-C, compare elav/Y and elav/+). For Aβ40 flies, learning defects were observed only in males but not in females (Fig. 3 A-C), suggesting a much higher level of Aβ40 than Aβ42 is required to affect learning ability (see Fig. 1 A and text). We also examined odor avoidance and electric-shock reactivity, two sensorimotor activities necessary for performing the learning task. There was no significant difference among all groups for 14- to 15-day-old flies for shock reactivity and avoidance of the odor methylcyclohexanol (Fig. 3D). Avoidance of octanol is slightly lower in flies expressing either Aβ40 or Aβ42 as compared to the UASAβ42/+ and UAS-Aβ40/+ controls but not significant compared to the elav controls (Fig. 3D). This slight difference should not contribute to observed learning defects, because Aβ40 female flies showed a normal learning score (Fig. 3 A-C, elav/+;;UAS-Aβ40/+). That learning was normal in Aβ40 female flies also suggests that progressive learning defects are a result of Aβ toxicity rather than of genetic background or stress imposed by the expression of peptides.
Fig. 3.
Progressive loss of learning ability in Aβ flies assayed by a Pavlovian olfactory associative learning paradigm. (A-C) Learning abilities at 2-3 (A), 6-7 (B), and 14-15 days old (C) are presented in mean ± SEM. The numbers of experiments are indicated on top of the bars. Asterisks show statistical difference from controls [(α<0.05, Tukey-Kramer honestly significant difference (HSD)]. (D) No statistical difference in olfactory acuity and shock reactivity between experimental genotype and appropriate control genotypes at 14-15 days old (n = 8; except n = 6 for octanol olfactory acuity for elav/Y;UASAβ42/+ and shock reactivity for UAS-Aβ40/+) at the level of α = 0.05 (Tukey-Kramer HSD).
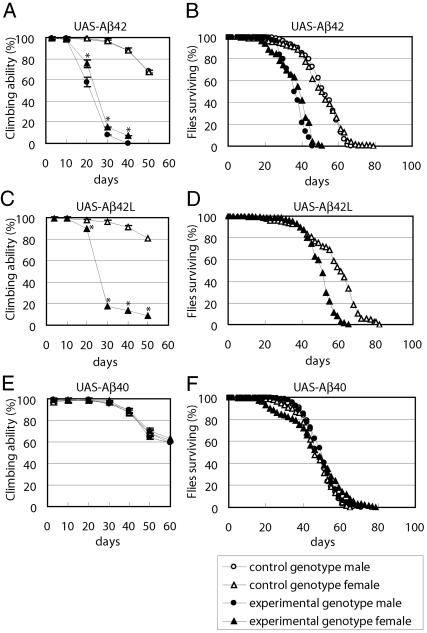
Climbing Disability and Shortened Life Span in Aβ42 but Not in Aβ40 Flies. Aβ42 flies started to show locomotor dysfunction after 3 weeks of age. The climbing ability of flies in response to light tapping (34) began to decline significantly after 20 days in Aβ42 flies but not in Aβ40 flies (Figs. 4 A and E). The presence of alterations in motor activity prevents us from examining learning ability after 3 weeks. Even older Aβ42 flies stayed at the bottom of the vial and could not climb up the wall. Accompanying this locomotor defect, the life span of Aβ42 flies was also much shorter, whereas Aβ40 flies were not affected (Figs. 4 B and F). To confirm the effect of Aβ42, we examined another independently isolated Aβ42 line, which has a lower level of Aβ42 expression (UAS-Aβ42L). Similar results were obtained regarding both climbing ability and life span (Fig. 4 C and D). We could examine only females of this line, because both UAS-Aβ42L and elav-GAL4c155 transgenes are located on the X chromosome.
Fig. 4.
Progressive climbing disability and shortened life span in Aβ42 flies. (A, C, and E) Climbing ability in Aβ42 flies (A and C, asterisks, P < 0.001, Student's t test) and Aβ40 flies (E). The SDs of 10 trials are within the symbols. (B, D, and F) Survival rate of Aβ42 flies (B and D) and Aβ40 flies (F).
Late-Onset Progressive Neurodegeneration Caused by Aβ42 but Not by Aβ40. Anatomical analysis by confocal microscope revealed extensive neurodegeneration in aged Aβ42 but not in Aβ40 flies. In 45-day-old Aβ42 flies, we observed severe neuronal loss, as indicated by the number of vacuoles in the Kenyon cell layer (Fig. 5D, arrowheads), a brain region crucial for olfactory learning (35-37). Degeneration was also seen in other brain regions (Figs. 5 E and F). In contrast, age-matched Aβ40 or control flies did not show obvious cell loss (Figs. 5 G and H). To eliminate the possibility that observed neurodegeneration is a nonspecific effect due to fly death, we analyzed 55-day-old Aβ40 or control flies. We did not see much degeneration in either group of brains (see Table 1, which is published as supporting information on the PNAS web site). To determine the time of onset of degeneration, we examined 3-, 14-, and 30-day-old Aβ42 fly brains. There was no detectable abnormality in 3- and 14-day-olds (Figs. 5 A and B), whereas a small amount of cell loss started to appear in 30-day-old brains (Fig. 5C, arrowheads; see also Table 1). The average number and area of vacuoles are summarized in Table 1. Similar results were also observed when using a different Gal4 driver, OK107, which drives peptide expression preferentially in the mushroom body structure (Fig. 5I, arrowheads; data not shown). Cell death in the Kenyon cell layer of Aβ42 flies was analyzed in the ultrastructural level. The degenerating neurons were readily identified, and vacuoles were detected as cell loss. The majority of dying neurons showed the typical features of necrotic-type cell death: digested cytoplasm (electron-lucent) with swollen mitochondria (Fig. 5J, arrows), whereas nuclei were relatively intact (Fig. 5J, indicated by N). Besides Aβ depositions, another characteristic lesion in AD patient brains is intracellulary formed protein aggregates called NFTs. The major component of NFTs is hyperphosphorylated τ protein, which is assembled into paired helical filament (PHF) structure (38-40). The pathological interaction between Aβ depositions and NFT formation remains to be elucidated, because none of the AD mouse models carrying abundant amyloid deposits developed NFTs (6, 7, 9-12, 14, 15). Therefore, we were motivated to determine whether accumulation of Aβ42 leads to the formation of NFT and/or PHF structure with fly endogenous τ protein (41). PHF τ was not detected by either immunoblotting or electron microscopy in Aβ42 fly head tissues.
Fig. 5.
Late-onset progressive neurodegeneration in Aβ42 brains. (A-H) Progressive neuronal loss occurred in Aβ42 (A-F, arrowheads) but not in Aβ40 or control brains (G and H). (A-D, G, and H) The Kenyon cell region. (E) Medial brain. (F) Lateral brain. Green, neuropil structure; red, nuclei. Arrows in D indicate the aggregates, presumably amyloid deposits. Kn, Kenyon cell layer; Ca, calyx; PB, protocerebral bridge; OL, optic lobe. [Bars (C and H) = 50 μm.] (I) Neuronal loss induced by different Gal4 line, OK107. (Bar = 50 μm.) (J) Ultrastructural analysis of degenerating neurons with digested cytoplasm (electron-lucent) and swollen mitochondria (arrows). N, nucleus. (Bar = 1 μm.)
Discussion
In this study, we have established accumulation of either Aβ40 or Aβ42 peptides in the Drosophila brain induces progressive learning defects, but only Aβ42 is capable of causing the formation of diffused Aβ deposits, locomotor dysfunction, neurodegeneration, and premature death. It is remarkable to note that in an organism with a life span of 2-3 months, accumulation of Aβ42 induces the sequential progression of pathological symptoms resembling those in mouse AD models (6-15) and AD patients (1, 42). Intriguingly, the onset of learning defects by Aβ42 occurs much earlier than that of degeneration in the flies, similar to that observed in mouse AD models and AD patients (7, 8, 43-46). Furthermore, that both Aβ40 and Aβ42 affect learning but only Aβ42 causes degeneration leads to the speculation that neuronal dysfunction and neurodegeneration may be mediated by different mechanisms.
We have concluded that most amyloid deposits in Aβ42 fly brains are not cored (mature) plaques but diffused (immature) deposits, because we could not detect clear amyloid fibrils at the ultrastructural level. The lack of mature plaques may be due to the short life span of Aβ42 flies (within 50 days) and/or the absence of potential cofactors needed to form cored plaques in the fly brain. On the other hand, this result indicates that the cored plaque formation is not necessary to induce any of the pathological phenotypes observed in the Aβ42 flies.
Aβ42 flies exhibit severe neurodegeneration in the absence of cored plaques containing clear amyloid fibril structures and the formation of NFTs. This has a parallel in studies that show polyglutamine-(47) or τ-induced (48, 49) neurodegeneration can be dissociated from the formation of nuclear inclusion or NFTs, respectively. These facts support the notion that an ordered prefibrillar oligomer, or protofibril, but not the fibrillar form, may be responsible for cell death (50).
It has been reported that cognitive defects and Aβ deposits were not well correlated in AD patients (43, 46). In AD mouse models, the development of synaptic dysfunction and/or behavioral deficits precedes the formation of amyloid deposits (13, 51, 52).
These facts are reminiscent of that Aβ40 flies showed learning defects without amyloid deposits. As for the Aβ42 flies, we cannot conclude whether the deposits contribute to behavioral defects, because the 3-day-old flies already developed small amounts of deposits. Recently, soluble oligomeric forms of Aβ peptides have been suggested to be responsible for synaptic dysfunctions (53). We detected putative oligomeric forms of both Aβ40 and Aβ42 in the fly brain (Fig. 1 A), whereas the pathological roles of these oligomers remain elusive.
Conclusion
This study strongly supports the idea that excessive accumulation of Aβ42 is sufficient to cause memory defects and neurodegeneration resembling AD and suggests that the molecular basis underlying Aβ toxicity is conserved over different organisms. Our Aβ flies may serve as a model for the genetic and pharmacological screening system for AD therapeutics targeting Aβ-induced neurotoxicity and Aβ clearance, as well as for the understanding of the molecular and cellular basis of AD pathogenesis.
Supplementary Material
Acknowledgments
We thank Dr. M. E. Fortini (National Cancer Institute, Frederick, MD) for the gift of the Drosophila τ antibody DT-2; M. Myers for mass spectrometric analysis; and D. Kretzschmar, M. Saitoe, S. Xia, and K. A. Iijima for helpful discussions. We also thank Drs. F. Hannan and C. Margulies for critical reading and Drs. T. Tully, J. Dubnau, H. Guo, and Y. Wang for comments on the manuscript. This work was supported by Alzheimer's Association Grant NIRG-03-5239 (to K.I.), by the National Institutes of Health (Y.Z.), and by grants from the Brain Research Center of the University System of Taiwan, National Science Council, and Technology Development Program of Ministry of Economy to (to A.-S.C.).
Abbreviations: AD, Alzheimer's disease; Aβ, amyloid-β; NFTs, neurofibrillary tangles; APP, β-amyloid precursor protein; FA, formic acid.
References
- 1.Selkoe, D. J. (2001) Physiol. Rev. 81, 741-766. [DOI] [PubMed] [Google Scholar]
- 2.Hardy, J. & Allsop, D. (1991) Trends Pharmacol. Sci. 12, 383-388. [DOI] [PubMed] [Google Scholar]
- 3.Hardy, J. & Selkoe, D. J. (2002) Science 297, 353-356. [DOI] [PubMed] [Google Scholar]
- 4.Price, D. L., Tanzi, R. E., Borchelt, D. R. & Sisodia, S. S. (1998) Annu. Rev. Genet. 32, 461-493. [DOI] [PubMed] [Google Scholar]
- 5.Sisodia, S. S. & St George-Hyslop, P. H. (2002) Nat. Rev. Neurosci. 3, 281-290. [DOI] [PubMed] [Google Scholar]
- 6.Games, D., Adams, D., Alessandrini, R., Barbour, R., Berthelette, P., Blackwell, C., Carr, T., Clemens, J., Donaldson, T., Gillespie, F., et al. (1995) Nature 373, 523-527. [DOI] [PubMed] [Google Scholar]
- 7.Hsiao, K., Chapman, P., Nilsen, S., Eckman, C., Harigaya, Y., Younkin, S., Yang, F. & Cole, G. (1996) Science 274, 99-102. [DOI] [PubMed] [Google Scholar]
- 8.Chen, G., Chen, K. S., Knox, J., Inglis, J., Bernard, A., Martin, S. J., Justice, A., McConlogue, L., Games, D., Freedman, S. B., et al. (2000) Nature 408, 975-979. [DOI] [PubMed] [Google Scholar]
- 9.Mucke, L., Masliah, E., Yu, G. Q., Mallory, M., Rockenstein, E. M., Tatsuno, G., Hu, K., Kholodenko, D., Johnson-Wood, K. & McConlogue, L. (2000) J. Neurosci. 20, 4050-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duff, K., Eckman, C., Zehr, C., Yu, X., Prada, C. M., Perez-tur, J., Hutton, M., Buee, L., Harigaya, Y., Yager, D., et al. (1996) Nature 383, 710-713. [DOI] [PubMed] [Google Scholar]
- 11.Borchelt, D. R., Ratovitski, T., van Lare, J., Lee, M. K., Gonzales, V., Jenkins, N. A., Copeland, N. G., Price, D. L. & Sisodia, S. S. (1997) Neuron 19, 939-945. [DOI] [PubMed] [Google Scholar]
- 12.Citron, M., Westaway, D., Xia, W., Carlson, G., Diehl, T., Levesque, G., Johnson-Wood, K., Lee, M., Seubert, P., Davis, A., et al. (1997) Nat. Med. 3, 67-72. [DOI] [PubMed] [Google Scholar]
- 13.Hsia, A. Y., Masliah, E., McConlogue, L., Yu, G. Q., Tatsuno, G., Hu, K., Kholodenko, D., Malenka, R. C., Nicoll, R. A. & Mucke, L. (1999) Proc. Natl. Acad. Sci. USA 96, 3228-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sturchler-Pierrat, C., Abramowski, D., Duke, M., Wiederhold, K. H., Mistl, C., Rothacher, S., Ledermann, B., Burki, K., Frey, P., Paganetti, P. A., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 13287-13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quon, D., Wang, Y., Catalano, R., Scardina, J. M., Murakami, K. & Cordell, B. (1991) Nature 352, 239-241. [DOI] [PubMed] [Google Scholar]
- 16.Finelli, A. L., Kelkar, A., Song, H.-J., Yang, H. & Konsolaki, M. (2004) Mol. Cell. Neurosci., in press. [DOI] [PubMed]
- 17.Takasugi, N., Tomita, T., Hayashi, I., Tsuruoka, M., Niimura, M., Takahashi, Y., Thinakaran, G. & Iwatsubo, T. (2003) Nature 422, 438-441. [DOI] [PubMed] [Google Scholar]
- 18.Fossgreen, A., Bruckner, B., Czech, C., Masters, C. L., Beyreuther, K. & Paro, R. (1998) Proc. Natl. Acad. Sci. USA 95, 13703-13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo, L., Tully, T. & White, K. (1992) Neuron 9, 595-605. [DOI] [PubMed] [Google Scholar]
- 20.Torroja, L., Packard, M., Gorczyca, M., White, K. & Budnik, V. (1999) J. Neurosci. 19, 7793-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torroja, L., Chu, H., Kotovsky, I. & White, K. (1999) Curr. Biol. 9, 489-492. [DOI] [PubMed] [Google Scholar]
- 22.Gunawardena, S. & Goldstein, L. S. (2001) Neuron 32, 389-401. [DOI] [PubMed] [Google Scholar]
- 23.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401-415. [DOI] [PubMed] [Google Scholar]
- 24.Cescato, R., Dumermuth, E., Spiess, M. & Paganetti, P. A. (2000) J. Neurochem. 74, 1131-1139. [DOI] [PubMed] [Google Scholar]
- 25.Wang, R., Sweeney, D., Gandy, S. E. & Sisodia, S. S. (1996) J. Biol. Chem. 271, 31894-31902. [DOI] [PubMed] [Google Scholar]
- 26.Tully, T. & Quinn, W. G. (1985) J. Comp. Physiol. A 157, 263-277. [DOI] [PubMed] [Google Scholar]
- 27.Guo, H. F., Tong, J., Hannan, F., Luo, L. & Zhong, Y. (2000) Nature 403, 895-898. [DOI] [PubMed] [Google Scholar]
- 28.Chiang, A. S., Liu, Y. C., Chiu, S. L., Hu, S. H., Huang, C. Y. & Hsieh, C. H. (2001) J. Comp. Neurol. 440, 1-11. [DOI] [PubMed] [Google Scholar]
- 29.Fay, D. S., Fluet, A., Johnson, C. J. & Link, C. D. (1998) J. Neurochem. 71, 1616-1625. [DOI] [PubMed] [Google Scholar]
- 30.Kretzschmar, D., Hasan, G., Sharma, S., Heisenberg, M. & Benzer, S. (1997) J. Neurosci. 17, 7425-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gravina, S. A., Ho, L., Eckman, C. B., Long, K. E., Otvos, L., Jr., Younkin, L. H., Suzuki, N. & Younkin, S. G. (1995) J. Biol. Chem. 270, 7013-7016. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, N., Iwatsubo, T., Odaka, A., Ishibashi, Y., Kitada, C. & Ihara, Y. (1994) Am. J. Pathol. 145, 452-460. [PMC free article] [PubMed] [Google Scholar]
- 33.Iwatsubo, T., Odaka, A., Suzuki, N., Mizusawa, H., Nukina, N. & Ihara, Y. (1994) Neuron 13, 45-53. [DOI] [PubMed] [Google Scholar]
- 34.Ganetzky, B. & Flanagan, J. R. (1978) Exp. Gerontol. 13, 189-196. [DOI] [PubMed] [Google Scholar]
- 35.de Belle, J. S. & Heisenberg, M. (1994) Science 263, 692-695. [DOI] [PubMed] [Google Scholar]
- 36.Connolly, J. B., Roberts, I. J., Armstrong, J. D., Kaiser, K., Forte, M., Tully, T. & O'Kane, C. J. (1996) Science 274, 2104-2107. [DOI] [PubMed] [Google Scholar]
- 37.Grotewiel, M. S., Beck, C. D., Wu, K. H., Zhu, X. R. & Davis, R. L. (1998) Nature 391, 455-460. [DOI] [PubMed] [Google Scholar]
- 38.Goedert, M., Wischik, C. M., Crowther, R. A., Walker, J. E. & Klug, A. (1988) Proc. Natl. Acad. Sci. USA 85, 4051-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondo, J., Honda, T., Mori, H., Hamada, Y., Miura, R., Ogawara, M. & Ihara, Y. (1988) Neuron 1, 827-834. [DOI] [PubMed] [Google Scholar]
- 40.Wischik, C. M., Novak, M., Thogersen, H. C., Edwards, P. C., Runswick, M. J., Jakes, R., Walker, J. E., Milstein, C., Roth, M. & Klug, A. (1988) Proc. Natl. Acad. Sci. USA 85, 4506-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heidary, G. & Fortini, M. E. (2001) Mech. Dev. 108, 171-178. [DOI] [PubMed] [Google Scholar]
- 42.Davis, K. L. & Samuels, S. C. (1998) Pharmacological Management of Neurological and Psychiatric Disorders (McGraw-Hill, New York).
- 43.Dickson, D. W., Crystal, H. A., Bevona, C., Honer, W., Vincent, I. & Davies, P. (1995) Neurobiol. Aging 16, 285-304. [DOI] [PubMed] [Google Scholar]
- 44.Morris, J. C., Storandt, M., McKeel, D. W., Jr., Rubin, E. H., Price, J. L., Grant, E. A. & Berg, L. (1996) Neurology 46, 707-719. [DOI] [PubMed] [Google Scholar]
- 45.Selkoe, D. J. (2002) Science 298, 789-791. [DOI] [PubMed] [Google Scholar]
- 46.Terry, R. D., Masliah, E., Salmon, D. P., Butters, N., DeTeresa, R., Hill, R., Hansen, L. A. & Katzman, R. (1991) Ann. Neurol. 30, 572-580. [DOI] [PubMed] [Google Scholar]
- 47.Warrick, J. M., Chan, H. Y., Gray-Board, G. L., Chai, Y., Paulson, H. L. & Bonini, N. M. (1999) Nat. Genet. 23, 425-428. [DOI] [PubMed] [Google Scholar]
- 48.Wittmann, C. W., Wszolek, M. F., Shulman, J. M., Salvaterra, P. M., Lewis, J., Hutton, M. & Feany, M. B. (2001) Science 293, 711-714. [DOI] [PubMed] [Google Scholar]
- 49.Jackson, G. R., Wiedau-Pazos, M., Sang, T. K., Wagle, N., Brown, C. A., Massachi, S. & Geschwind, D. H. (2002) Neuron 34, 509-519. [DOI] [PubMed] [Google Scholar]
- 50.Caughey, B. & Lansbury, P. T. (2003) Annu. Rev. Neurosci. 26, 267-298. [DOI] [PubMed] [Google Scholar]
- 51.Holcomb, L., Gordon, M. N., McGowan, E., Yu, X., Benkovic, S., Jantzen, P., Wright, K., Saad, I., Mueller, R., Morgan, D., et al. (1998) Nat. Med. 4, 97-100. [DOI] [PubMed] [Google Scholar]
- 52.Koistinaho, M., Ort, M., Cimadevilla, J. M., Vondrous, R., Cordell, B., Koistinaho, J., Bures, J. & Higgins, L. S. (2001) Proc. Natl. Acad. Sci. USA 98, 14675-14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walsh, D. M., Klyubin, I., Fadeeva, J. V., Cullen, W. K., Anwyl, R., Wolfe, M. S., Rowan, M. J. & Selkoe, D. J. (2002) Nature 416, 535-539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.