SUMMARY
Mitochondrial respiratory chain disorders are characterized by loss of electron transport chain (ETC) activity. Although the causes for many such diseases are known, there is a lack of effective therapies. To identify genes that confer resistance to severe ETC dysfunction when inactivated, we performed a genome-wide genetic screen in haploid human cells with the mitochondrial complex III inhibitor, antimycin. This screen revealed that loss of ATPIF1 strongly protects against antimycin-induced ETC dysfunction and cell death by allowing for maintenance of mitochondrial membrane potential. ATPIF1 loss protects against other forms of ETC dysfunction and is even essential for the viability of human ρ0 cells lacking mitochondrial DNA, a common system for studying ETC dysfunction. Importantly, inhibition of ATPIF1 ameliorates complex III blockade in primary hepatocytes, a cell type afflicted in severe mitochondrial disease. Taken together, these results suggest that inhibition of ATPIF1 can ameliorate severe ETC dysfunction in mitochondrial pathology.
INTRODUCTION
Defects in the activity of the electron transport chain (ETC) are the causative pathology in a diverse family of genetic diseases known as mitochondrial respiratory chain disorders. Patients with these diseases can often present with abnormalities in multiple organ systems (Pfeffer et al., 2012, DiMauro and Schon, 2003, Spinazzola et al., 2006). Although some mitochondrial respiratory chain disorders cause relatively mild abnormalities, such as exercise intolerance, there are severe forms of respiratory chain disorders that can lead to life-threatening loss of tissue parenchyma and organ failure (Morris, 1999, Lee and Sokol, 2007, DiMauro and Schon, 2003, Spinazzola et al., 2006). Yet despite an extensive characterization of the mechanisms underlying these diseases, there is a paucity of effective therapies to ameliorate severe respiratory chain dysfunction. Indeed, most efforts to date, such as dietary supplementation with small molecules and vitamins that can increase ETC activity or decrease reactive oxygen species have not demonstrated any clear efficacy across clinical trials, thus underscoring the need for novel therapeutic strategies (Pfeffer et al., 2012, Schon et al., 2010).
Genetic and chemical screens in mammalian cells have previously identified modulators of mitochondrial dynamics (Lefebvre et al., 2013, Kitami et al., 2012, Gohil et al., 2010, Yoon et al., 2010), but to our knowledge there have been no genetic screens carried out in mammalian cells to identify gene products that, when inactivated, increase survival under ETC dysfunction. Beginning with a positive-selection screen in human cells using the mitochondrial complex III inhibitor antimycin, we find that loss of ATPIF1 is protective against complex III blockade, as well as a multitude of other insults to the ETC, leading us to propose inhibition of ATPIF1 as a strategy for ameliorating severe mitochondrial respiratory chain disorders.
RESULTS AND DISCUSSION
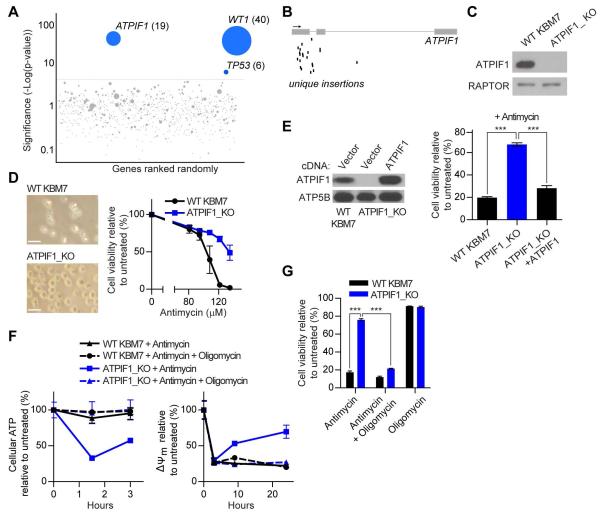
To identify genes whose products modulate sensitivity to ETC inhibition, we performed a genome-wide, insertional mutagenesis screen in the near-haploid KBM7 human cell line with antimycin, a complex III inhibitor of the ETC. This technology has been used successfully in the past to identify numerous proteins essential for the cytotoxicity of microbial factors (Carette et al., 2011b, Guimaraes et al., 2011), as well as transporters for toxic small molecules (Birsoy et al., 2013). In brief, we generated a library of mutagenized haploid KBM7 cells harboring approximately 70 million insertions that encompass more than 95% of all genes expressed in KBM7 cells (Carette et al., 2011a). Mutagenized cells were then treated with antimycin for three weeks and the surviving cells expanded and pooled. Insertions in the surviving population were mapped to the human genome using massively parallel sequencing. To identify genomic loci enriched for gene-trap insertions, we performed a proximity index analysis and identified several candidate genes: ATPIF1 (P = 3.04 × 10−43), WT1 (P = 1.78 × 10−40), and TP53 (P = 6.93 × 10−8) (Figure 1A). Because both WT1 and TP53 are tumor suppressors (Sherr, 2004) and would therefore be less attractive therapeutic targets, we focused our attention on ATPIF1. ATPIF1 is a highly-conserved mitochondrial protein that inhibits the ATPase activity of the F1-F0 ATP synthase and has been found to affect a variety of metabolic parameters, such as aerobic glycolysis (Sánchez-Cenizo et al., 2010), ATP synthase dimerization (García et al., 2006), and mitochondrial cristae density (Campanella et al., 2008, Campanella et al., 2009).
Figure 1. Haploid genetic screen identifies loss of ATPIF1 as protective against complex III inhibition.
(A) Mutagenized KBM7 cells were treated with antimycin and resistant cells were pooled. Gene-trap insertions were identified by massively parallel sequencing and mapped to the human genome. The y-axis represents the statistical significance of a given gene, while the x-axis represents the collection of genes with insertions. The red line indicates the cut-off of statistical significance chosen to determine whether a gene scored as a hit in the screen. For ATPIF1, WT1, and TP53, the number of unique insertions per gene is given in parentheses. (B) Map of unique insertions in ATPIF1 in the resistant cell population. The arrow denotes 5′-3′ directionality, boxes represent exons, and black bars indicate insertions. (C) Immunoblots for indicated proteins in WT and ATPIF1_KO KBM7 cells. (D) Micrographs (left) and viability (right) of WT and ATPIF1_KO KBM7 cells treated with antimycin for 4 days. Error bars are ± s.e.m. (n = 3). Scale bars, 20 μm. (E) Immunoblots for indicated proteins in WT, ATPIF1_KO, and ATPIF1_KO KBM7 cells with restored ATPIF1 expression (left) and viability of cells treated with antimycin (135 μM) for 2 days (right). Error bars are ± s.e.m. (n = 3). ***P < 0.001. (F) Cellular ATP (left) and ΔΨm (right) in WT and ATPIF1_KO KBM7 cells treated with antimycin (135 μM) and oligomycin (1 μM). Error bars are ± s.e.m. (n = 3). (G) Viability of WT and ATPIF1_KO KBM7 cells treated with antimycin (135 μM) and oligomycin (1 μM) for 2 days. Error bars are ± s.e.m. (n = 3). ***P < 0.001. See also Figures S1, S2, S3, and Table S1.
To investigate the role of ATPIF1 loss in protecting cells against complex III blockade, we isolated a KBM7 clone harboring a gene-trap insertion of ATPIF1 (ATPIF1_KO) and confirmed that it did not express detectable amounts of ATPIF1 protein (Figures 1B and 1C). Consistent with the results of our screen, ATPIF1_KO cells were substantially more resistant to antimycin-induced cell death than their wildtype (WT) counterparts (Figure 1D). Additionally, re-expression of ATPIF1 in ATPIF1_KO cells almost completely restored their sensitivity to antimycin (Figure 1E). As an independent confirmation of our findings, WT KBM7 cells expressing shRNAs targeting ATPIF1 also exhibited increased resistance to antimycin as well (Figure S1).
To probe the mechanism by which ATPIF1 loss can confer resistance to complex III inhibition, we examined the effects of antimycin on the metabolism and mitochondrial function of WT and ATPIF1_KO KBM7 cells. Upon inhibition of the ETC, mitochondrial membrane potential (ΔΨm) decreases and the F1-F0 ATP synthase reverses, consuming ATP to pump protons into the intermembrane space (Campanella et al., 2008, Campanella et al., 2009, Lefebvre et al., 2013, Lu et al., 2001). Normally an inactive tetramer, ATPIF1 dissociates into active dimers upon a large decrease in ΔΨm and subsequently inhibits reversal of the F1-F0 ATP synthase, an adaptive mechanism to prevent ATP consumption during periods of nutrient and oxygen deprivation (Cabezon et al., 2001, Fujikawa et al., 2012, Lu et al., 2001, Campanella et al., 2008, Campanella et al., 2009). However, in short-term experiments, decreased ATPIF1 activity during ETC dysfunction allows for maintenance of ΔΨm at the expense of ATP via reversal of the F1-F0 ATP synthase, but it is unclear whether maintenance of ΔΨm or conservation of ATP is the more important process for survival under ETC dysfunction (Campanella et al., 2008, Campanella et al., 2009, Lefebvre et al., 2013). Consistent with the F1-F0 ATP synthase operating in reverse, we observed that ATPIF1_KO cells had decreased ATP but increased ΔΨm upon antimycin treatment, as compared to WT KBM7 cells (Figure 1F). Metabolite profiling of ATPIF1_KO cells under antimycin treatment also revealed a greater depletion of glycolytic intermediates, in agreement with the increased ATP demand under conditions of ETC inhibition (Figure S2, Table S1). Importantly, the differences seen in ATP, ΔΨm, and overall survival under antimycin could be eliminated by co-treatment with oligomycin, a potent inhibitor of the F1-F0 ATP synthase (Figures 1F and 1G). It is unlikely that the effects of oligomycin on antimycin-treated cells were a result of additive toxicity because oligomycin itself had no effect on the viability of either WT KBM7 or ATPIF1_KO cells (Figure 1G). Of note, the addition of oligomycin to antimycin-treated ATPIF1_KO cells decreased ΔΨm, increased ATP levels, but led to decreased survival, suggesting that maintenance of ΔΨm is more important than preservation of ATP in ameliorating complex III blockade in KBM7 cells. To rule out any effects of ATPIF1 loss on general mitochondrial metabolism and cellular physiology, we also examined the mitochondrial mass, mitochondrial DNA (mtDNA) copy number, mitochondrial ultrastructure, and resting ΔΨm, ATP, viability, and oxygen consumption of WT and ATPIF1_KO KBM7 cells, but found no significant differences (Campanella et al., 2008) (Figure S3). Collectively, these data demonstrate that ATPIF1 loss confers resistance to complex III blockade through maintenance of ΔΨm via reversal of the F1-F0 ATP synthase.
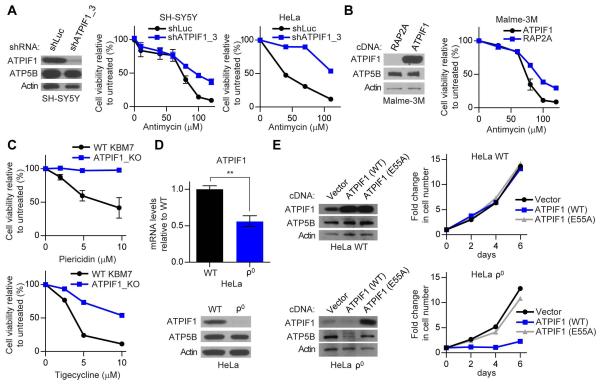
We next sought to determine if the effects of ATPIF1 loss on KBM7 cells were generalizable to other cell lines and additional forms of ETC dysfunction. Consistent with the results in KBM7 cells, SH-SY5Y and HeLa cells expressing an shRNA targeting ATPIF1 were more resistant to antimycin than cells expressing a control hairpin (Figure 2A). In addition, we found that overexpression of ATPIF1 in Malme-3M, a cell line with low endogenous levels of ATPIF1, increased their sensitivity to antimycin (Figure 2B). To investigate whether the protective effect of ATPIF1 loss was limited to only complex III inhibition, we tested a variety of pharmacological and genetic models of ETC dysfunction. ATPIF1_KO KBM7 cells were substantially more resistant to both piericidin, an inhibitor of complex I (Darrouzet et al., 1998), and tigecycline, an inhibitor of mitochondrial translation (škrtić et al., 2011), when compared to their WT counterparts (Figure 2C). Taken together, these data demonstrate that the levels of ATPIF1 can modulate sensitivity to different forms of ETC dysfunction in various human cell lines.
Figure 2. Loss of ATPIF1 is beneficial in both pharmacological and genetic models of ETC dysfunction.
(A) Immunoblots for indicated proteins in SH-SY5Y cells expressing a control shRNA against Luciferase (shLuc) or an shRNA against ATPIF1 (shATPIF1_3) (left). Viability of SH-SY5Y (middle) and HeLa (right) cells treated with antimycin for 4 days. Error bars are ± s.e.m. (n = 3). (B) Immunoblots for indicated proteins in Malme-3M cells overexpressing control RAP2A or ATPIF1 (left). Viability of Malme-3M cells treated with antimycin for 4 days (right). Error bars are ± s.e.m. (n = 3). (C) Viability of WT and ATPIF1_KO KBM7 cells treated with piericidin (top) or tigecycline (bottom) for 4 days. Error bars are ± s.e.m. (n = 3). (D) Relative ATPIF1 mRNA levels (top) and immunoblots for indicated proteins (bottom) in HeLa WT and ρ0 cells. Error bars are ± s.e.m. (n = 3). **P < 0.01. (E) Immunoblots (left) and relative proliferation (right) of HeLa WT and ρ0 cells transduced with control vector, ATPIF1 (WT), or ATPIF1 (E55A) constructs. Error bars are ± s.e.m. (n = 3).
The observation that ATPIF1_KO KBM7 cells were more resistant to inhibition of complex I, complex III, and mitochondrial protein synthesis raised the possibility that ATPIF1 loss could ameliorate the effects of dysfunction in multiple components of the ETC. To test this genetically, we examined ρ0 cells, which are devoid of any mtDNA and consequently have defects in complexes I, III, and IV, resulting in undetectable ETC activity (Jazayeri et al., 2003). To our surprise, we found that HeLa ρ0 cells intrinsically possess low mRNA and protein levels of ATPIF1, when compared to their WT counterparts (Figure 2D). Previous work has shown that ρ0 cells maintain ΔΨm by using the electrogenic exchange of ATP and ADP, coupled to ATP hydrolysis by an F1-F0 ATP synthase defective in pumping protons, and that this activity is important for cellular health (Buchet and Godinot, 1998, Appleby et al., 1999). We therefore hypothesized that there could be a strong selective pressure to decrease ATPIF1 levels under severe ETC dysfunction in order to facilitate reversal of the F1-F0 ATP synthase. A reduction of ATPIF1 in 143b ρ0 cells was observed recently, although the functional significance of this reduction on cell viability was not investigated (Lefebvre et al., 2013). To address this, we overexpressed WT ATPIF1 or a mutant ATPIF1 harboring an E55A substitution that renders the protein unable to interact with the F1-F0 ATP synthase (Ichikawa et al., 2001). Overexpression of WT ATPIF1, but not E55A ATPIF1, strongly impaired proliferation in HeLa ρ0 cells but not in HeLa WT cells (Figure 2E). The differences observed between WT and E55A ATPIF1 were not simply a result of E55A ATPIF1 protein instability because both variants of ATPIF1 were overexpressed to a similar degree, as seen in the immunoblots of HeLa WT cells (Figure 2E). Intriguingly, at the time of collection, we found that the surviving HeLa ρ0 cells infected with virus expressing WT ATPIF1 had lower amounts of ATPIF1 than those infected with virus expressing E55A ATPIF1, which is consistent with a selection against ATPIF1 activity on the F1-F0 ATP synthase in the ρ0 state (Figure 2E). Collectively, these data demonstrate that reduced ATPIF1 activity is essential for the viability of human ρ0 cells lacking a mitochondrial genome.
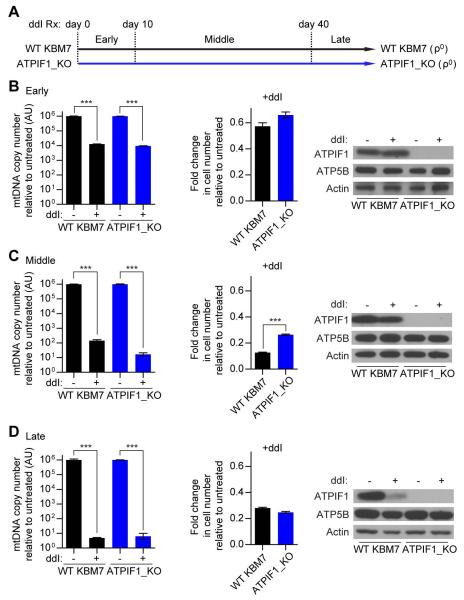
Failure to maintain proper amounts of the mitochondrial genome is a distinctive feature of a class of severe respiratory chain disorders known as mtDNA depletion syndromes (Lee and Sokol, 2007). Because of our interest in ATPIF1 inhibition as a potential strategy for ameliorating severe ETC dysfunction, we asked if loss of ATPIF1 alone was sufficient to improve cell viability during progressive mtDNA depletion. To do this, we cultured WT and ATPIF1_KO KBM7 cells with 2′,3′-dideoxyinosine (ddI), an inhibitor of mtDNA replication (Lewis et al., 2003, Walker et al., 2002), for approximately 51 days and monitored cellular behavior at defined time points (Figure 3A: see Experimental Procedures for exact time points at which phenotypes were assayed). ddI led to an immediate decrease in mtDNA copy number in the initial days of treatment, concomitant with a decrease in cell proliferation that was roughly equivalent between WT and ATPIF1_KO KBM7 cells (Figure 3B). It has been observed from previous studies that this amount of mtDNA depletion still allows for residual ETC function (Jazayeri et al., 2003) and so it is unlikely that ATPIF1 was maximally activated in the WT KBM7 cells at this point. mtDNA was progressively depleted with each successive week of ddI treatment and, by day 25, both WT and ATPIF1_KO KBM7 cells had trace amounts of mtDNA (Figure 3C). While both WT and ATPIF1_KO KBM7 cells proliferated slower than their untreated counterparts, ATPIF1_KO cells demonstrated a significantly faster rate of proliferation than WT KBM7 cells, consistent with loss of ATPIF1 improving cell viability under conditions of severe ETC dysfunction (Figure 3C). Taken together, these data demonstrate that loss of ATPIF1 is sufficient to improve cell viability during progressive mtDNA depletion.
Figure 3. Loss of ATPIF1 improves cell viability during progressive mtDNA depletion.
(A) Schematic depicting experimental paradigm of long-term treatment of WT and ATPIF1_KO KBM7 cells with ddI. Early (days 0 – 10), middle (days 10 – 40), and late (days 40 +) periods of ddI treatment are indicated and demarcated by dotted lines. During each period, mtDNA copy number, cell proliferation over four days, and ATPIF1 expression were analyzed. (B – D) mtDNA copy number (left), cell proliferation (middle), and immunoblots for indicated proteins (right) of WT and ATPIF1_KO KBM7 cells treated with ddI during the early (B), middle (C), and late (D) periods. Error bars are ± s.e.m. (n = 3). ***P < 0.001.
Given that low ATPIF1 levels were necessary for viability in HeLa ρ0 cells, we hypothesized that WT ρ0 KBM7 cells would express low amounts of ATPIF1 as well. In accordance with this, we observed a gradual decrease in ATPIF1 over 50 days of ddI treatment, with WT KBM7 cells exhibiting substantially reduced amounts of ATPIF1 and undetectable quantities of mtDNA (i.e. ρ0 state) at the end of the time course (Figures 3B-3D). Furthermore, WT ρ0 KBM7 cells with reduced ATPIF1 expression proliferated to a similar extent as ATPIF1_KO ρ0 KBM7 cells, suggesting that complete loss of ATPIF1 activity had no additional benefit on cell proliferation in the terminal ρ0 state (Figure 3D).
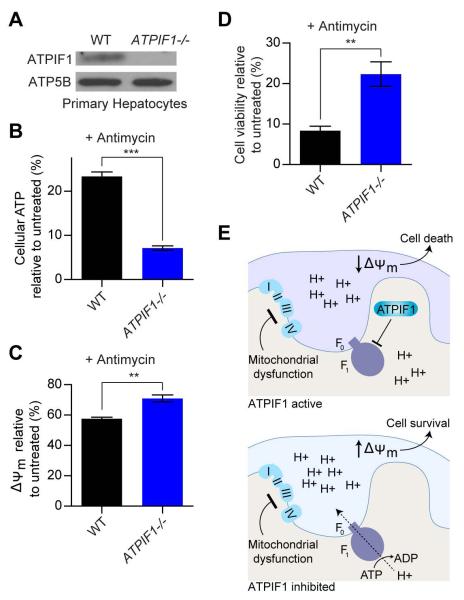
Because severe forms of mitochondrial respiratory chain disorders can lead to cell death and loss of tissue parenchyma in organs such as the liver (Morris, 1999, Lee and Sokol, 2007), we transitioned to a more physiological context and asked if loss of ATPIF1 in hepatocytes could ameliorate ETC dysfunction and improve cell viability. WT and ATPIF1 -/- mice were obtained from the International Knockout Mouse Consortium (Brown and Moore, 2012) (Figures S4A and S4B) and primary hepatocytes isolated from ATPIF1 -/- mice had undetectable amounts of ATPIF1 (Figure 4A). Consistent with the results seen in cell lines, antimycin treatment led to a greater decrease in cellular ATP (Figure 4B) and a greater increase in ΔΨm (Figure 4C) in ATPIF1 -/- hepatocytes than in WT hepatocytes, indicating that there was greater reversal of the F1-F0 ATP synthase in ATPIF1 -/- hepatocytes. Importantly, ATPIF1 -/- hepatocytes had increased cell viability relative to WT hepatocytes following treatment with antimycin (Figure 4D), which demonstrates that the beneficial effects of ATPIF1 loss under severe ETC dysfunction are not limited to rapidly proliferating cancer cell lines, but can also occur in post-mitotic, differentiated cells that better recapitulate the metabolism of tissues affected in severe mitochondrial respiratory chain disorders (Vander Heiden et al., 2009). The smaller effects of ATPIF1 loss on hepatocyte viability during antimycin treatment, as compared to KBM7 cells, are partially due to the frailty of primary mouse hepatocytes when cultured ex vivo (Edwards et al., 2013, Klaunig et al., 1981). Taken together, these data demonstrate that ATPIF1 loss in primary hepatocytes can ameliorate the effects of complex III blockade.
Figure 4. Inhibition of ATPIF1 ameliorates the effects of complex III blockade in primary hepatocytes.
(A) Immunoblots for indicated proteins of primary hepatocytes derived from WT and ATPIF1 -/- mice. (B) Cellular ATP of WT and ATPIF1 -/- primary hepatocytes treated with antimycin (0.625 μM) for 1.5 hours. Error bars are ± s.e.m. (n = 3). ***P < 0.001. (C) ΔΨm of WT and ATPIF1 -/- primary hepatocytes treated with antimycin (10 μM) for 1.5 hours. Error bars are ± s.e.m. (n = 3). **P < 0.01. (D) Viability of WT and ATPIF1 -/- primary hepatocytes treated with antimycin (1.25 μM) for 2 days. Error bars are ± s.e.m. (n = 3). **P < 0.01. (E) Schematic diagramming the behavior of cells with ETC dysfunction under conditions where ATPIF1 is active or inhibited. Inhibition of ATPIF1 is depicted by absence of the protein but represents any strategy to block ATPIF1 activity on the F1-F0 ATP synthase. See also Figure S4.
Our results suggest that ATPIF1 inhibition can be a strategy for ameliorating severe ETC dysfunction in mitochondrial respiratory chain disorders. Impaired ETC function in the presence of normal ATPIF1 activity leads to a persistent loss of ΔΨm, which hinders mitochondrial import of proteins (Neupert, 1997) and eventually promotes apoptosis (Gottlieb et al., 2003). However, upon ATPIF1 inhibition, increased reversal of the F1-F0 ATP synthase can bolster ΔΨm and improve mitochondrial and cellular health (Figure 4E), although it remains to be seen whether ATPIF1 loss is beneficial to all cell types in vivo since the ratio of ATPIF1 to F1-F0 ATP synthase expression, the amount of glycolysis, and the consumption of ATP can vary substantially between different tissues. Regardless, because several mitochondrial respiratory chain disorders, such as Alpers-Huttenlocher Syndrome and Pearson’s Syndrome, lead to progressive liver failure (Lee and Sokol, 2007), our findings in primary hepatocytes at least suggest that hepatic delivery of RNAi constructs targeting ATPIF1 either via adeno-associated virus or lipid nanoparticles, both of which have seen clinical efficacy in gene therapy of the liver, may have therapeutic value (Nathwani et al., 2011, Fitzgerald et al.). Notably, ATPIF1 -/- mice appear phenotypically normal and their hepatocytes exhibit no significant alterations in ATP synthase activity or mitochondrial structure (Nakamura et al., 2013). In agreement with these findings, we did not observe any significant differences in the mitochondrial mass of WT and ATPIF1 -/- primary hepatocytes (Figure S4C). Taken together, these data suggest that ATPIF1 inhibition is relatively well-tolerated.
In conclusion, we have used a positive-selection screening method in human cells to identify loss of ATPIF1 as protective against complex III blockade. We have further shown that ATPIF1 inhibition protects different cell types against numerous insults to the ETC. In particular, our work demonstrates that loss of ATPIF1 activity is essential for the viability of human ρ0 cells, a widely used system to study mitochondrial dysfunction, and that inhibition of ATPIF1 can ameliorate the effects of complex III blockade in primary hepatocytes, a cell type that is often affected in severe respiratory chain disorders. Given the lack of therapies for severe mitochondrial respiratory chain disorders, we thus believe that inhibition of ATPIF1 is a promising approach that warrants further investigation.
EXPERIMENTAL PROCEDURES
FACS assays
For measurements of ΔΨm, 100,000 cells were incubated with TMRM (25 nM) and the indicated amounts of drugs for the indicated amounts of time before collection. For measurements of mitochondrial mass, 100,000 cells were incubated with MitoTracker Green FM (50 nM) for one hour. For primary hepatocytes, cells were assayed in suspension immediately after harvest from the liver and incubated with verapamil (20 μM) to facilitate retention of TMRM and MitoTracker Green FM signals. Afterwards, cells were collected by centrifugation, washed once with PBS, and resuspended in PBS with 7-AAD (2 μg/mL) for analysis. KBM7 cells and primary hepatocytes were centrifuged at 2000 rpm for 5 minutes and 500 rpm for 5 minutes at 4°C, respectively.
Cell viability assays
For KBM7 cells, 100,000 cells were seeded and treated with the indicated drugs for the indicated amounts of time before then being analyzed with 7-AAD staining and FACS according to the manufacturer’s instructions (BD Pharmingen), unless indicated otherwise. For HeLa, SH-SY5Y, and Malme-3M cells, 500 – 2,000 cells were seeded per well of white, clear-bottom 96-well plates (Greiner Bio-One), treated with drugs, and then analyzed using CellTiter-Glo according to the manufacturer’s instructions (Promega). For primary hepatocytes, 100,000 cells were seeded per well of a 24-well TPP plate (Light Labs), treated with antimycin, and then analyzed using CellTiter-Glo.
Cell proliferation assays
For KBM7 cells, cell proliferation was assessed with a Beckman Z2 Coulter Counter using a size range of 8 – 30 μm. For all other cell lines, cell proliferation was assessed using CellTiter-Glo according to the manufacturer’s instructions and by normalizing all readings to initial values measured at the start of the experiment.
Long-term exposure to ddI
mtDNA copy number analysis was performed on cells treated with ddI for 8 days, 26 days, and 41 days. Rates of proliferation were measured for cells treated with ddI for 5 days, 31 days, and 51 days. Immunoblot analysis was done on cells treated with ddI for 5 days, 11 days, and 50 days.
Statistical analysis
All data are expressed as mean ± s.e.m. Significance was determined using the two-tailed unpaired Student’s t test. Differences with a P value less than 0.05 were considered significant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Sabatini laboratory and H.S.T. for all of their assistance and support. In particular, we thank D.K., T.W., and Z.Y.T. for careful revisions of the manuscript. This work was supported by grants from the US National Institutes of Health (NIH; CA103866, CA129105, AI07389), the David H. Koch Institute for Integrative Cancer Research, and The Alexander and Margaret Stewart Trust Fund to D.M.S. and fellowships from the US National Institute of Aging to W.W.C., the Jane Coffin Childs Memorial Fund and Leukemia and Lymphoma Society to K.B., the Damon Runyon Cancer Research Foundation to M.M.M. D.M.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS W.W.C., K.B., and D.M.S. conceived the project. W.W.C. and K.B. designed and performed most experiments and data analyses with input from D.M.S. M.M.M., B.Y., and E.C.B. assisted with experiments. J.E.C. and T.R.B. assisted with haploid genetic screening. C.B.C. performed metabolite profiling and analysis. H.S., I.S., and D.D.S. performed electron microscopy analysis. W.W.C., K.B., and D.M.S. wrote and edited the manuscript.
The authors declare that no competing financial interests exist.
REFERENCES
- Appleby RD, Porteous WK, Hughes G, James AM, Shannon D, Wei Y-H, Murphy MP. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. European Journal of Biochemistry. 1999;262:108–116. doi: 10.1046/j.1432-1327.1999.00350.x. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Possemato R, Yilmaz OH, Koch CE, Chen WW, Hutchins AW, Gultekin Y, Peterson TR, Carette JE, Brummelkamp TR, Clish CB, Sabatini DM. MCT1-mediated transport of a toxic molecule is an effective strategy for targeting glycolytic tumors. Nat Genet. 2013;45:104–108. doi: 10.1038/ng.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SM, Moore M. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mammalian Genome. 2012;23:632–640. doi: 10.1007/s00335-012-9427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchet K, Godinot C. Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted ρ° cells. Journal of Biological Chemistry. 1998;273:22983–22989. doi: 10.1074/jbc.273.36.22983. [DOI] [PubMed] [Google Scholar]
- Cabezon E, Runswick MJ, Leslie AGW, Walker JE. The structure of bovine IF1, the regulatory subunit of mitochondrial F-ATPase. EMBO J. 2001;20:6990–6996. doi: 10.1093/emboj/20.24.6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella M, Casswell E, Chong S, Farah Z, Wieckowski MR, Abramov AY, Tinker A, Duchen MR. Regulation of mitochondrial structure and function by the F1F0-ATPase inhibitor protein, IF1. Cell metabolism. 2008;8:13–25. doi: 10.1016/j.cmet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Campanella M, Parker N, Tan CH, Hall AM, Duchen MR. IF1: setting the pace of the F1F0-ATP synthase. Trends in biochemical sciences. 2009;34:343–350. doi: 10.1016/j.tibs.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Carette JE, Guimaraes CP, Wuethrich I, Blomen VA, Varadarajan M, Sun C, Bell G, Yuan B, Muellner MK, Nijman SM, Ploegh HL, Brummelkamp TR. Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat Biotech. 2011a;29:542–546. doi: 10.1038/nbt.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Cin PD, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011b;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrouzet E, Issartel J-P, Lunardi J, Dupuis A. The 49-kDa subunit of NADH-ubiquinone oxidoreductase (Complex I) is involved in the binding of piericidin and rotenone, two quinone-related inhibitors. FEBS Letters. 1998;431:34–38. doi: 10.1016/s0014-5793(98)00719-4. [DOI] [PubMed] [Google Scholar]
- Dimauro S, Schon EA. Mitochondrial respiratory-chain diseases. New England Journal of Medicine. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- Edwards M, Houseman L, Phillips I, Shephard E. Isolation of mouse hepatocytes. In: PHILLIPS IR, SHEPHARD EA, ORTIZ DE MONTELLANO PR, editors. Cytochrome P450 Protocols. Humana Press; 2013. [Google Scholar]
- Fitzgerald K, Frank-Kamenetsky M, Shulga-Morskaya S, Liebow A, Bettencourt BR, Sutherland JE, Hutabarat RM, Clausen VA, Karsten V, Cehelsky J, Nochur SV, Kotelianski V, Horton J, Mant T, Chiesa J, Ritter J, Munisamy M, Vaishnaw AK, Gollob JA, Simon A. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. The Lancet. doi: 10.1016/S0140-6736(13)61914-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa M, Imamura H, Nakamura J, Yoshida M. Assessing actual contribution of IF1, inhibitor of mitochondrial F0F1, to ATP homeostasis, cell growth, mitochondrial morphology, and cell viability. Journal of Biological Chemistry. 2012;287:18781–18787. doi: 10.1074/jbc.M112.345793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García JJ, Morales-Ríos E, Cortés-Hernández P, Rodríguez-Zavala JS. The inhibitor protein (IF1) promotes dimerization of the mitochondrial F1F0-ATP Synthase. Biochemistry. 2006;45:12695–12703. doi: 10.1021/bi060339j. [DOI] [PubMed] [Google Scholar]
- Gohil VM, Sheth SA, Nilsson R, Wojtovich AP, Lee JH, Perocchi F, Chen W, Clish CB, Ayata C, Brookes PS, Mootha VK. Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nat Biotech. 2010;28:249–255. doi: 10.1038/nbt.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Armour SM, Harris MH, Thompson CB. Mitochondrial membrane potential regulates matrix configuration and cytochrome c release during apoptosis. Cell Death Differ. 2003;10:709–717. doi: 10.1038/sj.cdd.4401231. [DOI] [PubMed] [Google Scholar]
- Guimaraes CP, Carette JE, Varadarajan M, Antos J, Popp MW, Spooner E, Brummelkamp TR, Ploegh HL. Identification of host cell factors required for intoxication through use of modified cholera toxin. The Journal of Cell Biology. 2011;195:751–764. doi: 10.1083/jcb.201108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa N, Karaki A, Kawabata M, Ushida S, Mizushima M, Hashimoto T. The region from phenylalanine-17 to phenylalanine-28 of a yeast mitochondrial ATPase inhibitor is essential for its ATPase inhibitory activity. Journal of Biochemistry. 2001;130:687–693. doi: 10.1093/oxfordjournals.jbchem.a003035. [DOI] [PubMed] [Google Scholar]
- Jazayeri M, Andreyev A, Will Y, Ward M, Anderson CM, Clevenger W. Inducible expression of a dominant negative DNA polymerase-γ depletes mitochondrial DNA and produces a ρ0 phenotype. Journal of Biological Chemistry. 2003;278:9823–9830. doi: 10.1074/jbc.m211730200. [DOI] [PubMed] [Google Scholar]
- Kitami T, Logan DJ, Negri J, Hasaka T, Tolliday NJ, Carpenter AE, Spiegelman BM, Mootha VK. A chemical screen probing the relationship between mitochondrial content and cell size. PLoS ONE. 2012;7:e33755. doi: 10.1371/journal.pone.0033755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaunig J, Goldblatt P, Hinton D, Lipsky M, Trump B. Mouse liver cell culture. In Vitro. 1981;17:926–934. doi: 10.1007/BF02618289. [DOI] [PubMed] [Google Scholar]
- Lee WS, Sokol RJ. Liver disease in mitochondrial disorders. Semin Liver Dis. 2007;27:259–273. doi: 10.1055/s-2007-985071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Du Q, Baird S, Ng AC-H, Nascimento M, Campanella M, Mcbride HM, Screaton RA. Genome-wide RNAi screen identifies ATPase inhibitory factor 1 (ATPIF1) as essential for PARK2 recruitment and mitophagy. Autophagy. 2013;9:1770–1779. doi: 10.4161/auto.25413. [DOI] [PubMed] [Google Scholar]
- Lewis W, Day BJ, Copeland WC. Mitochondrial toxicity of NRTI antiviral drugs: an integrated cellular perspective. Nat Rev Drug Discov. 2003;2:812–822. doi: 10.1038/nrd1201. [DOI] [PubMed] [Google Scholar]
- Lu Y-M, Miyazawa K, Yamaguchi K, Nowaki K, Iwatsuki H, Wakamatsu Y, Ichikawa N, Hashimoto T. Deletion of mitochondrial ATPase Inhibitor in the Yeast Saccharomyces cerevisiae decreased cellular and mitochondrial ATP levels under non-nutritional conditions and induced a respiration-deficient cell-type. Journal of Biochemistry. 2001;130:873–878. doi: 10.1093/oxfordjournals.jbchem.a003060. [DOI] [PubMed] [Google Scholar]
- Morris A, M. Mitochondrial respiratory chain disorders and the liver. Liver. 1999;19:357–368. doi: 10.1111/j.1478-3231.1999.tb00063.x. [DOI] [PubMed] [Google Scholar]
- Nakamura J, Fujikawa M, Yoshida M. IF1, a natural inhibitor of mitochondrial ATP synthase, is not essential for the normal growth and breeding of mice. Bioscience Reports. 2013:33. doi: 10.1042/BSR20130078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani AC, Tuddenham EGD, Rangarajan S, Rosales C, Mcintosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, O’beirne J, Smith K, Pasi J, Glader B, Rustagi P, Ng CYC, Kay MA, Zhou J, Spence Y, Morton CL, Allay J, Coleman J, Sleep S, Cunningham JM, Srivastava D, Basner-Tschakarjan E, Mingozzi F, High KA, Gray JT, Reiss UM, Nienhuis AW, Davidoff AM. Adenovirus-Associated Virus vector–mediated gene transfer in Hemophilia B. New England Journal of Medicine. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annual Review of Biochemistry. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Pfeffer G, Majamaa K, Turnbull DM, Thorburn D, Chinnery PF. Treatment for mitochondrial disorders. Cochrane Database of Systematic Reviews [Online] 2012 doi: 10.1002/14651858.CD004426.pub3. Available: http://onlinelibrary.wiley.com/doi/10.1002/14651858.CD004426.pub3/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Cenizo L, Formentini L, Aldea M, Ortega ÁD, García-Huerta P, Sánchez-Aragó M, Cuezva JM. Up-regulation of the ATPase Inhibitory Factor 1 (IF1) of the mitochondrial H+-ATP Synthase in human tumors mediates the metabolic shift of cancer cells to a warburg phenotype. Journal of Biological Chemistry. 2010;285:25308–25313. doi: 10.1074/jbc.M110.146480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA, Dimauro S, Hirano M, Gilkerson RW. Therapeutic prospects for mitochondrial disease. Trends in Molecular Medicine. 2010;16:268–276. doi: 10.1016/j.molmed.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- škrtić M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z, Hurren R, Jitkova Y, Gronda M, Maclean N, Lai, Courteney k., Eberhard Y, Bartoszko J, Spagnuolo P, Rutledge, Angela C, Datti A, Ketela T, Moffat J, Robinson, Brian h., Cameron, Jessie h., Wrana J, Eaves, Connie j., Minden, Mark d., Wang, Jean c. Y., Dick, John e., Humphries K, Nislow C, Giaever G, Schimmer, Aaron d. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2011;20:674–688. doi: 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinazzola A, Viscomi C, Fernandez-Vizarra E, Carrara F, D’adamo P, Calvo S, Marsano RM, Donnini C, Weiher H, Strisciuglio P, Parini R, Sarzi E, Chan A, Dimauro S, Rotig A, Gasparini P, Ferrero I, Mootha VK, Tiranti V, Zeviani M. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat Genet. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker UA, Setzer B, Venhoff N. Increased long-term mitochondrial toxicity in combinations of nucleoside analogue reverse-transcriptase inhibitors. AIDS. 2002;16:2165–2173. doi: 10.1097/00002030-200211080-00009. [DOI] [PubMed] [Google Scholar]
- Yoon JC, Ng A, Kim BH, Bianco A, Xavier RJ, Elledge SJ. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes & Development. 2010;24:1507–1518. doi: 10.1101/gad.1924910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.