Abstract
Bradyrhizobium japonicum Irr is a conditionally stable transcriptional activator and repressor that accumulates in cells under iron- limited, manganese-replete conditions, but degrades in a heme-dependent manner under high iron conditions, manganese limitation or upon exposure to H2O2. Here, we identified Irr-regulated genes that were relatively unresponsive to factors that promote Irr degradation. The promoters of those genes bound Irr with at least 200-fold greater affinity than promoters of the responsive genes, resulting in maintenance of promoter occupancy over a wide cellular Irr concentration range. For Irr-repressible genes, promoter occupancy correlated with transcriptional repression, resulting in differential levels of expression based on Irr affinity for target promoters. However, inactivation of positively controlled genes required neither promoter vacancy nor loss of DNA-binding activity by Irr. Thus, activation and repression functions of Irr may be uncoupled from each other under certain conditions. Abrogation of Irr activation function was heme-dependent, thus heme has two functionally separable roles in modulating Irr activity. The findings imply a greater complexity of control by Irr than can be achieved by conditional stability alone. We suggest that these regulatory mechanisms accommodate the differing needs for Irr regulon genes in response to the prevailing metabolic state of the cell.
INTRODUCTION
The ability of bacteria to sense nutrient availability and adapt accordingly contribute to their success in diverse environments. Iron is an essential nutrient required for many cellular processes. Bioavailability of iron is low in aerobic environments because it is mostly oxidized, and therefore insoluble. High affinity iron acquisition systems are expressed under iron limitation to scavenge the metal. Iron can also be toxic, as it catalyzes the generation of reactive oxygen species. Thus, metal homeostasis must be maintained.
Bradyrhizobium japonicum lives as a free-living soil organism or as the endosymbiont of soybean, where it fixes atmospheric nitrogen to ammonia to fulfill the nitrogen requirements of the host. B. japonicum belongs to the Alphaproteobacteria, a large taxonomic group that occupies diverse niches, including within eukaryotic cells in a symbiotic or pathogenic context. B. japonicum serves as a model system to understand metal metabolism and homeostasis in many Alphaproteobacterial species (Small et al., 2009a). Soils are highly variable ecosystems, and symbiosis represents a niche with specific nutritional requirements. Thus, B. japonicum and other rhizobia must be able to accommodate changes in metal availability.
The iron response regulator (Irr) is the major transcriptional regulator of iron-responsive gene expression in B. japonicum, and the breadth of its role in other alpha-Proteobacteria varies. Some rhizobia employ RirA in addition to Irr (Todd et al., 2002), and Rhodobacter capsulatus HbrL may control a larger iron-responsive regulon than Irr (Zappa and Bauer, 2013). B. japonicum Irr accumulates in iron-limited, manganese replete cells, and serves as both a positive and negative regulator of gene expression (Qi and O’Brian, 2002, Yang et al., 2006b). Irr recognizes and binds to the iron control cis-acting element (ICE) within the promoters of target genes (Rudolph et al., 2006, Yang et al., 2006b, Todd et al., 2006).
Irr senses iron through the status of heme by binding to ferrochelatase, a heme biosynthesis enzyme that uses iron as a substrate (Qi et al., 1999). When the iron level is sufficient, heme binds directly to Irr leading to its degradation in B. japonicum and Brucella abortus (Anderson et al., 2011, Martinez et al., 2005), whereas the Rhizobium leguminosarum Irr level is not substantially altered by the iron status, but its binding activity is affected by heme, at least in vitro (Singleton et al., 2010). Irr is an atypical member of the Fur family of metalloregulators in that it is active when the metal is limiting, it degrades or is inactivated in response to binding to its regulatory ligand, and it serves as a positive regulator of a large number of genes within its regulon.
Irr integrates the control of iron homeostasis and metabolism with other cellular phenomena such as the status of manganese and oxidative stress. In B. japonicum, manganese stabilizes Irr by interfering with heme binding, thereby increasing the heme concentration threshold that triggers Irr degradation (Puri et al., 2010). As a result, manganese limitation creates an iron deficiency, possibly to counter the reduced anti-oxidant activity of manganese by attenuating iron acquisition. B. japonicum Irr degrades in response to H2O2 exposure, thereby linking peroxide stress to iron-responsive gene expression (Yang et al., 2006a).
The Irr regulon is large and includes genes involved in a wide variety of cellular processes. Although they share a common iron-dependent regulatory mechanism, it is likely that additional levels of control are necessary in some cases to accommodate unique functions. In principle, this could be achieved by additional regulatory proteins or small RNAs that target a subset of the Irr regulon genes. Indeed, the heme biosynthesis genes hemA and hemB are negatively-regulated by Irr and activated by the FixL/FixJ/FixK2 regulatory cascade in response to O2 limitation (Page and Guerinot, 1995, Chauhan and O’Brian, 1997). Those genes are activated under low O2, low iron conditions, suggesting that FixK2-dependent activation overrides Irr-dependent repression. HmuP is a co-activator of the Irr-dependent heme utilization operon in B. japonicum, whereas utilization of other iron chelates do not require HmuP (Escamilla-Hernandez and O’Brian, 2012, Amarelle et al., 2010). The regulatory rationale for this is unknown, but perhaps it is because heme can be both an iron source and is also functional intact as the prosthetic group of heme enzymes. Irr also serves as an antirepressor of manganese-dependent repression of the irr gene by occluding binding of the Mur repressor to the irr promoter (Hohle and O’Brian, 2010).
In the current study, we identified two novel mechanisms of Irr-mediated control of gene expression that allow differential regulation of Irr operon genes without the apparent need for additional regulatory proteins.
RESULTS
In vivo occupancy of target promoters by Irr is strongly manganese-dependent for only some genes
Irr accumulates under iron limitation to positively or negatively regulate target genes. We found previously that manganese limitation results in substantial diminution of Irr levels in cells grown in low iron media (Puri et al., 2010)(Fig. 1A). As a result, manganese affects promoter occupancy by Irr accordingly, and gene expression responds to manganese as well. In the current work, we examined six additional different genes within the Irr regulon for their responsiveness to manganese under iron limitation. fhuE and bll7968 encode known and putative ferric siderophore receptors, respectively (Small and O’Brian, 2011, Small et al., 2009b). These genes are positively controlled by Irr, thus promoter occupancy and expression are high under iron limitation, and low in an irr mutant (Small et al., 2009b). The bfr, blr7895, hemB and leuC genes are repressed by Irr under iron limitation, and remain high independently of iron in an irr mutant (Hamza et al., 1998, Sangwan et al., 2008, Yang et al., 2006b). The bfr (bll6680) gene encodes a putative bacterioferritin. The function of blr7895 is not known, but its homolog in Agrobacterium tumefaciens contributes to resistance to hydrogen peroxide stress by an unknown mechanism (Ruangkiattikul et al., 2012). hemB encodes 5-aminolevlinic acid dehydratase, an enzyme within the pathway that synthesizes heme, an iron containing prosthetic group (Chauhan and O’Brian, 1993). leuC is predicted to encode 3-isopropylmalate dehydratase large subunit, an iron-sulfur protein required for leucine biosynthesis. The rationale for choosing these genes is based on the fact that two of them (bfr and blr7895) did not respond to manganese as expected, and the others were useful for comparisons. This is described in detail below.
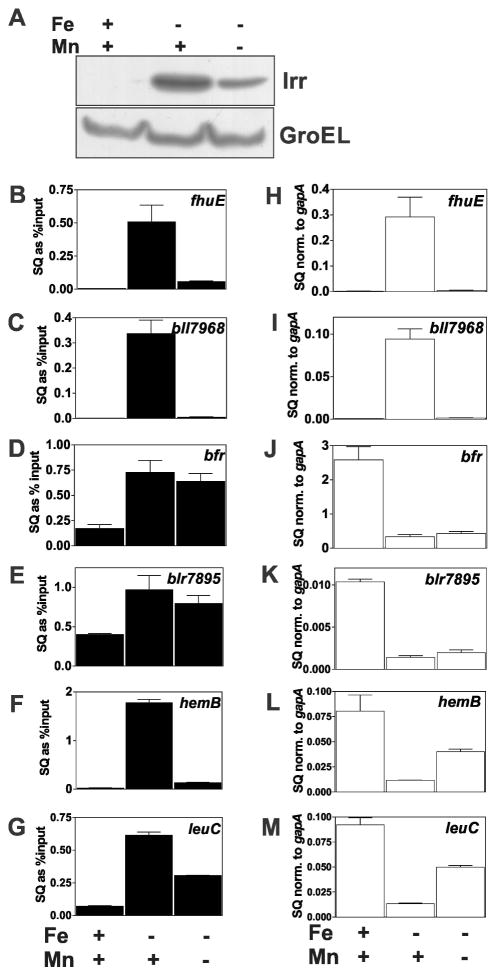
Fig. 1.
Effects of manganese on promoter occupancy and expression of Irr-regulated genes. Cells were grown in media supplemented with 20 μM FeCl3 (Fe) or 50 μM MnCl2 (Mn) (+) or not supplemented with metal (−). The actual Fe and Mn concentrations in the unsupplemented media were 0.3 μM and 0.4 μM, respectively. (A) Steady state levels of Irr were detected by immunoblotting using anti-Irr antibodies. GroEL was used as a control for an unregulated protein, and was detected using anti-GroEL antibodies. Fifteen micrograms of protein was loaded per lane. (B–G) In vivo promoter occupancy of the respective genes in cells grown under different metal conditions was measured by crosslinking and immunoprecipitation using anti-Irr antibodies as described in Materials and Methods. The precipitated DNA was analyzed by qPCR using primers delimiting the promoter regions of the respective genes. The data are expressed as the relative starting quantities (SQ) of immunoprecipitated DNA normalized to the input DNA and are presented as the average of three replicates ± the standard deviation. (H–M) Steady-state transcript levels of the respective genes obtained from cells grown under different metal conditions were analyzed by qPCR. The data are expressed as relative starting quantities (SQ) of respective mRNAs normalized to the housekeeping gene gapA, and presented as average of three replicates ± the standard deviation.
The effects of manganese on Irr occupancy of target gene promoters were examined using cross-linking and co-immunoprecipitation as previously described (Hohle and O’Brian, 2010) using anti-Irr antibodies. Co-precipitated DNA was examined by quantitative real time PCR (qPCR). Under low iron conditions, Irr occupancy of the fhuE, bll7968, hemB and leuC promoters were high in wild type cells when the medium was supplemented with manganese, and were low in cells grown without added manganese (Fig. 1B, C, F, G), although less so for the leuC promoter. These observations correlated with Irr levels in cells (Fig. 1A), and are consistent with that observed previously (Puri et al., 2010).
Interestingly, diminution of cellular Irr levels by removal of manganese from low iron media did not result in a corresponding decrease in promoter occupancy of the bfr and blr7895 genes (Fig. 1D, E). These findings show that not all genes within the Irr regulon are responsive to manganese, and Irr occupancy correlates with cellular Irr levels for only some promoters. Moreover, substantial occupancy of the bfr and blr7895 promoters by Irr persists even in cells grown under high iron, suggesting that they are less responsive to that metal as well.
The affinity of Irr for target promoters varies greatly in vitro
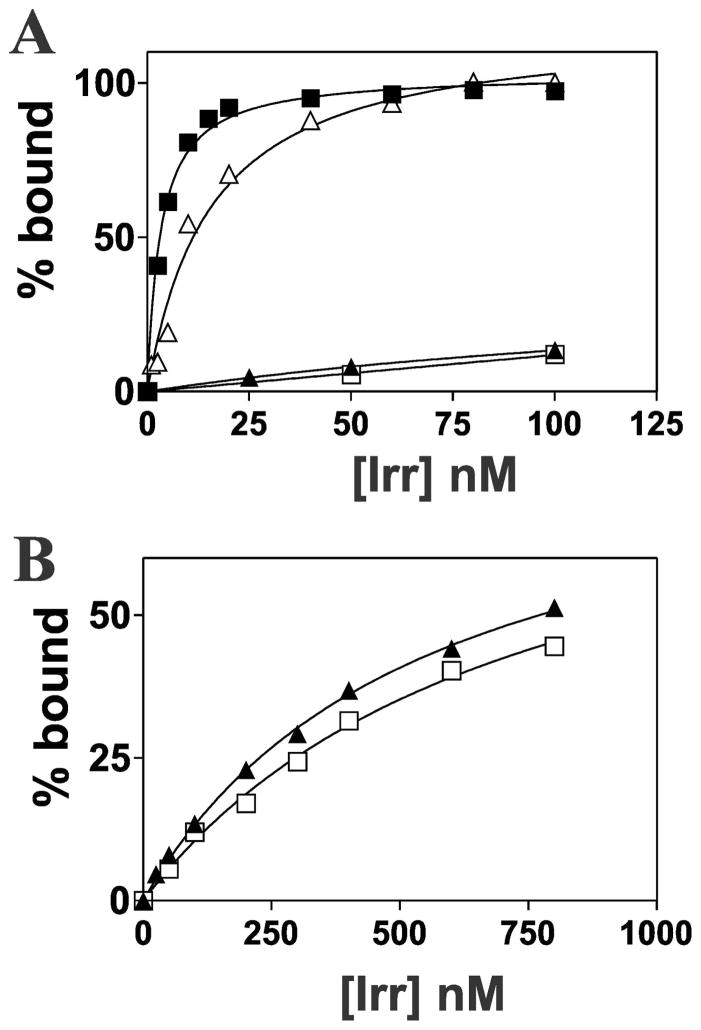
The simplest explanation for the high occupancy of the bfr or blr7895 promoter by Irr when the cellular Irr level is low is that Irr has a higher affinity for those promoters than for the fhuE, bll7968, hemB or leuC promoters. This variability could be due to an accessory protein that affects affinity, or Irr may simply have differing affinities for the promoters examined. We measured the affinity of Irr for 32P-labeled 45 bp double stranded DNA probes in vitro that correspond to each of the six promoter regions by electrophoretic mobility shift analysis (EMSA). Each DNA probe contained the 21 bp ICE motif within each promoter flanked by an additional 12 bp on each side (Fig. S1). Binding curves were generated by titrating the reactions with varying amounts of Irr, and the fraction bound was determined by the mobility of the bands (Fig. 2, Fig. S2). Dissociation binding constants (Kd) values derived from then are listed in Table 1. The Kd values for the bfr and blr7895 promoters were 3.3 nM and 1.5 nM, respectively. The Kd values for the fhuE and bll7968, hemB and leuC promoters were estimated to be greater than 440 nM, 320 nM, 1000 nM and 1000 nM, respectively. These latter 4 values are only approximations because binding did not saturate at 1000 nM Irr, which was the highest concentration that could be achieved in the reactions. Nevertheless, the data show a very large range of Irr binding affinities for target promoters. The high affinities of the bfr and blr7895 promoters for Irr can explain high occupancy even when Irr levels are low, hence less responsive to manganese. Similarly, weak binding of Irr to the other promoters is consistent with low promoter occupancy under low manganese conditions where the cellular Irr level is low, resulting in responsiveness to manganese.
Fig. 2.
Analysis of Irr-binding to the promoters of Irr-regulated genes. (A) EMSA was carried out using 0.1 nM 32P-labelled promoter DNA of bfr (closed squares), fhuE (closed triangles), PfhuE/ICEbfr (open triangles) and Pbfr/ICEfhuE (open squares) titrated with various concentrations of Irr. Bound and unbound DNA was resolved on a 5% non-denaturing polyacrylamide gel and visualized by autoradiography. Autoradiographs were scanned, and bands were quantified to determine bound and unbound DNA. (B) Binding curves of fhuE (closed triangles) and Pbfr/ICEfhuE (open squares) at higher Irr concentrations.
TABLE 1.
Dissociation binding constants (Kd values) of Irr for ICE motif sequencesa
| ICE motif | Kd (nM) |
|---|---|
| bfr | 3.3 ± 0.3 |
| blr7895 | 1.5 ± 0.3 |
| fhuE | >440b |
| bll7968 | >320b |
| Pbfr/ICEfhuE | >730b |
| PfhuE/ICEbfr | 16.3 ± 3.1 |
| P7895/ICE7968 | >340 |
| P7968/ICE7895 | 4.6 ± 0.7 |
| leuC | >1000b |
| hemB | >1000b |
| controlc | NBd |
Kd values were determined by measuring bound and unbound 32P-labeled DNA fragments 39 bp in length in EMSA analysis as described in the text.
Values are approximate because binding did not saturate at 1000 nM Irr, which was the highest concentration that could be achieved in the binding reactions.
The control is a 39 bp DNA sequence corresponding to the multiple cloning site of pBluescript SK+.
No binding was detected.
The iron control element (ICE) motif is sufficient to confer binding affinity of Irr to target promoters in vitro
The ICE motif is a 21 bp imperfect inverted repeat consensus sequence necessary for Irr binding (Rudolph et al., 2006, Yang et al., 2006b). We wanted to determine whether differences in the ICE motif sequence within each promoter was sufficient to explain the wide range of Irr binding affinities. The ICE motifs within the promoters of fhuE and bfr differ from each other by 6 bp. Therefore, substitutions were made so that the weak binding fhuE promoter contained the ICE motif of the strong binding bfr promoter (PfhuE/ICEbfr) and vice versa (Pbfr/ICEfhuE) (Fig. S1). These oligonucleotides were analyzed by EMSA to determine the dissociation binding constants (Fig. 2; Fig. S2, Table 1). The wild type bfr promoter had a Kd value of 3.3 nM, but the Kd for the Pbfr/ICEfhuE promoter was >730 nM. Correspondingly, the Kd values of the wild type fhuE promoter and the PfhuE/ICEbfr promoter were >550 nM and 16 nM, respectively.
Binding experiments were also carried out between the respective high and low affinity promoters of blr7895 and bll7968, which contain ICE motifs that differ from each other by 6 bp (Fig. S1; Table 1). Insertion of the blr7895 ICE into bll7968 promoter sequence (P7968/ICE7895) increased the Irr binding affinity from >320 nM to 4.6 nM. In addition, insertion of the bll7968 ICE motif into the blr7895 promoter (P7895/ICE7968) significantly reduced binding, from 1.5 nM to >340 nM.
Collectively, the in vitro binding experiments show that the ICE motif is a major determinant in promoter affinity for Irr.
Responsiveness of Irr occupancy to hydrogen peroxide in vivo depends on promoter affinity for Irr
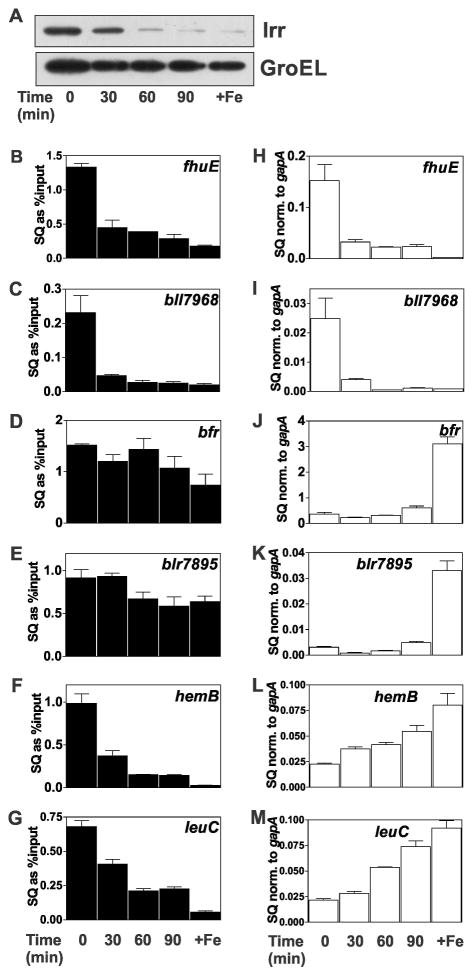
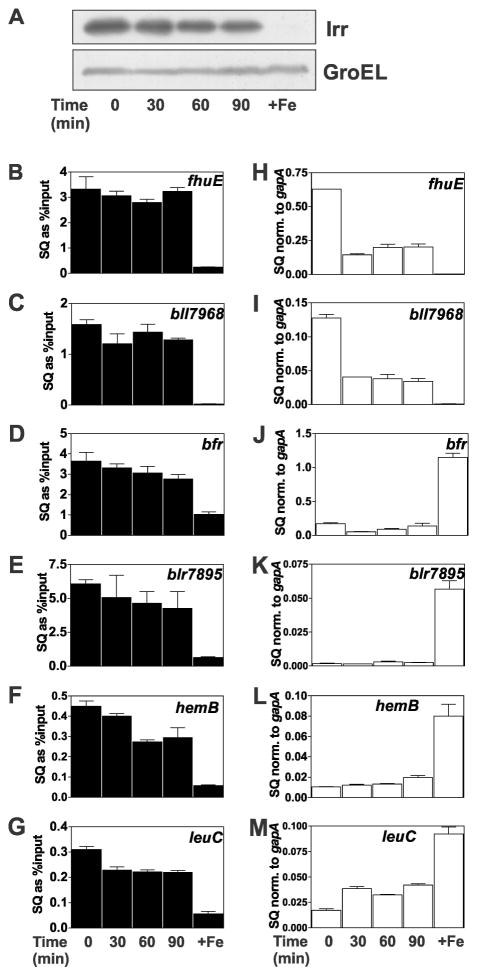
The data presented thus far suggest that the status of promoter occupancy by Irr in response to manganese is dependent on the affinity of Irr to the target gene promoter. As a result, diminution of the cellular Irr level has less effect on the occupancy of high affinity promoters compared with those of low affinity for Irr. This idea predicts that factors other than manganese that affect cellular Irr levels will also differentially influence promoter occupancy. We found previously that Irr degrades in response to short term exposure to exogenous H2O2 in cells grown with 2 μM FeCl3, an iron concentration that is not sufficient to fully degrade Irr (Yang et al., 2006a). The Irr level in cells was followed over 90 min after H2O2 exposure by western blotting (Fig. 3A). Irr decreased with time after H2O2 addition, similar to what was reported previously (Yang et al., 2006a). Corresponding decreases in promoter occupancy of the fhuE, bll7968, hemB and leuC genes were observed (Fig. 3B, C, F, G). By contrast, occupancy of the high Irr affinity promoters of the bfr and blr7895 genes remained relatively high upon exposure to H2O2 even though Irr decreased over that time period (Fig. 3D,E). These observations support the conclusion that environmental factors that affect the cellular Irr level will result in differential occupancy of target genes based on promoter affinity for Irr, and that these responses are not specific to manganese.
Fig. 3.
Effects of H2O2 on promoter occupancy and expression of Irr-regulated genes. Cultures were grown to mid-log phase in media supplemented with 2 μM FeCl3 and 50 μM MnCl2. At time 0, 2 mM H2O2 was added to the cultures and were continually aerated at 29°C. Cells were grown in 20 μM (+Fe) without H2O2 treatment as a control. Aliquots were harvested at indicated time points for analysis. (A) Steady-state levels of Irr and GroEL were detected by immunoblotting as described in Fig. 1. (B–G) Promoter occupancy of respective genes by Irr in cells treated with H2O2 were carried out as described in the Fig. 1 legend. The data are expressed as the relative starting quantities (SQ) of immunoprecipitated DNA normalized to the input DNA and are presented as average of three replicates ± the standard deviation. (H–M) Steady-state transcript levels of the respective genes obtained from cells treated with H2O2 were analyzed by qPCR as described in the Fig. 1 legend. The data are expressed as relative starting quantities (SQ) of respective mRNAs normalized to the housekeeping gene gapA, and presented as average of three replicates ± the standard deviation.
Expression of negatively-regulated genes in response to environmental factors correlates with Irr occupancy of their promoters
We sought to address whether environmental factors that affect Irr levels regulate gene expression in a manner consistent with occupancy of their promoters. Gene expression was monitored by measuring mRNA abundance by quantitative real time PCR (qPCR). The negatively regulated genes bfr and blr7895 showed low mRNA abundance under iron limitation that was essentially independent of the manganese status (Figs. 1J, K), which is consistent with high occupancy of their promoters under low or high manganese exposure (Fig. 1D, E). Expression of the leuC and hemB genes were more responsive than bfr or blr7895 to manganese in iron-limited cells, showing partial derepression when manganese is limiting (Fig. 1L, M). This derepression correlated with decreased Irr occupancy of the leuC and hemB promoters (Fig. 1F, G).
Transcript abundance was also measured in cells grown in 2 μM iron that were exposed to 2 mM H2O2 over a 90 minute time course (Fig. 3H–M). The bfr and blr7895 genes remained repressed over that time compared to the 20 μM iron control as discerned by the low mRNA abundance (Fig. 3J, K). This is consistent with the maintenance of high Irr occupancy over that period (Fig. 3D, E). By contrast, leuC and hemB mRNA increased with time upon H2O2 exposure (Fig. 3L, M) concomitant with decreasing promoter occupancy (Fig. 3F, G), which in turn correlates with low affinity of Irr for those promoters.
To further address the idea that promoter affinity for Irr strongly influences gene expression in response to environmental factors that affect Irr levels, we examined the consequences of replacing the ICE motif in the bfr promoter with a low affinity one. As noted above, the ICE motifs within the bfr and fhuE promoters differ by 6 bp, and changes in those residues were sufficient to drastically alter the affinity of the respective promoters to Irr in vitro (Table 1). We constructed a mutant strain in which the bfr gene contains the ICE motif of the fhuE promoter (bfr/ICEfhuE), and the 6 bp substitutions are the only changes in the genome.
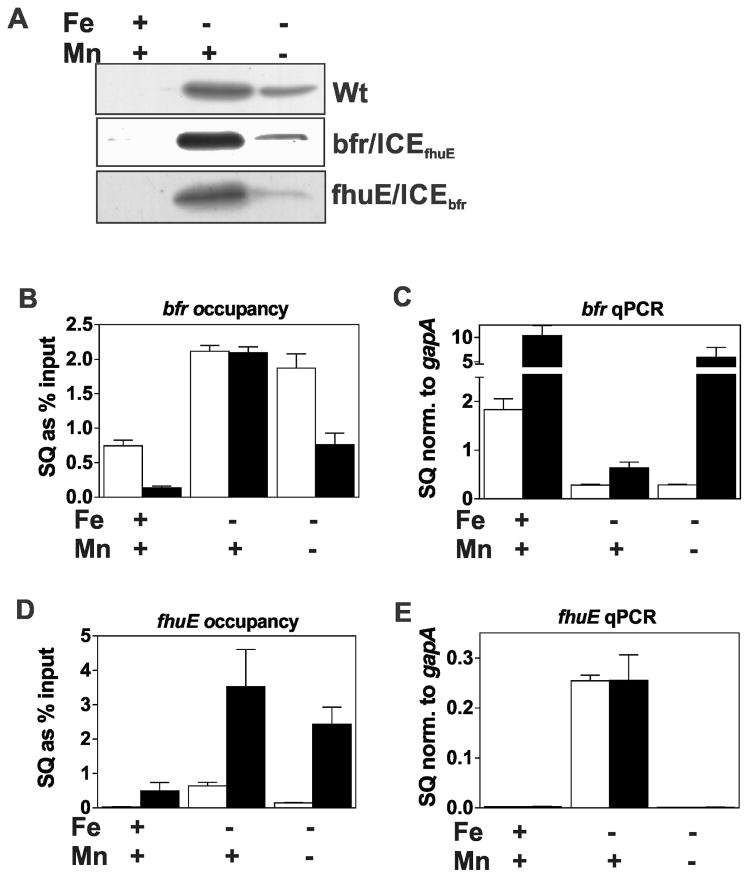
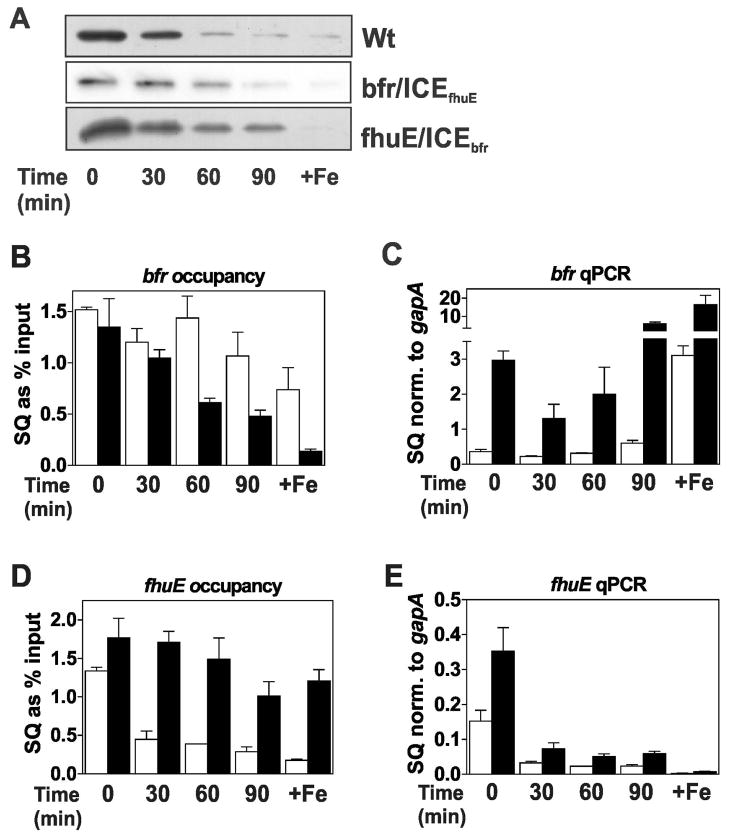
Cells of the wild type and bfr/ICEfhuE mutant were grown in manganese replete or depleted media under iron limitation. In the presence of manganese, where Irr levels are high (Fig. 4A), promoter occupancy of the wild type and mutant were similar (Fig. 4B), and the genes were likewise similarly repressed in each strain (Fig. 4C). Under low manganese conditions, occupancy of the wild type promoter was essentially the same as in manganese-replete cells, but the bfr/ICEfhuE promoter occupancy by Irr was lower (Fig. 4B). Correspondingly, bfr expression from the mutant promoter was derepressed resulting in expression even higher than that found under high iron conditions of the wild type (Fig. 4C). These data show that introduction of a low affinity Irr binding site into the bfr gene promoter is sufficient to confer strong manganese responsiveness to expression from that promoter. Even under iron replete conditions where Irr levels are extremely low, there was a large difference in promoter occupancy and expression between the wild type and the mutant (Figs. 4B, C). This shows that bfr is not fully derepressed in wild type cells even when grown in 20 μM Fe due to having a promoter with very high affinity for Irr.
Fig. 4.
Effects of ICE motif substitutions on promoter occupancy by Irr and gene expression in response to manganese. Cultures of wild type and the ICE mutant strains, bfr/ICEfhuE and fhuE/ICEbfr, were grown in media supplemented with 20 μM FeCl3 (Fe) or 50 μM MnCl2 (Mn) (+) or not supplemented with metal (−). The actual Fe and Mn concentrations in the unsupplemented media were 0.3 μM and 0.4 μM, respectively. (A) Steady state levels of Irr in wild-type and the ICE mutants were detected by immunoblotting as described in Fig. 1. (B) Promoter occupancy of bfr (open bars) and Pbfr/ICEfhuE gene (closed bars). (C) Steady state transcript levels of bfr (open bars) and Pbfr/ICEfhuE gene (closed bars). (D) Promoter occupancy of fhuE (open bars) and PfhuE/ICEbfr gene (closed bars). (E) Steady state transcript levels of the fhuE (open bars) and PfhuE/ICEbfr genes (closed bars).
The effects of H2O2 exposure on occupancy of, and expression from, the wild type and bfr/ICEfhuE promoters were examined (Fig. 5A–C). Cells grown in media supplemented with 2 μM FeCl3 were exposed to 2 mM H2O2 over 90 minutes (Fig. 5B, C), similar to that described in Fig. 3. Occupancy of the bfr/ICEfhuE promoter was more dependent on H2O2, decreasing throughout the time course of the experiment, as did Irr levels (Fig. 5A, B). Also, bfr mRNA levels expressed from the wild type promoter remained very low through 90 minutes, but expression from the bfr/ICEfhuE promoter was strongly responsive to H2O2 (Fig. 5C), showing a 10-fold increase in bfr transcripts relative to the wild type after 90 minutes exposure (Fig. 5C). Thus, introduction of a low affinity ICE motif into the bfr promoter conferred H2O2 responsiveness on that gene.
Fig. 5.
Effects of ICE motif substitutions on promoter occupancy by Irr and gene expression in response to H2O2. Cultures of wild type and the ICE mutant strains, bfr/ICEfhuE and fhuE/ICEbfr, were grown to mid-log phase in media supplemented with 2 μM FeCl3 and 50 μM MnCl2. At time 0, 2 mM H2O2 was added to the cultures and were continually aerated at 29°C. Cells were grown in 20 μM (+Fe) without H2O2 treatment as a control. (A) Steady-state levels of Irr in wild-type and the ICE mutants were detected by immunoblotting as described in Fig. 1. (B) Promoter occupancy of bfr (open bars) and Pbfr/ICEfhuE gene (closed bars). (C) Steady state transcript levels of bfr (open bars) and Pbfr/ICEfhuE gene (closed bars). (D) Promoter occupancy of fhuE (open bars) and PfhuE/ICEbfr gene (closed bars). (E) Steady state transcript levels of the fhuE (open bars) and PfhuE/ICEbfr genes (closed bars).
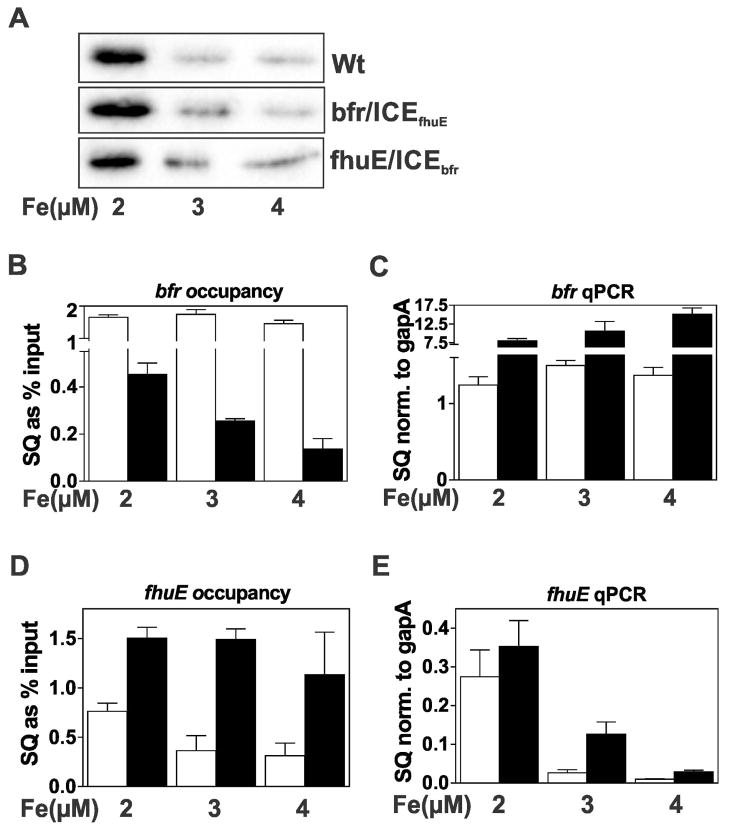
Finally, we examined the effects of iron on substituting the high affinity ICE motif in the bfr promoter for a low affinity one. Irr regulon genes with low or high affinity promoters respond to iron when comparing cells in iron deplete media compared with >10 μM supplementation. We chose a narrow range from 2 to 4 μM of iron where Irr levels vary but remain detectable (Fig. 6A) to better gauge sensitivity. The wild type bfr gene was mostly unresponsive to iron in this range both in terms of promoter occupancy (Fig. 6B) and mRNA abundance level that remained repressed (Fig. 6C). In the mutant, however, occupancy of the bfr/ICEfhuE promoter was iron responsive (Fig. 6B), and expression was strongly derepressed (Fig. 6C).
Fig. 6.
Effects of ICE motif substitutions on promoter occupancy by Irr and gene expression in response to iron. Cultures of wild type and the ICE mutant strains, bfr/ICEfhuE and fhuE/ICEbfr, were grown in media supplemented with 2 μM, 3 μM or 4 μM FeCl3 and with 50 μM MnCl2. (A) Steady-state levels of Irr in wild type and the ICE mutants were detected by immunoblotting as described in Fig. 1. (B) Promoter occupancy of bfr (open bars) and Pbfr/ICEfhuE gene (closed bars). (C) Steady state transcript levels of bfr (open bars) and Pbfr/ICEfhuE gene (closed bars). (D) Promoter occupancy of fhuE (open bars) and PfhuE/ICEbfr gene (closed bars). (E) Steady state transcript levels of the fhuE (open bars) and PfhuE/ICEbfr genes (closed bars).
Collectively, the evidence shows that, for genes that are negatively regulated by Irr, expression correlates strongly with promoter occupancy, which in turn is strongly influenced by the affinity of Irr to target promoters.
Inactivation of a positively regulated gene can occur without Irr vacancy of its promoter
The fhuE and bll7986 genes have promoters with low affinity for Irr which renders occupancy sensitive to the manganese status under low iron conditions (Fig. 1B,C). We found that expression of fhuE and bll7986 as measured by mRNA abundance was high in cells grown in low iron, high manganese media, but not in cells grown in low manganese or high iron media (Fig. 1H, I). Thus, expression of these genes was manganese-responsive.
Expression of the fhuE and bll7986 genes were also followed in response to H2O2 in cells grown with 2 μM iron as described above for the promoter occupancy experiments. Transcript levels of both genes dropped sharply by 30 minutes after H2O2 exposure (Fig. 3H, I). Thus, factors that promote Irr degradation lead to inactivation of positively-controlled genes.
The sensitivity of fhuE and bll7968 expression to the status of H2O2 and manganese appears to have a causal relationship with promoter occupancy, and the findings for the Irr-repressible genes predict that a positively regulated gene expressed from a promoter with high affinity for Irr would be less sensitive to those environmental factors. However, experiments described below show that this is not the case.
Based on microarray data (Yang et al., 2006b), binding studies (Small et al., 2009b) and bioinformatics predictions (Rodionov et al., 2006), we identified 8 genes or operons that are positively controlled by Irr and also have a known or predicted ICE motif. Both published (Small et al., 2009b) and unpublished studies failed to find ICE motifs of these genes with high affinity for Irr as estimated by those that are at least half-bound by 250 nM Irr in EMSA analysis. Nevertheless, we constructed a mutant strain containing an fhuE gene derivative with a high affinity promoter. To do this, we made a 6 bp change within the ICE motif of fhuE promoter so that it is identical to the ICE motif from the bfr promoter (fhuE/ICEbfr). These are the only changes in the genome of the fhuE/ICEbfr mutant strain.
Occupancy of the fhuE/ICEbfr promoter was very high in iron limited cells compared with the wild type promoter grown in either high or low manganese media (Fig. 4D), and thus high Irr affinity resulted in greater occupancy. However, fhuE transcript levels in the wild type and mutant strains were similar in low iron, high manganese cells despite the large difference in Irr occupancy of their promoters (Fig. 4E). Moreover, removal of manganese from the low iron medium resulted in nearly undetectable fhuE transcripts even though Irr occupancy of the mutant promoter remained very high. Finally, occupancy of the mutant promoter by Irr in iron replete cells was only modestly lower than that found in the wild type promoter in iron-limited cells. Thus, expression of fhuE in the mutant strain was manganese-responsive even though occupancy of its promoter by Irr remained and high was relatively unresponsive to the metal.
We then compared the effects of H2O2 exposure on wild type and fhuE/ICEbfr cells grown in media supplemented with 2 μM FeCl3 (Fig. 5A, D, E). Whereas Irr occupancy of the wild type fhuE promoter decreased over time upon H2O2 exposure, occupancy of the fhuE/ICEbfr promoter remained high throughout the time course. Nevertheless, transcript levels in the mutant strain declined rapidly in response to H2O2 exposure as was also observed in the wild type.
Responses to iron were also examined in the narrow range of 2 to 4 μM as described above (Fig. 6A, D, E). Again, introduction of a high affinity Irr binding site into the fhuE promoter yielded very high Irr occupancy that changed little from 2 to 4 μM iron (Fig. 6D). Despite the high Irr occupancy, fhuE transcripts decreased in the mutant strain with increasing iron concentrations (Fig. 6E).
Collectively, the findings show that environmental factors that decrease the cellular Irr level can deactivate expression of target genes by a mechanism that does not require promoter vacancy. This differs from Irr-repressible genes, where repression correlates with promoter occupancy.
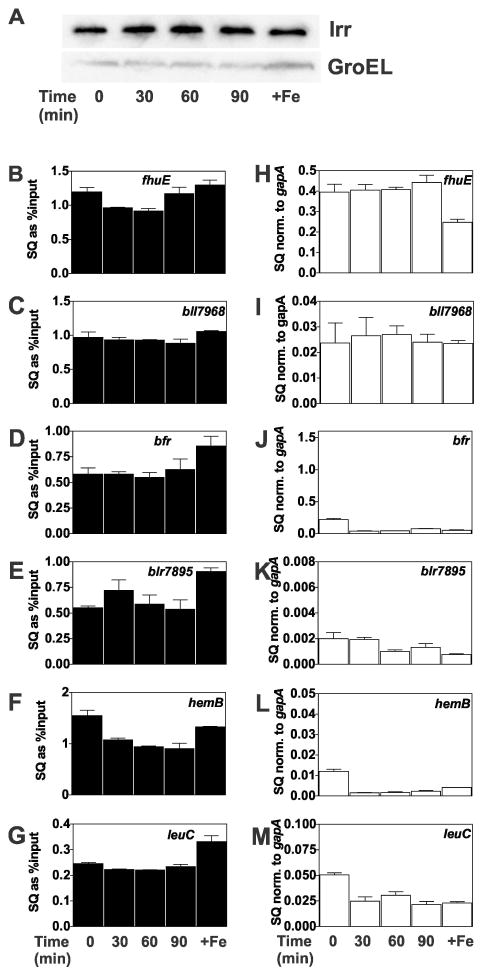
Exposure of iron-limited cells to hydrogen peroxide inactivates expression of Irr target genes, but does not affect cellular Irr levels, promoter occupancy or repression
Whereas Irr degrades in response to H2O2 in cells grown in 2 μM iron, very little degradation occurs upon short term exposure of iron-limited cells to H2O2 (Yang et al., 2006a), which was reproduced here (Fig. 7A). In vitro, H2O2 can oxidize Irr in the presence of heme (Yang et al., 2006a), and thus we wanted to address whether H2O2 exposure affected Irr occupancy or expression of target genes in vivo.
Fig. 7.
Effect of H2O2 on promoter occupancy by Irr and expression of Irr-regulated genes in wild type cells grown in iron-limited media. Cultures were grown to mid-log phase in media not supplemented with iron. The actual iron concentration was 0.3 μM. At time 0, 2 mM H2O2 was added to the cultures and were continually aerated at 29°C and harvested at the times shown. Cells were grown in 20 μM (+Fe) as a control. (A) Steady-state levels of Irr and GroEL were detected by immunoblotting as described in Fig. 1. (B–G) Promoter occupancy of respective genes by Irr in cells treated with H2O2 were carried out as described in the text. The data are expressed as the relative starting quantities (SQ) of immunoprecipitated DNA normalized to the input DNA and are presented as average of three replicates ± the standard deviation. (H–M) Steady-state transcript levels of the respective genes obtained from cells treated with H2O2 were analyzed by qPCR. The data are expressed as relative starting quantities (SQ) of respective mRNAs normalized to the housekeeping gene gapA, and presented as average of three replicates ± the standard deviation.
Promoter occupancy of genes positively or negatively regulated by Irr remained high throughout the 90 minute exposure to H2O2 (Figs. 7B–G), consistent with the maintenance of a high level of cellular Irr over that time period (Fig. 7A). Correspondingly, the negatively-regulated genes blr7895, bfr, leuC and hemB remained repressed under those conditions compared with a high iron control (Fig. 7J–M), showing that Irr remains functional as a repressor. However, transcript levels of the positively-controlled genes fhuE and bll7968 decreased by 30 minutes after H2O2 exposure and remained low even though occupancy by Irr of those promoters remained high. Thus, loss of activation function of Irr can occur without loss of DNA-binding activity or repressor activity in wild type cells.
Loss of Irr-dependent activation of target genes is heme-dependent
To determine whether the effects of H2O2 on Irr activity in iron-limited cells depended on heme, we measured promoter occupancy and expression of Irr-responsive genes in a heme-deficient strain in response to H2O2 exposure. Irr is stable in a heme-deficient strain under conditions where it degrades in the wild type (Puri et al., 2010, Qi and O’Brian, 2002, Yang et al., 2006a), and thus Irr levels remained high in iron-replete control cells of the mutant (Fig. 8A). As in the wild type, cellular Irr levels remained high throughout the 90 exposure of H2O2 in the heme-deficient strain (Fig. 8A). Consistent with this, promoter occupancy by Irr remained high in the six genes examined (Fig. 8B–G), and expression of the negatively controlled genes blr7895, bfr, leuC and hemB remained repressed (Fig. 8J–M). However, unlike the wild type, expression of fhuE and bll7968 did not diminish in response to H2O2 exposure (Fig. 8H, I). These findings show that H2O2-dependent loss of Irr activation function requires heme by a mechanism that is distinct from heme-dependent degradation of Irr.
Fig. 8.
Effect of H2O2 on promoter occupancy by Irr and expression of Irr-regulated genes in heme-deficient strain ΔhemAH grown in iron-limited media. Cultures were grown to mid-log phase in media not supplemented with iron and supplemented with 150 nM hemin to satisfy heme auxotrophy. The actual iron concentration was 0.3 μM. At time 0, 2 mM H2O2 was added to the cultures and were continually aerated at 29°C, and harvested at the times shown. (A) Steady-state levels of Irr and GroEL were detected by immunoblotting as described in Fig. 1. (B–G) Promoter occupancy of respective genes by Irr in cells treated with H2O2 were carried out as described in the Fig. 1 legend. The data are expressed as the relative starting quantities (SQ) of immunoprecipitated DNA normalized to the input DNA and are presented as average of three replicates ± the standard deviation. (H–M) Steady state transcript levels of the respective genes obtained from cells treated with H2O2 were analyzed by qPCR. The data are expressed as relative starting quantities (SQ) of respective mRNAs normalized to the housekeeping gene gapA, and presented as average of three replicates ± the standard deviation.
DISCUSSION
In previous work, we showed that B. japonicum Irr is a conditionally stable protein that accumulates in cells under iron limitation and when manganese is sufficient, but degrades in a heme-dependent manner under high iron conditions, manganese limitation or upon exposure to H2O2. Conditions that degrade Irr result in the inactivation of positively controlled genes and derepression of negatively regulated genes. In the present study, we identified two additional levels of control, one which relies on differential affinity of Irr for target promoters, and another which abrogates the activation function of Irr without loss of DNA-binding activity or repressor function. Heme is required for the loss of activation function by a mechanism that does not require degradation. We suggest that these additional levels of control allow differential regulation among negatively regulated genes, and the uncoupling of activation from repression under certain conditions. Thus, the Irr regulon shows a greater complexity of control than can be achieved by conditional stability alone.
The Kd values of Irr for target promoters spanned from low nanomolar to the micromolar range. Irr levels in cells were estimated by quantifying protein levels in cells by Western blots using pure protein as a standard (data not shown), and also estimating the cell volume to be 0.6–1 μm3 (Kanbe et al., 2007), yielding a cellular Irr concentration of 0.7 to 1.1 μM under iron limitation. The comparison between Kd values and Irr concentration is imperfect because supercoiled DNA is the target for Irr in vivo, purified Irr may not be as robust as the in vivo protein, and cell volume estimates include the periplasm, but Irr is cytosolic. Nevertheless, the estimations imply that the cellular Irr concentration is sufficiently high to occupy promoters with low affinity for it, as was confirmed by the in vivo analyses described here. A broad range of binding affinities of the Fur family proteins Zur from Streptomyces coelicolor (Shin et al., 2011) and Fur from Neisseria gonorrhoeae (Yu and Genco, 2012) for target promoters has been observed, but the concentration of those proteins do not vary greatly in response to their respective regulatory metal. Instead, the metal primarily controls DNA-binding activity.
Irr occupancy of target promoters correlated well with its affinity for the promoter, and for the ICE motif in particular. We suggest that, for negatively regulated genes, the broad range of affinities of Irr for target promoters allows different levels of repression at a given Irr concentration to accommodate the physiological function of each gene product and the cellular status (Fig. 9). The large subunit 3-isopropylmalate dehydrogenase encoded by leuC is an iron-sulfur protein, and the hemB product 5-aminolevulinic acid dehydrogenase is involved in the synthesis of heme, an iron containing prosthetic group. Repression of these genes under low iron conditions ensures that the synthesis of the proteins does not exceed iron availability. However, B. japonicum cells grow aerobically under that condition, hence some expression of housekeeping genes such as hemB and leuC must be maintained. Thus, a weaker affinity promoter may allow some level of expression (derepression) under a low or moderate iron availability where Irr levels are high. By contrast, bacterioferritin and perhaps Blr7895 are likely to be important in managing stress associated with iron-dependent chemistry, hence strong repression is appropriate under low or moderate iron levels, which can be achieved by having those genes controlled by promoters with a high affinity Irr-binding site. Similarly, manganese limitation creates an iron deficiency (Puri et al., 2010), and repression of bfr and blr7895 is maintained even though Irr levels are diminished under that condition.
Fig. 9.
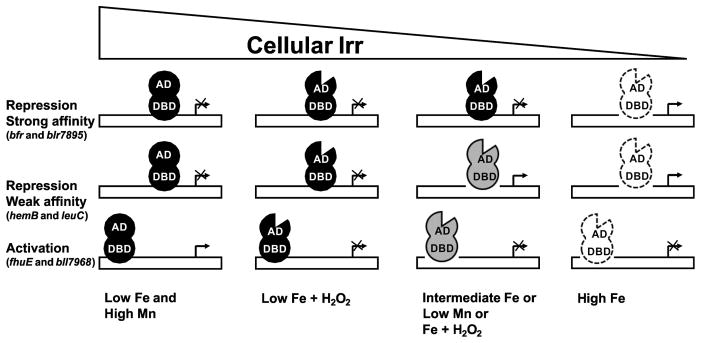
Representation of the effects of environmental factors on cellular Irr levels, promoter occupancy by Irr and gene expression. AD and DBD are activation domains and DNA-binding domains of Irr, respectively. The notch in the AD depicts heme-dependent loss of activation function of Irr. The lighter shading of Irr denotes decreasing occupancy on the Irr promoter.
For genes positively regulated by Irr, we showed that abrogation of activation can occur by a process that is distinct from Irr degradation. As a result, diminution of gene expression can occur even when promoter occupancy by Irr remains high (Fig. 9). Experiments with the fhuE/ICEbfr mutant show that the low level of Irr remaining in cells after exposure to iron, low manganese conditions or H2O2 plus 2 μM Fe is not functional as an activator. Similarly, fhuE and bll7986 transcripts declined in iron-limited wild type cells in response to H2O2 even though cellular Irr levels and promoter occupancy remained high. Retention of promoter occupancy under these conditions shows that loss of activation function of Irr does not require loss of DNA binding activity.
The retention of DNA binding activity of Irr by treatments that modify activation function or stability may explain why Irr was able to repress gene expression under those conditions. It is likely that DNA binding is sufficient to occlude the promoter from RNA polymerase, and therefore repression by Irr is not affected by modifications that do not change promoter occupancy. This is consistent with previous work showing that Irr binding to a target promoter is sufficient to repress transcription in vitro (Sangwan et al., 2008). We suggest that loss of repression function by Irr occurs primarily through its diminution in cells via degradation resulting in promoter vacancy. The extent of this vacancy is dictated by the affinity of the promoter for the regulator (Fig. 9). Irr-dependent activation also requires promoter occupancy, hence Irr stability, but loss of activity need not be accompanied by promoter vacancy. As a result, inactivation is not sensitive to affinity of Irr to the target promoter. It is plausible that the activated genes have promoters with low affinity for Irr because there is no evolutionary selection for a high affinity binding site.
The loss of activation function of Irr without degradation uncouples activation from repression in a manner that control by degradation alone cannot, and this may be advantageous under certain conditions. For example, cells exposed to H2O2 under low iron conditions where Irr functions may respond by down regulating genes involved in iron transport (e.g. fhuE, bll7968) as a means to mitigate the formation of reaction oxygen species through the Fenton reaction. Under these conditions, a concomitant derepression of genes that encode iron-containing proteins would not be appropriate because there is no increase in the iron level. More generally, this control may be appropriate for any cellular state that does not call for simultaneous inactivation and derepression of the respective Irr regulon genes.
The decrease in fhuE or bll7968 in iron-limited wild type cells in response to H2O2 was not observed in the heme-defective mutant strain, showing that heme is involved in Irr-dependent gene expression that is distinct from its role in degradation. Specifically, heme is required for abrogation of the activation function of Irr. The data suggest that Irr has DNA-binding and activation domains that can be functionally distinguished based on the ability of the latter to be specifically altered by heme (Fig. 9). This alteration may interfere with Irr binding to RNA polymerase or to another regulatory protein. Heme is known to catalyze Irr oxidation in vitro, and evidence indicates that this is important for degradation in vivo (Yang et al., 2005, Yang et al., 2006a). Oxidation may also play a role in inactivation, but if so degradation must require at least one additional event since heme-dependent degradation and inactivation are functionally separable.
Heme binds to Irr to inactivate it, but this does not abrogate DNA-binding in vivo. This conclusion differs from that reported for Rhizobium leguminosarum Irr, where heme inhibits DNA binding in vitro (Singleton et al., 2010). It is also at variance with findings for essentially all other characterized Fur family proteins, where in those cases the protein-binding regulatory ligand greatly diminishes DNA binding activity (Bsat et al., 1998, Gaballa and Helmann, 1998, Patzer and Hantke, 1998, Ahn et al., 2006, Hohle and O’Brian, 2009, Platero et al., 2007).
The auxiliary regulator HmuP was identified in Irr-dependent control of the heme transport cluster of B. japonicum (Escamilla-Hernandez and O’Brian, 2012), and control of the irr gene by Irr proceeds by anti-repression of the Mur protein (Hohle and O’Brian, 2010). Although we cannot rule out additional regulatory factors that mediate control of Irr regulon genes, the observations described here do not mandate one, and can be explained in terms of differential promoter affinity and abrogation of its activation function while keeping repressor activity intact.
MATERIALS AND METHODS
Strains and media
Bradyrhizobium japonicum USDA I110 is the parent strain used in this study. Strain ΔhemAH is a double mutant defective in hemA and hemH genes, encoding heme biosynthesis enzymes (Qi and O’Brian, 2002). B. japonicum strains were routinely grown at 29°C in glycerol-salts-yeast (GSY) medium as previously described (Frustaci et al., 1991). Strain ΔhemAH was grown in media supplemented with 50 μg/ml kanamycin, 50 μg/ml streptomycin, 100 μg/ml spectinomycin, with 1 μM hemin hydrochloride to fulfill its heme auxotrophy. For experiments requiring low heme conditions, the medium was supplemented with 0.15 μM heme, which allows growth comparable to the wild type but retains other heme-defective phenotypes (Qi and O’Brian, 2002).
For low iron or low manganese conditions, modified GSY was used, which contains 0.5 g per liter of yeast extract instead of 1 g per liter, with no exogenous iron or manganese added. The actual iron and manganese concentrations in the medium were 0.3 and 0.4 μM, respectively, as determined with a PerkinElmer model 1100B atomic absorption spectrophotometer. Media for growth under high iron or high manganese conditions are supplemented with 20 μM FeCl3 or 50 μM MnCl2, respectively.
Hydrogen peroxide treatments
Cells were grown to mid log phase in modified GSY medium supplemented with no iron or 2 μM FeCl3 and with 50 μM MnCl2. Cultures were then treated with 2 mM hydrogen peroxide (Sigma, MO) with continuous shaking at 29°C. Aliquots were collected at specified time points for analysis.
Construction of the ICE mutant strains
The promoter regions of bfr and fhuE were modified so that the ICE motif sequence within one gene promoter was substituted for the other. To do this, DNA fragments containing the 21bp ICE motif flanked on either side by 700bp DNA were amplified by PCR using genomic DNA as template, and the products were ligated into pBluescript SK+. The 6 bp substitutions in each construct were made by site-directed mutagenesis (Stratagene, La Jolla, CA). The mutated constructs were introduced into pLO1 (Lenz et al., 1994), mobilized into strain USDA I110, and recombinants were selected as described previously (Escamilla-Hernandez and O’Brian, 2012). Mutants were confirmed by DNA sequencing.
Immunoblotting
Cultures were harvested, washed and re-suspended in phosphate-buffered saline (10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl [pH 7.4]). Total protein concentrations were determined by BCA protein assay (Pierce, Rockford, IL). Fifteen μg of total cell protein were boiled in loading buffer and resolved by electrophoresed through 15% SDS-polyacrylamide gels. Immunoblotting was carried out with Irr or GroEL (Stressgen, Vancouver, Canada) polyclonal antibodies and HRP-conjugated goat anti-rabbit IgG (Southern Biotech, Birmingham, AL) with detection using the Immobion system (Millipore, Billerica, MA).
Overexpression and purification of Irr
The irr gene was amplified by PCR and cloned into pETM-11 vector containing an N-terminal 6xHis tag. The vector with insert was transformed into chemical competent BL21 (DE3) E. coli cells. Cells were inoculated from an overnight culture grown in Luria-Bertani media containing 20 μg/ml kanamycin and 25 μg/ml chloramphenicol into 1 l of fresh 2×YT medium (Sambrook et al., 1989) containing the same antibiotics and 50 μM MnCl2. Overexpression was induced in cells at the midlog phase by the addition of 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) at 37 °C for 4 h with shaking. Cells were pelleted by centrifugation, washed in lysis buffer (5 mM Tris-HCl, pH 8.0). The pellet was further washed in lysis buffer containing 4% Thesit (Sigma, MO) followed by a wash in lysis buffer with 2M urea, before finally solubilizing in lysis buffer with 8M urea. Cells were disrupted by passage through a French pressure cell at 1,200 psi and clarified by centrifugation at 13,000 × g for 10 min. Two milliliters of Ni-NTA slurry (Qiagen) was added to the cleared lysate and rocked for 60 min at room temperature (22 °C).
The Ni-NTA slurry-protein mixture was poured into a column and washed with ten column volumes of lysis wash buffer (5 mM Tris-HCl, 8 M Urea, and 30 mM imidazole, pH 8.0). His-Irr was eluted using lysis elution buffer (5 mM Tris-HCl, 8 M Urea, and 250 mM Imidazole, pH 8.0). The purified protein was run through an FPLC buffer exchange column so that the final buffer consisted of 10 mM Tris-HCl, 100 μM MnCl2, pH 8.0.
EMSA
Electrophoretic mobility shift assays (EMSA) were used to analyze the binding of purified Irr to various DNA probes in vitro as described previously (Friedman and O’Brian, 2004). The negative-control DNA corresponds to the sequence found in the multiple-cloning site of pBluescript SK+. The test DNA probes were 45 bp in length and contained 21-bp ICE (underlined in the sequences below) flanked either by the original genomic flanks or the genomic flanks of a different ICE motif. The sequences of the probes are shown in Fig. S1. EMSA reactions were analyzed as autoradiograms of 5% non-denaturing polyacrylamide gels. Autoradiograms were scanned using GS-700 densitometer (Bio-Rad), and signal intensities were determined and quantified by Quantity One software (Bio-Rad). To determine the dissociation binding constant (Kd), binding reactions containing 0.1 nM 32P-labelled DNA were titrated with various concentrations of Irr. Bound and unbound DNAs were quantified by comparing relative signal intensities and analyzed using GraphPad Prism software (GraphPad software Inc., San Diego, CA)
Analysis of RNA by quantitative real-time PCR
Steady state transcript levels of selected genes were determined by qPCR with SsoAdvanced SYBR Green supermix (Bio-Rad) using iCycler thermal cycler (Bio-Rad). RNA was isolated from B. japonicum cells using a hot phenol method as described previously (Yang et al., 2006a). cDNA was synthesized from 1 μg total RNA using iScript cDNA synthesis kit (Bio-Rad). qPCRs were carried out as previously described (Hohle and O’Brian, 2009). Data were normalized to gapA and are expressed as average of triplicates, with standard deviation represented by the error bars.
Quantitative in vivo cross-linking and immunoprecipitation
Cells were grown to mid-log phase. Twenty-five-milliliter aliquots were spun down and the pellets were saved at −80°C until further use. For cross-linking, pellets were re-suspended in phosphate-buffered saline (10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4) and formaldehyde was added to 1% (vol/vol) and the samples were gently rocked at room temperature for 10 min. To quench the cross-linking, glycine was added to a final concentration of 10 mg/ml, and the cells were shaken gently at 4°C for 30 min. Cells were collected by centrifugation, and the pellets were resuspended and washed twice with phosphate-buffered saline. Washed cells were re-suspended in 2 ml lysis buffer (100 mM Tris pH 8.0, 300 mM NaCl, and 10 mM EDTA) and lysed with a French pressure cell as described previously. The cell extracts were aliquoted and frozen at −80°C until further use. Protein-A agarose beads, pre-blocked with salmon sperm DNA and bovine serum albumin (Millipore, Billerica, MA), were washed with lysis buffer and resuspended to a slurry of 33%. The lysates were pre-cleared by adding 20 μl of bead slurry to 200 μl cell lysate and mixed slowly at room temperature for 1 h with a rotation mixer, followed by centrifugation for 1 min at 1,000 × g. One microliter of the lysate was diluted with 9 μl of the dilution buffer and the mixture was saved at −80 °C, to serve as the input-control (1% of input DNA). One hundred microliters of the remaining supernatant was diluted with 900 μl of dilution buffer. Serum containing anti-Irr poly- clonal antibodies was added to a dilution of 0.08 μl for every 15 μg of total protein and the tube was left rotating slowly at 4°C overnight with a rotation mixer. Fifty microliters of washed bead slurry was added to the samples and mixed by rotation at room temperature for 1 h. The protein-DNA complex was washed and eluted from the beads at room temperature as per the manufacturer’s instructions. The input-control samples were also diluted with the elution buffer to the same final volume as the samples. The DNA from both the samples and the input controls was uncrosslinked from the protein by adding NaCl to 0.2 M to the eluted samples, and the samples were incubated at 65°C overnight. DNA was purified using the Qiagen PCR purification kit and eluted in 50 μl of elution buffer. Immunoprecipitated DNA of selected promoter regions was quantified using qPCR as described above, using 0.4 μl as template. The data were normalized to input DNA.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01 GM099667 to M.R.O’B.
References
- Ahn BE, Cha J, Lee EJ, Han AR, Thompson CJ, Roe JH. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol Microbiol. 2006;59:1848–1858. doi: 10.1111/j.1365-2958.2006.05065.x. [DOI] [PubMed] [Google Scholar]
- Amarelle V, Koziol U, Rosconi F, Noya F, O’Brian MR, Fabiano E. A new small regulatory protein, HmuP, modulates haemin acquisition in Sinorhizobium meliloti. Microbiology. 2010;156:1873–1882. doi: 10.1099/mic.0.037713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson ES, Paulley JT, Martinson DA, Gaines JM, Steele KH, Roop RM., II The iron-responsive regulator Irr Is required for wild-type expression of the gene encoding the heme transporter BhuA in Brucella abortus 2308. J Bacteriol. 2011;193:5359–5364. doi: 10.1128/JB.00372-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD. Bacillus subtilis contains multiple Fur homologs: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- Chauhan S, O’Brian MR. Bradyrhizobium japonicum δ-aminolevulinic acid dehydratase is essential for symbiosis with soybean and contains a novel metal-binding domain. J Bacteriol. 1993;175:7222–7227. doi: 10.1128/jb.175.22.7222-7227.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, O’Brian MR. Transcriptional regulation of δ-aminolevulinic acid dehydratase synthesis by oxygen in Bradyrhizobium japonicum and evidence for developmental control of the hemB gene. J Bacteriol. 1997;179:3706–3710. doi: 10.1128/jb.179.11.3706-3710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla-Hernandez R, O’Brian MR. HmuP is a co-activator of Irr-dependent expression of heme utilization genes in Bradyrhizobium japonicum. J Bacteriol. 2012;194:3137–3143. doi: 10.1128/JB.00071-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman YE, O’Brian MR. The ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum is an iron-responsive transcriptional repressor in vitro. J Biol Chem. 2004;279:32100–32105. doi: 10.1074/jbc.M404924200. [DOI] [PubMed] [Google Scholar]
- Frustaci JM, Sangwan I, O’Brian MR. Aerobic growth and respiration of a δ-aminolevulinic acid synthase (hemA) mutant of Bradyrhizobium japonicum. J Bacteriol. 1991;173:1145–1150. doi: 10.1128/jb.173.3.1145-1150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Helmann JD. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza I, Chauhan S, Hassett R, O’Brian MR. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J Biol Chem. 1998;273:21669–21674. doi: 10.1074/jbc.273.34.21669. [DOI] [PubMed] [Google Scholar]
- Hohle TH, O’Brian MR. The mntH gene encodes the major Mn2+ transporter in Bradyrhizobium japonicum and is regulated by manganese via the Fur protein. Mol Microbiol. 2009;72:399–409. doi: 10.1111/j.1365-2958.2009.06650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohle TH, O’Brian MR. Transcriptional control of the Bradyrhizobium japonicum irr gene requires repression by Fur and antirepression by Irr. J Biol Chem. 2010;285:26074–26080. doi: 10.1074/jbc.M110.145979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanbe M, Yagasaki J, Zehner S, Gottfert M, Aizawa S. Characterization of two sets of subpolar flagella in Bradyrhizobium japonicum. J Bacteriol. 2007;189:1083–1089. doi: 10.1128/JB.01405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz O, Schwartz E, Dernedde J, Eitinger M, Friedrich B. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J Bacteriol. 1994;176:4385–4393. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M, Ugalde RA, Almiron M. Dimeric Brucella abortus Irr protein controls its own expression and binds haem. Microbiology. 2005;151:3427–3433. doi: 10.1099/mic.0.28213-0. [DOI] [PubMed] [Google Scholar]
- Page KM, Guerinot ML. Oxygen control of the Bradyrhizobium japonicum hemA gene. J Bacteriol. 1995;177:3979–3984. doi: 10.1128/jb.177.14.3979-3984.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- Platero R, de Lorenzo V, Garat B, Fabiano E. Sinorhizobium meliloti fur-like (Mur) protein binds a fur box-like sequence present in the mntA promoter in a manganese-responsive manner. Appl Environ Microbiol. 2007;73:4832–4838. doi: 10.1128/AEM.00686-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri S, Hohle TH, O’Brian MR. Control of bacterial iron homeostasis by manganese. Proc Natl Acad Sci USA. 2010;107:10691–10695. doi: 10.1073/pnas.1002342107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Hamza I, O’Brian MR. Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc Natl Acad Sci USA. 1999;96:13056–13061. doi: 10.1073/pnas.96.23.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, O’Brian MR. Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol Cell. 2002;9:155–162. doi: 10.1016/s1097-2765(01)00431-2. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-Proteobacteria. PLoS Comput Biol. 2006;2:e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruangkiattikul N, Bhubhanil S, Chamsing J, Niamyim P, Sukchawalit R, Mongkolsuk S. Agrobacterium tumefaciens membrane-bound ferritin plays a role in protection against hydrogen peroxide toxicity and is negatively regulated by the iron response regulator. FEMS Microbiology Letters. 2012;329:87–92. doi: 10.1111/j.1574-6968.2012.02509.x. [DOI] [PubMed] [Google Scholar]
- Rudolph G, Semini G, Hauser F, Lindemann A, Friberg M, Hennecke H, Fischer HM. The Iron control element, acting in positive and negative control of iron-regulated Bradyrhizobium japonicum genes, is a target for the Irr protein. J Bacteriol. 2006;188:733–744. doi: 10.1128/JB.188.2.733-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a laboratory manual. 2. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sangwan I, Small SK, O’Brian MR. The Bradyrhizobium japonicum Irr protein is a transcriptional repressor with high-affinity DNA-binding activity. J Bacteriol. 2008;190:5172–5177. doi: 10.1128/JB.00495-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Jung HJ, An YJ, Cho YB, Cha SS, Roe JH. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc Natl Acad Sci U S A. 2011;108:5045–5050. doi: 10.1073/pnas.1017744108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C, White GF, Todd JD, Marritt SJ, Cheesman MR, Johnston AW, Le Brun NE. Heme-responsive DNA binding by the global iron regulator Irr from Rhizobium leguminosarum. J Biol Chem. 2010;285:16023–16031. doi: 10.1074/jbc.M109.067215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SK, O’Brian MR. The Bradyrhizobium japonicum frcB gene encodes a diheme ferric reductase. J Bacteriol. 2011;193:4088–4094. doi: 10.1128/JB.05064-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SK, Puri S, O’Brian MR. Heme-dependent metalloregulation by the iron response regulator (Irr) protein in Rhizobium and other Alpha-proteobacteria. Biometals. 2009a;22:89–97. doi: 10.1007/s10534-008-9192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SK, Puri S, Sangwan I, O’Brian MR. Positive control of ferric siderophore receptor gene expression by the Irr protein in Bradyrhizobium japonicum. J Bacteriol. 2009b;191:1361–1368. doi: 10.1128/JB.01571-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JD, Sawers G, Rodionov DA, Johnston AW. The Rhizobium leguminosarum regulator IrrA affects the transcription of a wide range of genes in response to Fe availability. Mol Genet Genomics. 2006;275:564–577. doi: 10.1007/s00438-006-0115-y. [DOI] [PubMed] [Google Scholar]
- Todd JD, Wexler M, Sawers G, Yeoman KH, Poole PS, Johnston AW. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology. 2002;148:4059–4071. doi: 10.1099/00221287-148-12-4059. [DOI] [PubMed] [Google Scholar]
- Yang J, Ishimori K, O’Brian MR. Two heme binding sites are involved in the regulated degradation of the bacterial iron response regulator (Irr) protein. J Biol Chem. 2005;280:7671–7676. doi: 10.1074/jbc.M411664200. [DOI] [PubMed] [Google Scholar]
- Yang J, Panek HR, O’Brian MR. Oxidative stress promotes degradation of the Irr protein to regulate haem biosynthesis in Bradyrhizobium japonicum. Mol Microbiol. 2006a;60:209–218. doi: 10.1111/j.1365-2958.2006.05087.x. [DOI] [PubMed] [Google Scholar]
- Yang J, Sangwan I, Lindemann A, Hauser F, Hennecke H, Fischer HM, O’Brian MR. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol Microbiol. 2006b;60:427–437. doi: 10.1111/j.1365-2958.2006.05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Genco CA. Fur-mediated activation of gene transcription in the human pathogen Neisseria gonorrhoeae. J Bacteriol. 2012;194:1730–1742. doi: 10.1128/JB.06176-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappa S, Bauer CE. The LysR-type transcription factor HbrL is a global regulator of iron homeostasis and porphyrin synthesis in Rhodobacter capsulatus. Mol Microbiol. 2013;90:1277–1292. doi: 10.1111/mmi.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.