Abstract
Salmonella enterica subsp. enterica serovar Heidelberg (S. Heidelberg) is one of the top serovars causing human salmonellosis. Recently, an antibiotic-resistant strain of this serovar was implicated in a large 2011 multistate outbreak resulting from consumption of contaminated ground turkey that involved 136 confirmed cases, with one death. In this study, we assessed the evolutionary diversity of 44 S. Heidelberg isolates using whole-genome sequencing (WGS) generated by the 454 GS FLX (Roche) platform. The isolates, including 30 with nearly indistinguishable (one band difference) Xbal pulsed-field gel electrophoresis patterns (JF6X01.0032, JF6X01.0058), were collected from various sources between 1982 and 2011 and included nine isolates associated with the 2011 outbreak. Additionally, we determined the complete sequence for the chromosome and three plasmids from a clinical isolate associated with the 2011 outbreak using the Pacific Biosciences (PacBio) system. Using single-nucleotide polymorphism (SNP) analyses, we were able to distinguish highly clonal isolates, including strains isolated at different times in the same year. The isolates from the recent 2011 outbreak clustered together with a mean SNP variation of only 17 SNPs. The S. Heidelberg isolates carried a variety of phages, such as prophage P22, P4, lambda-like prophage Gifsy-2, and the P2-like phage which carries the sopE1 gene, virulence genes including 62 pathogenicity, and 13 fimbrial markers and resistance plasmids of the incompatibility (Inc)I1, IncA/C, and IncHI2 groups. Twenty-one strains contained an IncX plasmid carrying a type IV secretion system. On the basis of the recent and historical isolates used in this study, our results demonstrated that, in addition to providing detailed genetic information for the isolates, WGS can identify SNP targets that can be utilized for differentiating highly clonal S. Heidelberg isolates.
Keywords: outbreak, antimicrobial resistance, plasmid, SNP analysis, trace-back
Introduction
Disease caused by nontyphoidal serotypes of Salmonella enterica is the leading cause of food-related death in the United States (Scallan et al. 2011). Salmonella enterica supspecies (supsp.) enterica serovar Heidelberg (S. Heidelberg) has been among the most frequently isolated serovars in clinical cases of salmonellosis, causing an estimated 84,000 illnesses in the United States annually (Foley et al. 2011; Han et al. 2011). It is frequently isolated from poultry and poultry meat (Zhao et al. 2008). S. Heidelberg was the seventh most common serovar isolated from humans in 2011 (CDC 2013) and was found in chicken breast, ground turkey, and eggs, the main sources of S. Heidelberg infections (Chittick et al. 2006; NARMS 2010). S. Heidelberg also tends to be associated with invasive diseases such as septicemia and myocarditis (Wilmshurst and Sutcliffe 1995). After S. enterica supsp. enterica serovar Typhimurium (S. Typhimurium), S. Heidelberg is the serovar of Salmonella most often associated with Salmonella-related deaths in the United States (Kennedy et al. 2004; Patchanee et al. 2008; Crump et al. 2011). The National Antimicrobial Resistance Monitoring System (NARMS), which is responsible for monitoring antimicrobial resistance in Salmonella estimated that 65% of the S. Heidelberg isolates from ground turkey in 2010 were resistant to ≥3 antimicrobial classes. Presently, the antimicrobial agents to which this serovar is most commonly resistant are ceftriaxone (a drug of choice for treatment), along with resistance to streptomycin, tetracycline, sulfamethoxazole, chloramphenicol, and trimethoprim-sulfamethoxazole (NARMS 2010).
The Centers for Disease Control and Prevention (CDC) investigated a multistate (34 states) outbreak of antimicrobial-resistant S. Heidelberg infections comprised of 136 confirmed cases between February 27 and September 13, 2011. Among the 94 case patients for which there was available information, 37 (39%) had been hospitalized and one patient died. Collaborative investigative efforts by state and federal officials implicated ground turkey as the source of this outbreak, and as a result, 36 million pounds of ground turkey meat were recalled (CDC 2011; Folster et al. 2012).
Recently, our investigative capabilities have been greatly enhanced with the development and increasing feasibility of whole-genome sequencing (WGS) as a molecular epidemiological tool to complement current foodborne outbreak investigation techniques. WGS is of particular interest because it provides definitive data for distinguishing outbreak isolates from nonoutbreak isolates in common and highly clonal populations (Allard et al. 2012, 2013). In this study, we sought to determine how effectively WGS and single nucleotide polymorphisms (SNPs) analysis would differentiate outbreak isolates of S. Heidelberg from nonoutbreak isolates that share the same Xbal and BlnI pulsed-field gel electrophoresis (PFGE) patterns. Using WGS data and virulence assays, we also wanted to determine whether the outbreak isolates belonged to a new strain with potentially higher pathogenicity or whether these isolates were typical members of a common PFGE type present in retail poultry. In addition, the data show the role of transmissible mobile genetic elements in the evolution of virulence and resistance among S. Heidelberg.
Materials and Methods
Bacterial Strains, Growth Condition, and Characterization
Isolates were chosen for the study based on similarity or dissimilarity by PFGE to S. Heidelberg isolated from a large ground turkey-associated outbreak in 2011 (supplementary fig. S1, Supplementary Material online) (CDC 2011; Folster et al. 2012). Forty-four isolates of S. Heidelberg were included in the study (table 1). Isolates were obtained from animal (n = 9), retail meat (n = 27), and human clinical (n = 7) sources in the United States, and one of the isolates was obtained from an unknown sample source in Brazil. Four isolates were collected from 1982 to 1987; 38 of the isolates were collected from 2002 to 2011, and collection dates for two of the isolates were unknown. Five of the isolates were from the Salmonella Reference Collection A (SARA) (Beltran et al. 1991), and nine of the isolates were collected in the course of the investigation of a ground turkey-associated outbreak in 2011 (Folster et al. 2012).
Table 1.
Metadata of S. Heidelberg Isolates Used in This Study
| Salmonella enterica susp. enterica serovar and strain | Source Country | Source State | Source Type | Isolated Date | Accession | Closed Plasmid | BioProject | Plasmid Profile | Phage Profile |
|---|---|---|---|---|---|---|---|---|---|
| Heidelberg str. 20752 | USA | NC | Animal | 2002 | AMNR00000000 | 78501 | mob1, IncHI2 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 21381 | USA | CT | Food | 2002 | AMMZ00000000 | 80505 | mob1, IncA/C | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 24359 | USA | MO | Animal | 2003 | AMNP00000000 | 78497 | mob1, IncI1, IncHI2 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 24388 | USA | MO | Animal | 2003 | AMNQ00000000 | 78499 | mob1 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 24390 | USA | MO | Animal | 2003 | AMMY00000000 | 78475 | mob1, IncI | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 24391 | USA | MO | Animal | 2003 | AMNO00000000 | 78495 | mob1, IncI | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 24393 | USA | MO | Animal | 2003 | AMNM00000000 | 78491 | IncHI2 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 29169 | USA | CT | Food | 2003 | APIX00000000 | 80527 | mob1 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 32507 | USA | GA | Food | 2003 | AMNL00000000 | 80525 | mob1, IncHI2 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 41563 | USA | OR | Human | 2011 | AJGX00000000a | pCFSAN000312_01 | 78487 | mob1, IncI1, VirB/D4 | P22, Gifsy-2 like, P4, P2-like |
| Heidelberg str. 41565 | USA | WA | Food | 2011 | AJHA00000000a | 80521 | mob1, IncHI2 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 41566 | USA | WA | Food | 2011 | AJGZ00000000a | 78489 | mob1, IncI1, VirB/D4 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 41567 | USA | WA | Food | 2011 | AMNG00000000 | 80511 | mob1, IncI1, VirB/D4 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 41573 | USA | OH | Human | 2011 | AJGY00000000a | pCFSAN000316_01 | 80513 | mob1, IncI1, VirB/D4 | P22, Gifsy-2 like, P4, P2-like |
| Heidelberg str. 41576 | USA | OH | Human | 2011 | AMNH00000000 | 80515 | mob1, IncI1, VirB/D4 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 41578 | USA | OH | Human | 2011 | CP004086 | pSEEH1578_01,_02,_03 | 78477 | mob1, IncI1, VirB/D4 | P22, Gifsy-2 like, P4, P2-like |
| Heidelberg str. 41579 | USA | OH | Food | 2011 | AJGW00000000a | 78479 | mob1, IncI1 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 41584 | USA | MI | Human | 2011 | AMNI00000000 | 80517 | mob1, IncI1, VirB/D4 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. 82-2052 | USA | ME | Human | 1982 | AMMX00000000b | 78473 | IncI1, VirB/D4 | P22, Gifsy-2 like, Fels-2, ST64B | |
| Heidelberg str. N1514 | USA | GA | Food | 2004 | AKYM00000000 | 69871 | VirB/D4 | P22, Gifsy-2 like | |
| Heidelberg str. N1536 | USA | GA | Food | 2004 | AKYN00000000 | 69869 | IncI, VirB/D4 | P22, Gifsy-2 like | |
| Heidelberg str. N15757 | USA | GA | Food | 2007 | AMND00000000 | 80509 | mob1, IncI1, VirB/D4, IncHI2 | P22, Gifsy-2 like | |
| Heidelberg str. N18393 | USA | CT | Food | 2008 | AMNF00000000 | 78485 | mob1, IncI1 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. N18413 | USA | GA | Food | 2008 | AMNS00000000 | 80529 | mob1, IncI1 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. N18440 | USA | MD | Food | 2008 | AMNT00000000 | 80531 | mob1, IncI1 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. N18453 | USA | MD | Food | 2008 | AMNU00000000 | 80533 | mob1, IncI1 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. N189 | USA | CT | Food | 2004 | AMMP00000000 | 69873 | mob2, IncI1, VB/D4 | P22, Gifsy-2 like | |
| Heidelberg str. N19871 | USA | NY | Food | 2008 | AMNC00000000 | 78481 | mob1, IncI1 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. N19992 | USA | CA | Food | 2009 | AMMC00000000 | 66709 | ColE1 | P22, Gifsy-2 like, P2-like | |
| Heidelberg str. N20134 | USA | GA | Food | 2009 | AMNN00000000 | 78493 | mob1, IncI1 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. N26457 | USA | CO | Food | 2010 | AMNE00000000 | 78483 | mob1, IncI1, IncA/C | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. N29341 | USA | MN | Food | 2011 | AMNJ00000000 | 80519 | mob1, IncI1, VirB/D4 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. N30678 | USA | NM | Food | 2011 | AMNK00000000 | 80523 | mob1, IncI1, VirB/D4, IncHI2 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. N418 | USA | NM | Food | 2004 | AMMB00000000 | 66707 | mob1, IncA/C | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. N4403 | USA | CA | Food | 2005 | AMMT00000000 | ` | 69861 | mob2, IncI1, VirB/D4 | P22, Gifsy-2 like |
| Heidelberg str. N4496 | USA | CO | Food | 2005 | AMMQ00000000 | 69867 | VirB/D4 | P22, Gifsy-2 like | |
| Heidelberg str. N4541 | USA | CT | Food | 2005 | AMMV00000000 | 69857 | VirB/D4 | P22, Gifsy-2 like | |
| Heidelberg str. N4630 | USA | GA | Food | 2005 | AMNA00000000 | 80507 | mob1, IncI1 | P22, Gifsy-2 like, P4, P2-like | |
| Heidelberg str. N653 | USA | OR | Food | 2004 | AMMW00000000 | pCFSAN000412_01, _02 | 69855 | mob2, IncI1, VirB/D4 | P22, Gifsy-2 like |
| Heidelberg str. SARA31 | USA | MD | Animal | 1987 | AMLV00000000 | pCFSAN000441_01 | 66679 | VirB/D4 | P22, Gifsy-2 like |
| Heidelberg str. SARA32 | USA | TX | Animal | 1986 | AMLU00000000 | pCFSAN000442_01 | 66677 | VirB/D4 | P22, Gifsy-2 like |
| Heidelberg str. SARA35 | Brazil | n/a | Unknown | n/a | AMLT00000000b | pCFSAN000443_01 | 66675 | ColE1, 8 kb resistance plasmid | P22, Gifsy-2 like |
| Heidelberg str. SARA37 | USA | CO | Animal | 1987 | AMLR00000000 | 66673 | IncI1, VirB/D4 | P22, Gifsy-2 like, Fels-2, ST64B | |
| Heidelberg str. SARA 39 | USA | NC | Human | n/a | AMMJ00000000 | 69887 | IncHI2 | P22, Gifsy-2 like, Fels-2 |
aFirst released (Hoffmann et al. 2012).
bFirst released (Timme et al. 2013).
Salmonella isolates were cultured on trypticase soy agar (TSA; Becton, Dickinson, NJ) and in trypticase soy broth (TSA; Becton) overnight at 37 °C. All isolates used for WGS were serotyped by conventional methods and tested for antimicrobial susceptibility according to the NARMS standard protocol as previously described (Zhao et al. 2008). Antimicrobial susceptibility testing, using a panel consisting of 15 antimicrobial agents (amikacin, ampicillin, amoxicillin-clavulanic acid, cefoxitin, ceftiofur, ceftriaxone, chloramphenicol, ciprofloxacin, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfisoxazole, tetracycline, and trimethoprim-sulfamethoxazole) was performed according to the NARMS methodology with the Sensititre automated antimicrobial susceptibility system (Trek Diagnostic Systems, Westlake, OH). Isolate antimicrobial resistance was determined by comparison of MICs to values established MICs by the Clinical and Laboratory Standards Institute (CLSI). PFGE was performed according to the CDC PulseNet protocol (http://www.cdc.gov/pulsenet/pathogens/index.html, last accessed April 23, 2014). Genomic DNA of each strain was isolated from overnight cultures using DNeasy Blood and Tissue Kit (Qiagen, CA). All S. Heidelberg isolates were stored in TSB containing 15% glycerol at −80 °C.
Genome Sequencing, Assembly, and Annotation
We performed shotgun sequencing of the 44 S. Heidelberg using the Genome Sequencer FLX 454 (Roche, Branford, CT) and the GS FLX Titanium Sequencing Kit XLR70 according to the manufacturer’s protocol to generate an average genome coverage of 23×. De novo assemblies were performed using Roche’s Newbler software (v.2.6) with the resulting contigs being annotated using the NCBIs Prokaryotic Genomes Automatic Annotation Pipeline (Klimke et al. 2009).
Using DNA from the 2011 clinical outbreak isolate 41578, we also sought to close the genome using the Pacific Biosciences (PacBio) sequencing platform. Specifically, we prepared a single 10-kb library that was sequenced using the C2 chemistry on eight single-molecule real-time cells with a 90-min collection protocol on the PacBio RS. The 10-kb continuous-long-read data were de novo assembled using the PacBio hierarchical genome assembly process/Quiver software package, followed by Minimus 2, and they were polished with Quiver.
Comparative and Phylogenetic Genome Analysis
For comparative analysis, in addition to the 44 isolates sequenced in this study, we included sequence data available for S. Heidelberg SL476 complete genome (NC_011083), two plasmids (NC_011081 and NC_011082), and the whole-genome shotgun sequence for S. Heidelberg SL486 (ABEL00000000) at the NCBI genome database.
Phylogenetic informative SNP sites (i.e., SNPs shared by two or more strains in the alignment) were identified by two different methods. The first involved mapping the 454 reads to the complete reference genome of S. Heidelberg strain SL476 using Roche Newbler software GS Reference Mapper (v.2.8.). SNP sites were then found using the SNP calling function of GS Reference Mapper. SNP positions were defined, where one or more isolates differed from strain SL476 with coverage ≥10× and with ≥95% of the reads containing the SNP after having filtered out the SNPs from homopolymer artifacts, followed by a custom pipeline to construct a SNP matrix (Allard et al. 2013). S. Heidelberg SL486 could not be included in this analysis as its raw sequence data were not available. The second SNP detection method was a reference free k-mer-based approach implemented in the program kSNP (Gardner and Slezak 2010). This is a collection of Perl scripts that aggregate the results from Jellyfish v1.1.3 (used for k-mer counting) (Marcais and Kingsford 2011) and MUMmer v3.22 (used to align k-mers and detect variable positions) (Kurtz et al. 2004). Analyses with kSNP were based on a k-mer length of 25.
To construct evolutionary relationships among the isolates, the maximum-likelihood (ML) method was implemented in the Genetic Algorithm for Rapid Likelihood Inference (GARLI) software (Zwickl 2006b). GARLI analyses were performed using a web service (Bazinet and Cummings 2011) that uses a special programming library and associated tools (Bazinet et al. 2007). All ML trees were constructed with GTR+I+G nucleotide substitution model. The 100 replicate runs of the nonbootstrapped data set were conducted to identify the most probable phylogeny based on our observed SNP matrix. We determined branch support by performing 1,000 bootstrap replicates.
We elucidated the relationships among isolates using the Bayesian clustering method implemented in STRUCTURE v2.3.2 (Pritchard et al. 2000; Falush et al. 2003). The analyses were based on the kSNP matrix, and we ran ten replicate analyses at K = 2–9 under the admixture model with correlated allele frequencies being performed and visualized with DISTRUCT v1.1. (Rosenberg 2004). Each independent run consisted of 50,000 generations serving as burnin followed by 100,000 generations. The ΔK statistic (Evanno et al. 2005) was used to identify the k value that best fits the data. Pairwise genetic distances, calculated as the number of nucleotide differences, were generated in MEGA5 (Tamura et al. 2007). Core genes were identified using the UCLUST algorithm (Edgar 2010) using 95% sequence identity as the cutoff. Alignments of core genes were accomplished using MUSCLE with default settings; these alignments were then used to identify variable core genes.
Prediction of Prophages, Plasmids, Resistance, and Virulence Genes
Prophages were identified using PHAST (Zhou et al. 2011). Sequences for intact phages were extracted from the original contig, in which they were found and mapped against the entire assembled data to determine whether the phage was present in an isolate at multiple sites on different contigs. Contigs that could not be aligned to the reference genome were evaluated using BLAST to identify plasmids. The plasmids were analyzed by comparative analysis using MAUVE algorithm (Darling et al. 2010) and with the comparative analysis tools of RAST (Aziz et al. 2008). For plasmid closure, a concatenated sequence was generated from a 500-bp sequence from each end of the contig. This artificial sequence from the single contig of the plasmid in study was used as a reference to map all the 454 raw reads, using runMapping from Newbler. If there were mapped reads that covered the conjunction point in the reference, the contig was a closed plasmid.
To determine the incompatibility (Inc) groups for plasmids, we used BLAST to find sequences described by Johnson and Nolan (2009) for specific Inc groups that would produce theoretical PCR amplicons for known Inc group sequences. Resistance and virulence genes were identified by mapping sequence data available at an in-house database, consisting of 1,379 resistance genes, and 107 previously characterized virulence genes (84 pathogenicity and 23 fimbrial markers; Huehn et al. 2009). Using a presence/absence matrix of genes conferring resistance to antibiotics and disinfectant agents, we constructed a similarity tree based on binary distances under neighbor-joining algorithm for tree construction; topological support was assessed based on 100 bootstrap replicates. The analyses were performed using the R package ape (Paradis et al. 2004; Liu et al. 2011).
Caenorhabditis elegans Assay
Pathogenicity was evaluated using the Caenorhabditis elegans survival assay. Caenorhabditis elegans strain SS104 glp-4 (bn2) was acquired from the Caenorhabditis Genetics Center. Worm cultures were maintained at 16 °C, which is the permissive temperature for this temperature-sensitive sterile mutant of C. elegans. Strains were cultured in C. elegans habitation media (CeHM) in tissue culture flasks on a platform shaker (Sprando et al. 2009). Nematodes were bleached (0.5 M NaOH, 1% hypochlorite) to collect eggs, which were incubated in M9 media for 24 h to bring them to synchronized L1 stage and then transferred to CeHM. To produce synchronized L4 stage worms, L1 worms were grown for an additional 72 h in CeHM.
Pathogen lawns for survival assays with test strains, as well as the food bacteria OP50, were prepared by inoculating Nematode Growth Media (in 6-cm Petri plates) with 50 μl of an overnight bacterial culture. Plates were incubated overnight at room temperature before worms were added. We used 60–80 synchronized L4 worms for each treatment group. Worms were scored every 24 h for survival (Aballay et al. 2000).
Animal survival was plotted using Kaplan–Meier survival curves and analyzed by log rank test using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). Survival assays were repeated at least two times. Survival curves resulting in P values < 0.05 relative to control were considered significantly different.
Results
Antimicrobial Resistance and PFGE
Two representative isolates from the 2011 ground turkey-associated outbreak were previously reported to be resistant to ampicillin, gentamicin, streptomycin, and tetracycline (Folster et al. 2012). Antimicrobial susceptibility testing of seven additional representative isolates showed resistance to ampicillin, gentamicin, and tetracycline among six; and streptomycin, tetracycline, and kanamycin resistance in one of the isolates. The susceptibility and PFGE (Xbal and BlnI) results for all nine of the outbreak representative isolates included in this study and the reference isolates (table 1) are shown in supplementary figure S1, Supplementary Material online. The nine outbreak associated isolates exhibited indistinguishable, or nearly indistinguishable (one band difference), Xbal PFGE patterns JF6X01.0032 or JF6X01.0058, and BlnI pattern JF6A26.0076 and one food isolate had Xbal PFGE pattern JF6X01.0058 and BlnI pattern JF6A26.0017. Of the 35 isolates not associated with the outbreak, 15 had composite Xbal/BlnI PFGE patterns indistinguishable from ground turkey outbreak isolates whereas 20 isolates had composite XbaI/BlnI PFGE patterns ≤98% similar to isolates from the 2011 outbreak (supplementary fig. S1, Supplementary Material online). Nine reference isolates showed the same PFGE pattern, as well as the same antibiotic resistance profile, noted for most of the outbreak isolates (supplementary fig. S1, Supplementary Material online).
Draft Genome Size
Shotgun sequencing produced 44 draft genomes with an average coverage of 23× and a minimum of 4,700 genes. The genomic size of the 44 S. Heidelberg isolates (including extrachromosomal DNA) ranged from 4.671 to 5.130 Mb (fig. 1). Differences in genomic size were due primarily to the presence or absence of mobile genetic elements, including phages and plasmids, especially the carriage of IncHI2 or IncA/C plasmids (fig. 1). Next to the genome size variation, figure 1 shows the estimated N50 sizes within S. Heidelberg draft genome sequences. The estimated N50 value is a rough estimate of the quality and coverage of the draft genomes, and it represents the average contig size after assembly with the Newbler software.
Fig. 1.—
The number of assembled bases (Mb) and N50 contig size (kb) for each sequenced S. Heidelberg isolate. Samples are colored according to the presence of antimicrobial resistance plasmids. No antimicrobial resistance plasmid, filled triangles; antimicrobial resistance Inc-I plasmid, filled diamonds; antimicrobial resistance Inc-H1/2 plasmid, filled circles; antimicrobial resistance plasmid Inc-I and H1/2, filled circles with borders; antimicrobial resistance plasmid Inc-AC, filled squares; antimicrobial resistance plasmid IncI and Inc-AC, filled squares with borders.
Phylogenetic Relationships among S. Heidelberg Using SNP Analysis
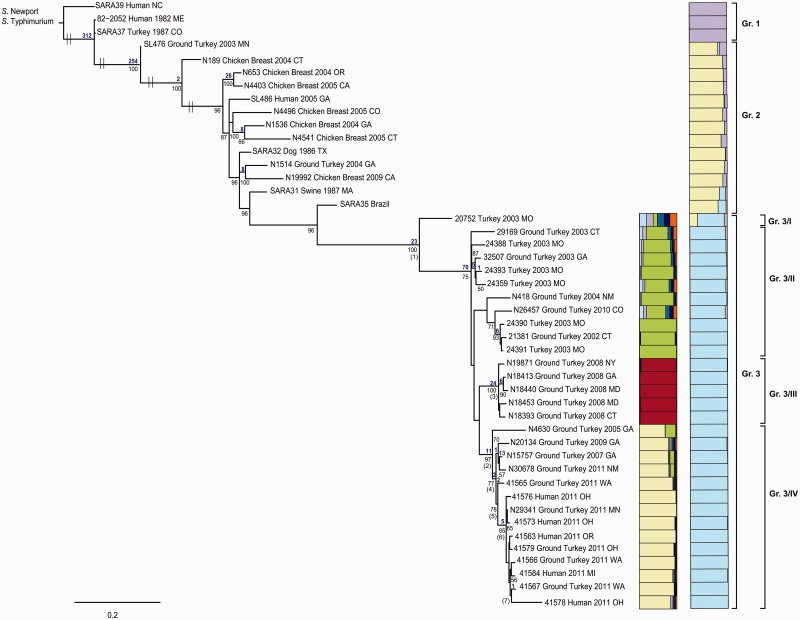
Phylogenetically informative SNP sites were identified using two independent methods. Figure 2 shows a ML tree based on SNP analysis of 44 S. Heidelberg isolates mapped to strain SL476. Among the collective 40,716 variable SNPs identified, 12,187 were determined to be “informative” (i.e., SNPs shared by at least two isolates). The ML tree shown in figure 2 clearly demonstrates that S. Heidelberg formed a monophyletic group distinct from the outgroup comprised Salmonella enterica supsp. enterica serovar Newport (S. Newport) and S. Typhimurium. The SNP divergence between S. Heidelberg and the two outgroups, S. Newport and S. Typhimurium, is 24,724 and 24,305 SNPs, respectively. The resultant tree shows two more interesting points. First, isolates having a similar Xbal PFGE pattern (supplementary fig. S1, Supplementary Material online) clustered together. Second, there is a tendency for isolates to cluster based on date of collection, such as noted for isolates from 2003, 2008, and 2011. In addition, the 2011 clinical and food isolates that were originally thought to be associated with the ground turkey outbreak isolated in different states clustered together with 80% bootstrap support (fig. 2), which could indicate that they are all members of a clonal community likely derived from the same source. Consistent with the trend for temporally isolated strains to cluster closely together, one isolate, N29341, that was isolated in Minnesota in 2011 from ground turkey by NARMS, clustered tightly together with the outbreak isolates received from CDC, whereas all other reference isolates having the same PFGE pattern not associated with the ground turkey outbreak are distinct from the isolates of the 2011 outbreak (fig. 2). These results suggest that N29341 might be epidemiologically related to the 2011 outbreaks strains, as Minnesota was one of the states involved in that outbreak.
Fig. 2.—
ML tree based on SNP analysis of 44 S. Heidelberg isolates and previously reported S. Heidelberg genome sequences SL476 (12). A total of 40,716 variable SNPs with 12,187 being informative were found using GS Reference Mapper followed by a custom pipeline. The ML tree was generated in GARLI v.2.0 (Zwickl 2006a) under the GTR+Γ model of nucleotide evolution and visualized using Figtree v1.3.1. Parameter space was searched for the best tree with simultaneous estimation for model parameters using a ML search. The best tree was identified from 100 runs on the nonbootstrapped data set. Measures of clade confidence are reported below each node in the form of bootstrap values (1,000 iterations). Bootstrap values <70% were not shown. The tree was rooted using S. Newport 637564 and S. Typhimurium AZ057. The taxa of source for each isolate, geographic location, and date were mapped onto the tree. Prophage observations are further depicted on the tree using colored bars, shown on the right.
The SNP matrix, including the same S. Newport and S. Typhimurium outgroups, built using the k-mer strategy of kSNP consisted of 41,432 variable SNP positions with 10,873 being informative. Using the k-mer strategy, we identified 716 more SNPs than were noted within the in-house pipeline, where 40,716 variable SNPs were observed. Notably, the SNP diversity within the highly clonal S. Heidelberg isolates was lower, only 4,053 SNPs were found with 1,394 being informative. The resulting ML tree (fig. 3) was well resolved and congruent with the tree topology in figure 2. To show that the tree topology is not an artifact of mobile elements that are not present in all isolates, we constructed a kSNP matrix using SNPs present only on core genes. This produced a matrix of 24,447 SNPs, of which 7,630 were informative and present in genes shared by all 46 S. Heidelberg isolates and the two outgroups. Only 718 SNPs, with 284 being informative, were found in genes shared only by S. Heidelberg isolates. The SNP matrix of the core genes was used to construct a ML phylogenetic tree (supplementary fig. S2, Supplementary Material online), which also confirms the tree topology in figures 2 and 3.
Fig. 3.—
ML tree based for the 44 S. Heidelberg isolates and two previously reported S. Heidelberg genome sequences SL476 and SL486 (Fricke et al. 2011). A total of 4,053 SNPs with 1,394 being informative were found based on k-mer analysis using kSNP. ML trees were generated as described in figure 2. Bootstrap values (1,000 iterations) are reported below each node. The numbers of unambiguous substitutions that mapped to the tree only once and are greater than zero are given above each node in blue. The numbers in parenthesis represent the nodes in table 3. To the right of the tree, two Distruct plots were reconstructed with the same SNP matrix—one including all 46 S. Heidelberg isolates and, adjacent to that, another with only those isolates from group 3—to present a fine-scale structure is shown. The Distruct plot was generated using a model-based Bayesian clustering method implemented in Structure v2.3.2 and visualized with DISTRUCT v1.1. 10 replicate analysis at K = 2–9 under the admixture model with correlated allele frequencies were performed. Each independent run consisted of 50,000 generations serving as burnin followed by 100,000 generations. Different colors represent the different clusters and each bar represents an individual isolate. The fraction of the bar that is a given color represents the coefficient of membership to that cluster (e.g., multicolored bars indicate membership to multiple groups indicative of admixture).
In this study, we determined that all 46 S. Heidelberg isolates, including S. Heidelberg SL476 and S. Heidelberg SL486, partitioned into three paraphyletic groups, each having 100% bootstrap support (fig. 3). Group 1 contains three historical isolates (two clinical and one turkey isolate) with a PFGE pattern, for which there was no assigned Pulse-Net designation (supplementary fig. S1, Supplementary Material online). Group 2 is comprised of 11 isolates, including clinical, food, and animal isolates, with XbaI PFGE pattern JF6X01.0022, as well as the isolates SL476 and N19992 having different XbaI patterns JF6X01.0133 and JF6X01.0326, respectively. Group 3 is composed of 30 isolates from food, animal, and clinical sources, including the outbreak isolates, all having a very similar XbaI pattern (JF6X01.0032, JF6X01.0034, JF6X01.0058).
The distribution of PFGE patterns on the tree topology based on SNPs differs between the two enzymes XbaI and BlnI. For example, N19992 (group 2) and N18393 (group 3) are quite distinct on the ML tree, even though both have the same BlnI pattern JF6A26.0015 and are adjacent to each other on the PFGE dendrogram (supplementary fig. S1, Supplementary Material online). Pairwise SNP variation between the three distinct S. Heidelberg lineages seen in figure 3 are presented in table 2 and support the phylogenetic partitions revealed above. Mean divergence among the three clades ranged from 200 to 712 nt differences, whereas the mean intragroup nucleotide differences ranged from 47 to 155 (table 2).
Table 2.
(Mean Pairwise Distance (Number of Nucleotide Differences) between the (A) Three Major S. Heidelberg Groups in figure 3 and the (B) Four S. Heidelberg Subgroups from Group 3 in figure 3
| S. Heidelberg Gr. 1 | S. Heidelberg Gr. 2 | S. Heidelberg Gr. 3 | ||
|---|---|---|---|---|
| (A) | ||||
| S. Heidelberg Gr. 1 | 155a | |||
| S. Heidelberg Gr. 2 | 535 | 101a | ||
| S. Heidelberg Gr. 3 | 712 | 200 | 47a |
| Gr. 3/I | Gr. 3/II | Gr. 3/III | Gr. 3/IV | |
|---|---|---|---|---|
| (B) | ||||
| S. Heidelberg Gr. 3/I | n/c | |||
| S. Heidelberg Gr. 3/II | 144 | 30a | ||
| S. Heidelberg Gr. 3/III | 139 | 42 | 15a | |
| S. Heidelberg Gr. 3/IV | 153 | 46 | 51 | 27a |
| Outbreak isolates | 151 | 46 | 65 | 36 |
aIntragroup mean pairwise distance (number of nucleotide differences).
Notably, the mean SNP variation within the outbreak isolates is only 17 SNPs. The phylogenetic analysis clustered the outbreak isolates together even though the PFGE profile (supplementary fig. S1, Supplementary Material online) is slightly different (XbaI pattern JF6X01.0032, JF6X01.0058; BlnI pattern JF6A26.0017 and JF6A26.0076). For example, isolates 41578 and 41579, sharing the same PFGE and antibiotic resistance profile with isolates N19871, N18413, N18440, and N18453 (isolated in 2008), are more closely related to the other outbreak isolates, even though they have a different XbaI pattern. We also found that the outbreak isolates 41578 and 41579 are quite distinct from isolates N19871, N18413, N18440, and N18453, which have 44 SNPs that allow complete discrimination between the two groups (fig. 3). Furthermore, isolate 41565, which has the same PFGE and antibiotic resistance profile as the 2003 isolates 32507, 24393, and 20752, is more closely related to the other isolates associated with the 2011 outbreak, even with having a different BlnI pattern and antibiotic resistance profile (fig. 3).
To provide an additional picture as to the number of distinct groups the sampled serovars segregate into, we constructed groups using the program STRUCTURE, the results of which were visualized using DISTRUCT (fig. 3). The results are consistent with the groupings formed through the phylogenetic analyses in that there are three distinct groups (each represented by a different color [fig. 3]).
Differences between Outbreak Isolates and Nonoutbreak Isolates in Group 3
To further investigate the relationships among outbreak isolates, we constructed the Distruct plot for group 3. Within that group, there is support for four subgroups; these four groups also are found in the phylogenetic analysis, each with a ≥75% bootstrap support. Pairwise SNP variation between the four sublineages also support the phylogenetic partitions described above (table 2 and fig. 3). Mean divergence among the four subclades ranged from 36 to 153 nt differences, whereas the mean intragroup nucleotide differences ranged from 15 to 30 (table 2). The 2008 strains seem to have quite a unique SNP profile when compared with the remaining isolates of group 3. The ML tree, the Distruct plot, and the pairwise SNP variation show the evolutionary changes occurring among isolates from 2003 to 2009 and 2011, revealing the apparent emergence of the genetically unique S. Heidelberg strain that was responsible for the 2011 outbreak.
Table 3 lists variable genes having unique nucleotide substitutions which defined specific sublineages that could be used to separate them from other highly clonal S. Heidelberg isolates in group 3. Table 3 also lists their SNP change and, if applicable, the gene carrying the SNPs and the resulting amino acid change. Twenty-three unique SNPs, with 14 being nonsynonymous (i.e., causing an amino acid change) SNPs, were found to be unique to group 3 and clustering together isolates with XbaI pattern (JF6X01.0032, JF6X01.0034, JF6X01.0058). These nucleotide substitutions were found on 20 different genes that could be useful for subtyping of strains belonging to group 3 (table 3 and fig. 3). Furthermore, 24 unique SNPs (11 being nonsynonymous) were found at the node that characterizes the 2008 ground turkey isolates, which seemed to have their own unique SNP profile distinct from that of the remaining group 3 isolates. Twelve of the 24 SNPs are located on the insertion sequence (IS) 2 element with five SNPs being within the IS2 TnpA transposase and seven SNPs on the IS2 TnpB transposase genes (table 3 and fig. 3).
Table 3.
Antibiotic Resistance and Plasmid Characteristics of S. Heidelberg Isolates Used in This Study
| Node on Tree | SNP Location | NT Change | AA Change | NT Position | Locus Tag |
|---|---|---|---|---|---|
| 1 | d-Mannonate oxidoreductase | A→G | D→G | 839 | SEEHRA37_03221 |
| 1 | Translocation protein TolB | T→A | P | 1152 | SEEHRA37_24108 |
| 1 | Glutathione S-transferase | C→A | W→Stopp | 233 | SEEHRA37_20055 |
| 1 | Nonannotated | C→T | R→C | n/a | SEEHRA37_contig25 |
| 1 | Hypothetical protein/putative transcriptional regulator | G→T | V→F | 376 | SEEHRA37_19825 |
| 1 | Ureidoglycolate dehydrogenase AllD | C→T | T→I | 20 | SEEHRA37_20355 |
| 1 | Starvation-inducible outer membrane lipoprotein | C→T | S | 63 | SEEHRA37_07830 |
| 1 | ATPase recombination and repair protein | G→T | L | 966 | SEEHRA37_05779 |
| 1 | DNA-specific endonuclease I | A→C | K→T | 341 | SEEHRA37_19810 |
| 1 | Scaffolding protein | C→A | V | 135 | SEEHRA37_21903 |
| 1 | Peptidase T | G→T | V→L | 976 | SEEHRA37_13506 |
| 1 | Predicted metal-dependent membrane protease | C→A | A | 108 | SEEHRA37_15707 |
| 1 | Hypothetical protein | A→G | N→D | 418 | SEEHRA37_04201 |
| 1 | Glycine/serine hydroxymethyltransferase GlyA | T→G | S→A | 145 | SEEHRA37_00365 |
| 1 | Preprotein translocase subunit SecF | G→A | A | 534 | SEEHRA37_22413 |
| 1 | Hypothetical protein | A→G | T→A | 1879 | SEEHRA37_01714 |
| 1 | Ribosomal protein L21 RplU | A→C | I→L | 220 | SEEHRA37_20845 |
| 1 | Nonannotated | C→G | Q→E | n/a | SEEHRA37_contig19 |
| 1 | Isoaspartyl peptidase | C→T | A→V | 380 | SEEHRA37_16329 |
| 1 | Lysine-N-methylase | C→T | G | 57 | SEEHRA37_07110 |
| 1 | Hypothetical protein | T→C | L | 162 | SEEHRA37_23678 |
| 1 | Hypothetical protein | A→C | K→T | 227 | SEEHRA37_07475 |
| 1 | Nonannotated | T→C | I→V | n/a | SEEHRA37_contig14 |
| 2 | Hypothetical protein | A→G | Q→R | 851 | SEEHRA37_20355 |
| 2 | Sensor kinase DpiB | A→T | I→F | 67 | SEEHRA37_10675 |
| 2 | Notannotated | C→T | P | 63 | SEEHRA37_contig6 |
| 2 | Hypothetical protein | G→A | F | n/a | SEEHRA37_contig6 |
| 2 | Nonannotated | G→A | G→S | n/a | SEEHRA37_contig5 |
| 2 | Proline dehydrogenase PutA | C→A | G | 975 | SEEHRA37_04036 |
| 2 | Electron transport complex protein RnfC | T→C | I | 276 | SEEHRA37_02971 |
| 2 | Protein involved in chromosome partitioning MukB | G→C | G→A | 1514 | SEEHRA37_14706 |
| 2 | N-ethylmaleimide reductase | A→T | G | 351 | SEEHRA37_02866 |
| 2 | Hypothetical protein | C→T | L | 1039 | SEEHRA37_16509 |
| 2 | Multidrug efflux system subunit MdtC | A→T | Q→H | 2067 | SEEHRA37_00060 |
| 3 | Putrescine/spermidine ABC transporter ATPase protein PotA | G→T | P→Q | 47 | SEEHRA37_13511 |
| 3 | Pilus assembly protein, porin PapC | G→T | R→S | 4 | SEEHRA37_18324 |
| 3 | IS2 repressor TnpA | G→A | T | 93 | SEEH8393_18082 |
| 3 | IS2 transposase TnpB | A→G | T | 195 | SEEH8393_18077 |
| 3 | IS2 transposase TnpB | A→G | N→D | 502 | SEEH8393_18077 |
| 3 | IS2 transposase TnpB | G→A | A | 483 | SEEH8393_18077 |
| 3 | Not annotated (plasmid sequence) | T→C | A→V | n/a | SEEH8393_contig56 |
| 3 | IS2 repressor TnpA | C→T | Y | 309 | SEEH8393_18082 |
| 3 | IS2 repressor TnpA | A→G | E | 195 | SEEH8393_18082 |
| 3 | IS2 transposase TnpB | C→A | V | 525 | SEEH8393_18077 |
| 3 | IS2 transposase TnpB | C→A | P | 192 | SEEH8393_18077 |
| 3 | IS2 transposase TnpB | A→G | A | 789 | SEEH8393_18077 |
| 3 | YacA | T→G | V | 123 | SEEH8393_01004 |
| 3 | Hypothetical protein | G→A | A→T | 232 | SEEHRA37_09669 |
| 3 | Multifunctional fatty acid oxidation complex subunit alpha FadJ | G→T | M→I | 2094 | SEEHRA37_15762 |
| 3 | tRNA uridine 5-carboxymethylaminomethyl modification enzyme GidA | G→A | G→S | 1199 | SEEHRA37_11040 |
| 3 | Permease DsdX | C→A | A→D | 1166 | SEEHRA37_02334 |
| 3 | IS2 transposase TnpB | T→C | R | 549 | SEEH8393_18077 |
| 3 | IS2 repressor TnpA | C→T | A | 225 | SEEH8393_18082 |
| 3 | IS2 transposase TnpB | C→A | P→T | 319 | SEEH8393_18077 |
| 3 | IS2 repressor TnpA | C→T | L | 102 | SEEH8393_18082 |
| 3 | Arginyl-tRNA synthetase ArgS | A→T | G | 237 | SEEHRA37_07325 |
| 3 | Nonannotated | C→A | A→N | n/a | SEEHRA37_contig18 |
| 3 | Hypothetical protein | G→A | D→N | 103 | SEEHRA_37_05617 |
| 4 | Not annotated | G→A | V→L | n/a | SEERA37_contig2 |
| 4 | Sodium/glucose cotransporter | G→A | S→N | 1037 | SEERA37_14011 |
| 5 | Phosphoserine phosphatase | C→A | P→Q | 653 | SEEHRA37_11265 |
| 5 | Gluconate transporter GntP | C→G | L→V | 187 | SEEHRA37_18800 |
| 6 | Flagellin and related hook-associated proteins | G→C | T→S | 282 | SEEHRA37_18390 |
| 6 | Electron transfer flavoprotein, beta subunit | G→A | G→S | 358 | SEEHRA37_02456 |
| 6 | Hypothetical protein (plasmid sequence) | C→A | L→M | 562 | SEEH8393_17461 |
| 6 | Not annotated (plasmid sequence) | G→A | G→R | n/a | SEEH8393_contig56 |
| 6 | Not annotated (plasmid sequence) | C→T | G→S | n/a | SEEHRA39_contig149 |
| 7 | Type III secretion protein SopE | T→A | H→Q | 560 | SEEHRA37_14371 |
Importantly, two nonsynonymous SNPs were found to be only present in all isolates associated with the 2011 outbreak. These two SNPs were located within two different variable core genes: a putative hydrolase with amino acid change P653Q, and the gluconate transporter GntP gene with amino acid change L187V (table 3 and fig. 3). Five SNPs, with four being nonsynonymous, were unique to isolates 41563, 41566, 41567, 41573, 42578, 41579, and N29341 (associated to the outbreak) and not observed in 41565 (table 3 and fig. 3). From these five SNPs, three were in plasmid sequences, whereas two were found on variable core genes: the flagellin CDS resulting in an amino acid change from threonine to serine at position 282 (ΔT282S), and within a putative hypothetical protein (possibly functioning as H+ gluconate symporter related to permeases) producing an amino acid change from glycine to serine at position 358 (ΔG358S) (table 3).
Genetic Variation among S. Heidelberg
At a minimum, 273 genes involved in a variety of different functions, such as DNA replication and repair, cell division, transcription, metabolism, and virulence, vary among the S. Heidelberg isolates observed in clinical, animals, and/or food samples, including the isolates associated with the 2011 outbreak. The variable core genes, including the number of SNPs and haplotypes are listed in supplementary table S1, Supplementary Material online. Figure 4 shows a histogram comparing strains based on: 1) the number of SNPs according to the number of core genes and 2) the haplotype diversity according to the number of core genes. Only a few mutation “hotspot” genes, such as tailspike (35 SNPs), scaffolding protein (22 SNPs), DNA polymerase V subunit umuD (20 SNPs), phage lysozyme (18 SNPs), gifsy-2 prophage protein (15 SNPs), terminase small subunit (14 SNPs), outer membrane lipoprotein Blc (7 SNPs), and alpha amylase (6 SNPs), were found to have a disproportionate number of SNPs that could be useful for rapid subtyping the serovar Heidelberg and/or as targets of resequencing. Interestingly, although these genes have several SNPs, the haplotype diversity is fairly low, with only a few haplotypes (2–4) among 46 isolates being identified. This fact suggests that, even though there are genes with up to 35 SNPs, there are only two to four unique sequences among S. Heidelberg that are congruent with our tree topology for three different groups of S. Heidelberg.
Fig. 4.—
(A) Histogram showing the number of SNPs per core genes. (B) Histogram showing haplotype diversity for all variable, core genes.
Complete Genome from Clinical Isolate 41578
In the course of performing this study, we demonstrated that it was possible to rapidly determine the complete sequence for the chromosome and three plasmids from a clinical isolate associated with the 2011 outbreak using the Pacific Biosciences (PacBio) system. A single contig of 4,793,479 bp (GC content 52.2%) representing the complete chromosome and three contigs of 4,773 bp, 35,297 bp, and 117,929 bp representing three plasmids were generated. The chromosome contains 12 different Salmonella pathogenicity islands and four prophages (prophage P22, lambda-like prophage Gifsy-2, prophage P4, and prophage P2-like; fig. 5). The largest plasmid pSEEH1578_01 is a resistance plasmid with incompatibility group IncI1 encoding the resistance to gentamicin (aacC), streptomycin (aadA1), tetracycline (tetA, tetR(A)), and ampicillin (blatem1) (fig. 5). The plasmid pSEEH1578_02 is a VirB/D4 plasmid that carries the type IV secretion system (T4SS; fig. 5), and plasmid pSEEH1578_03 is a mobilization plasmid that carries genes capable of promoting plasmid mobilization such as mbeA, mbeB, mbeC, and mbeD.
Fig. 5.—
Chromosome and plasmids features of a clinical S. Heidelberg isolate 41578. The circular map was drawn using dnaplotter. Different features are shown in different colored bars. (A) Chromosome: The coding sequence are shown in dark blue, rRNA is shown in green, tRNA is shown in yellow, prophages are shown in blue, and Salmonella pathogenicity islands are shown in red. Track 7 represents the GC content while Track 8 shows the GC skew [(G − C)/G + C]. (B) IncI1 antimicrobial resistance plasmid: The coding sequences are shown in dark blue and resistance genes are shown in red. Track 5 shows the GC skew [(G − C)/G + C]. Regions of GC content above average of the plasmid are drawn outside the ring in yellow, whereas regions below average are inside the ring in purple. (C) VirB/D4 virulence plasmid: The coding sequences are shown in dark blue, genes that carry the T4SS are shown in red, genes responsible for plasmid stability, and replication is shown in green. Track 6 shows the GC skew [(G − C)/G + C]. Regions of GC content above average of the plasmid are drawn outside the ring in yellow, whereas regions below average are inside the ring in purple.
Identification of Prophages
PHAST analysis identified six different phages among the S. Heidelberg isolates. Only those phages that PHAST determined as being intact were further analyzed. All 44 isolates contained prophage P22 and lambda-like prophage Gifsy-2. Group 1, which was composed of three historical isolates (SARA 39, SARA 37, and 82-2052), also contained prophage Fels-2, not observed in any of the group 2 or group 3 isolates. Additionally, SARA 37 and 82-2053 also contained prophage ST64B (table 1 and fig. 2).
Heidelberg isolates belonging to group 3, which includes the 2011 outbreak isolates, contained prophage P4 and P2-like, neither of which was seen in any of the isolates from group 1 and 2, excepting isolate N19992, a group 2 isolate that was confirmed to contain the P2-like prophage (table 1 and fig. 2). The phage presence and absence matrix shows correlation with the tree phylogeny shown in figure 2.
Identification of Virulence Genes among S. Heidelberg
From the 107 known and recognized Salmonella-associated virulence genes, we identified 62 pathogenicity and 13 fimbrial markers that were highly conserved among the 44 S. Heidelberg isolates (supplementary table S2, Supplementary Material online). One outer membrane fimbrial usher gene, safC, was only found in the three historical isolates (SARA 39, SARA 37, and 82-2052), all belonging to group 1. All other detected virulence genes, observed to be located on the Salmonella pathogenicity islands (SPI-1, SPI-2, SPI-3, SPI-4, and SPI-5), the lambda-like prophage Gifsy-2, and/or islets, were present in all S. Heidelberg isolates, suggesting that all isolates are pathogenic (supplementary table S2, Supplementary Material online). For example, all isolates carried a chromosomal msgA gene, which is essential to Salmonella mouse virulence (Gunn et al. 1995), and is common in S. Typhimurium (Huehn et al. 2010). Other genes present among all isolates encoded proteins involved in type III secretion system (T3SS) and adhesins. Remarkably, the sopE1 virulence gene, involved in the translocation of effector proteins into host cells, was found in all of our strains. Moreover, although sopE1 is known to be carried by P2-like phages, in this study we also identified this gene on lambda-like phage Gifsy-2, which is present in all S. Heidelberg isolates. Consequently, all S. Heidelberg isolates have the sopE1 virulence gene. Group 3 isolates, which also contain the P2-like phage, have the sopE1 gene twice in their chromosome; one gene is carried by lambda-like phage Gifsy-2 and the other one is located on the P2-like phage. Analysis confirmed that these two genes shared a high degree of similarity having no more than eight mutations between the two different sopE1 alleles. In contrast, virulence determinants known to be associated with SPI-7, prophage Gifsy-1, Gifsy-3, Fels-1, and Salmonella plasmid virulence (spv) were not identified in any of the 44 isolates examined in this study, confirming sequencing results that none of the strains carried any of these mobile genetic elements.
Description of VirB/D4 Plasmids
Several plasmids were found among the 44 S. Heidelberg isolates sequenced in this study (table 1). Electrophoresis of samples obtained using a basic alkaline plasmid purification procedure demonstrated the presence of endogenous plasmids that had not integrated into the host genome (data not shown). The plasmid sizes varied from 3.7 to over 200 kb. Twenty-one S. Heidelberg isolates, including all five human isolates and two ground turkey isolates associated with the 2011 outbreak, contained a plasmid ranging in size from 33 to 40 kb, which carried genes (virB1, virB2, virB3-4, virB5, virB6, virB7, virB8, virB9, virB10, virB11, virD2, and virD4) associated with the VirB/D4 T4SS (table 1). Three specific roles for the bacterial T4SS have been identified: 1) to facilitate translocation of DNA via a conjugative mechanism to recipient cells; 2) to facilitate translocation of effector molecules, such as proteins, to eukaryotic target cells; and 3) to function in DNA uptake or dissemination from or into the environment milieu (Christie et al. 2005; Alvarez-Martinez and Christie 2009).
To further study the diversity among all 22 identified VB/D4 plasmids, we identified SNPs that might be useful in differentiating these plasmids. The SNP matrix from the 22 plasmids and the reference plasmid built using the k-mer strategy of kSNP consisted of 338 informative SNP positions. Figure 6 shows a ML tree based on SNP analysis. The phylogeny placed the VB/D4 plasmids into three well-supported clades (95%, 100%, and 100% bootstrap support). Clade I contained two plasmids isolated from the historical isolates, SARA 37 and 82-2052, belonging to group 1 on the ML tree shown in figure 3. Clade III is composed of ten plasmids, including the reference plasmid pSARA30 and 9 isolates belonging to group 2 in figure 4. Clade II contains 11 plasmids, including 7 plasmids from 7 isolates associated with the 2011 outbreak. Clade II can be further separated into two subclades, one of which contains nine plasmids derived from isolates belonging to group 3 in figure 4, while subclade 2 contains one clinical isolate and isolate N653, the unique isolate found to carry two VB/D4 plasmids (the other plasmid partitioned into clade III). Moreover, using the primers designed by Johnson et al. (2012) for the relaxase gene (taxC) (VirD2 component) to screen for the four different IncX subgroups, we determined that the VB/D4 plasmids from clade I, subclade 1 from clade II, and clade III belonged to the IncX1 plasmid subgroup, whereas the two plasmids from clade II subclade 2 were deemed to be members of the IncX4 subgroup.
Fig. 6.—
ML tree based for the 22 identified VB/D4 plasmids, including the reference S. Heidelberg plasmid pSARA30. A total of 338 SNPs with all being informative, were found based on k-mer analysis using kSNP. The numbers of unambiguous substitutions that mapped to the tree only once are given above each node. ML trees were generated as described in figure 3. Bootstrap values (1,000 iterations) are reported below each node.
Interestingly, isolate N653 carried two plasmids, one being in the IncX1 and another in the IncX4 incompatibility groups. It appears that there are significant differences among these groups that allow simultaneous maintenance of two plasmids belonging to the same incompatibility group, a heretofore believed unachievable occurrence. Given the notion that two plasmids in the same Inc group cannot simultaneously coexist within the same bacterium, another plausible explanation could be that IncX4 might actually belong to a different incompatibility group other than that of IncX1, which was only previously experimentally confirmed to belong to IncX (Norman et al. 2008). Consistent with this hypothesis was the fact that a BLAST search with the IncX4 repA gene (present only in both putative IncX4 plasmids) sequence identified significant sequence similarity to only the Citrobacter rodentium ICC168 plasmid pCROD2 (previously grouped to IncX4 [Johnson et al. 2012]) and two plasmids from S. Heidelberg (pSH163_34, pSH696_34), suggesting that the IncX4 group is a new incompatibility group distinct from that of IncX1. If this is confirmed to the case, coexistence of these two “IncX” plasmids would be explained, although additional research needs to be conducted to confirm this possibility.
In this study, eight of the plasmids carrying the VirB/D4 T4SS were completely sequenced including the two plasmids from isolate, N653 (table 1). The annotated sequence demonstrated that the eight plasmids carry, in addition to the VirB/D4 T4SS systems, genes encoding other virulence-associated determinants and several proteins involved in a variety of metabolic processes, for example, hemolysin expression-modulating gene ymoA, DNA-binding protein genes (hns), (stpA), the toxin/antitoxin stability genes (stbE), (stbD), IncN plasmid Kika gene (kika), and DNA topoisomerase III (topB).
A progressive Mauve alignment using the data from the eight completely sequenced plasmids and the reference S. Heidelberg plasmid pSARA30 (supplementary fig. S3, Supplementary Material online) separated the plasmids into three distinct groups. As expected, the plasmids partitioned in different clades in the plasmid VirB/D4 ML tree (fig. 6) were also found by Mauve analysis to belong to different groups. Group 1 included plasmids isolated from S. Heidelberg SARA 31, SARA 32, N4403, and N653, all having a size of approximately ∼38.0 kb and showing the highest degree of sequence similarity to the S. Heidelberg plasmid pSARA30. Unique to this group are several hypothetical proteins: truncated transposase, DEAD/DEAH box helicase domain protein, and putative N-acetyltransferase. The second group is composed of the plasmids isolated from S. Heidelberg 41573 (∼38 kb) and 41563 (∼33 kb)—both involved in the 2011 outbreak. The best BLAST hit for them is Escherichia coli plasmid pDKX1-TEM-52 (GenBank accession number: JQ269336). Attributes unique to this group are several hypothetical proteins, a replicon initiation protein, and an ATPase central domain-containing protein. The third group contains plasmids isolated from S. Heidelberg 41578, a 2011 outbreak clinical isolate, and the chicken breast isolate N653. Both plasmids are ∼35 kb in size and members of the IncX4 subgroup. They share a high degree of sequence similarity to C. rodentium ICC168 plasmid pCROD2 (FN543504). Unique to this group are some metabolic genes, such as putative toxin–antitoxin system genes (hicA), (hicB), DnaJ-class molecular chaperone (dnaJ) (heat shock protein), plasmid replication gene (repA), site-specific recombinase (xerD), and acetyl-CoA carboxylase beta subunit (accD). With regards to the VirB/D4 gene clusters found on the IncX or IncX-like plasmids in this study, a considerable variation was observed, sometimes having <55% sequence identity between homologous genes (supplementary figs. S3 and S4, Supplementary Material online), a phenomenon that has been observed elsewhere among the Enterobacteriaceae (Johnson et al. 2012). Comparison of the plasmids from the three groups demonstrated that most of the T4SS-associated genes divergence was seen in plasmids derived from group 3 isolates.
Noteworthy was the observation that three of these eight plasmids, each isolated from different 2011 clinical outbreak strains (41563 OR Blood, 41578 OH stool, 41573 OH Urine), differed in DNA sequence and gene carriage, suggestive of extensive horizontal gene transfer (HGT) or a mixed subpopulation of S. Heidelberg isolates.
Caenorhabditis elegans System
To answer the question whether the isolates that contain the VirB/D4 plasmid are more virulent, we performed a survival assay using a C. elegans system to compare the virulence potential of our strains. This system has been used successfully as an invertebrate host model to assess the virulence determinants of human pathogens, such as Vibrio cholerae (Sahu et al. 2012) and S. Typhimurium (Aballay et al. 2000). We tested six different S. Heidelberg isolates (four ground turkey isolates, one swine isolate, and one clinical outbreak isolate) having different plasmid profiles. Strain 29169 singly carries a small mobilization plasmid; strain SARA 31 carries only the VirB/D4 T4SS plasmid; strain 418 carries a small mobilization plasmid and an IncA/C plasmid; strain 41565 carries a small mobilization plasmid and an IncHI2 plasmid; strain 41578 has a small mobilization plasmid, the VirB/D4 T4SS plasmid, and an IncI1 plasmid; and N30678 has four plasmids: the small mobilization plasmid, the VirB/D4 T4SS plasmid, the IncI1, and the IncHI1. Except SARA 31, in which phage P4 and P2-like were not present, all other isolates had the same phage profile.
The results, which showed that S. Heidelberg isolates are significantly (P < 0.0001) more pathogenic than the E. coli OP 50 control strain, failed to demonstrate any appreciable difference in pathogenicity between strains carrying the VirB/D4 T4SS plasmid and those strains that did not (fig. 7). However, we also identified some VirB/D4 T4SS components on the resistance plasmids IncA/C, IncHI2, and IncI1. Interestingly, isolate 29169, which did not carry any large plasmid and any of the T4SS components, was found to be significantly (P < 0.0001) less pathogenic using this assay than any of the other isolates evaluated. Isolate 41565, which carried the small mobility plasmid as well as additional IncHI2 plasmid, demonstrated the second weakest pathogenic potential. Isolate SARA 31, which carried the VB/D4 T4SS plasmid but no other resistance plasmid, showed a survival curve profile similar to that of isolate N418, which did not have the VB/D4 T4SS plasmid but carried the IncA/C plasmid.
Fig. 7.—
Caenorhabditis elegans survival data from six S. Heidelberg isolates. The figure shows that the six S. Heidelberg isolates (29169, SARA 31, 418, 41565, 41578, and N30678) are significantly (P < 0.0001) more pathogenic than the Escherichia coli OP 50 control strain. Further the figures show that isolates not carrying any T4SS components tend to be significant less pathogenic than those isolates that do carry them.
The highest degree of pathogenicity was seen in N30678, which carries four plasmids including resistance and VB/D4 plasmids, and it is significantly (P < 0.05) different from all other isolates except the clinical outbreak isolate 41578, which also consist resistance and VB/D4 plasmids (fig. 7). The data show that isolates not carrying any T4SS components tend to be significant less (P < 0.0001) pathogenic than those isolates that do carry the VB/D4 T4SS plasmid. Moreover, those strains that carry both the VB/D4 T4SS plasmid and an antibiotic resistance plasmid, which also carries the T4SS genes, demonstrated a pathogenic potential greater than isolates carrying only one of these plasmids.
Identification of Antimicrobial Resistance Genes and Plasmids
Analysis of WGS data showed 27 plasmid-associated antibiotic resistance genes among the 44 isolates and included genes expected to produce resistance to aminoglycosides, beta-lactams, phenicols, folate pathway inhibitors, tetracyclines, and cephems. These genes were not detected in the eight pan susceptible isolates that also did not carry any of the antibiotic resistance plasmids. Resistance phenotypes correlated with genotypes in all strains examined (table 4). However, some strains with the same genotype exhibited different antimicrobial resistance patterns. For example, both strains N18440 and N18413 contain the aadA1 gene but only isolate N18440 showed resistance to streptomycin. In this study, streptomycin resistance was defined using the MIC value >32 mg/l determined by CLSI. However, studies have shown that strains harboring an aadA gene cassette can have MICs of 16 mg/l, which would classify these strains as streptomycin-susceptible (Sunde and Norström 2005).
Table 4.
Antibiotic Resistance and Plasmid Characteristics of S. Heidelberg Isolates Used in This Study
| Isolates | Antimicrobial Resistance Profile | MDR Plasmids | Resistance Genes on Plasmids |
|---|---|---|---|
| SARA37 | STR, TET | IncI1 | aph, sph, strA, strB, tetC, ble |
| SARA35 | AMP | ca 8 kb | blaTEM1 |
| SARA32 | Susceptible | None | None |
| SARA31 | Susceptible | None | None |
| N418 | AMC,AMP,FOX,AXO,CHL,GEN,KAN,STR,SUL,TET,TIO | IncA/C | aac3-VI, aadA1, aadA2, aadB, aphA1, strA, strB, blaCMY2, blaTEM1, sul1, sul2, cmlA1, floR, tetA, tetR(A), qacΔE, sugE2 |
| N19992 | Susceptible | None | None |
| SARA39 | SUL | IncHI2 | aadA1, sul1, tetG, tetR(G), qacΔE |
| N189 | AMC,AMP,FOX,AXO,TIO | IncI1 | blaCMY2, sugE2 |
| N1514 | Susceptible | None | None |
| N1536 | AMP, GEN,STR,SUL | IncI1 | aac3-VI, aadA1, blaTEM1, sul1, qacΔE |
| N4496 | Susceptible | None | None |
| N4403 | GEN,STR,SUL | IncI1 | aac3-VI, aadA1, sul1, qacΔE |
| N4541 | Susceptible | None | None |
| N653 | GEN,STR,SUL | IncI1 | aac3-VI, aadA1, sul1, qacΔE |
| 82-2052 | KAN, STR, TET | IncI1 | aph, sph, strA, strB, tetC, ble |
| 24390 | GEN,STR,SUL | IncI1 | aac3-VI, aadA1, sul1, qacΔE |
| 21381 | AMC,AMP,FOX,AXO,GEN,KAN,STR,SUL,TET,TIO | IncI1; IncA/C | aac3-VI, aadA1, aphA1, strA, strB, blaCMY2, sul1, sul2, tetA, tetR(A), qacΔE, sugE2 |
| N4630 | GEN,STR,SUL | IncI1 | aac3-VI, aadA1,sul1, qacΔE |
| N19871 | AMP,GEN,STR,TET | IncI1 | aacC, aadA1, blaTEM1, tetA, tetR(A) |
| N15757 | AMP,KAN,STR,TET | IncI1; IncHI2 | aph, sph, strA, strB, blaTEM1, tetC, tetD, ble |
| N26457 | AMC,AMP,FOX,AXO,GEN,KAN,STR,SUL,TET,TIO | IncI1; IncA/C | aac3-VI, aadA1, aphA1, strA, strB, blaCMY2, sul1, sul2, tetA, tetR(A), qacΔE, sugE2 |
| N18393 | AMP,GEN,TET | IncI1 | aacC, aadA1, blaTEM1, tetA, tetR(A) |
| N29341 | AMP,GEN,STR,TET | IncI1 | aacC, aadA1, strA, strB, blaTEM1, tetA, tetR(A) |
| 41578 | AMP,GEN,STR,TET | IncI1 | aacC, aadA1, blaTEM1, tetA, tetR(A) |
| 41563 | AMP,GEN,STR,TET | IncI1 | aacC, aadA1, strA, strB, blaTEM1, tetA, tetR(A) |
| 41567 | AMP,GEN,STR,TET | IncI1 | aacC, aadA1, strA, strB, blaTEM1, tetA, tetR(A) |
| 41573 | AMP,GEN,TET | IncI1 | aacC, aadA1,blaTEM1, tetA, tetR(A) |
| 41576 | AMP,GEN,STR,TET | IncI1 | aacC, aadA1, strA, strB, blaTEM1, tetA, tetR(A) |
| 41584 | AMP,GEN,STR,TET | IncI1 | aacC, aadA1, strA, strB, blaTEM1, tetA, tetR(A) |
| 41579 | AMP,GEN,TET | IncI1 | aacC, aadA1, blaTEM1, tetA, tetR(A) |
| 41566 | AMP,GEN,STR,TET | IncI1 | aacC, aadA1, strA, strB,blaTEM1, tetA, tetR(A) |
| 41565 | KAN,STR,TET | IncHI2 | aph, sph, strA, strB, tetC, tetD, ble |
| N30678 | AMP,GEN,KAN,STR,SUL,TET | IncI1; IncHI2 | aac3-VI, aadA1, aph, sph, strA, strB, blaTEM1, sul1, tetC, tetD, ble, qacΔE |
| 32507 | KAN,STR,TET | IncHI2 | aph, sph, strA, strB, tetC, tetD, ble |
| 24393 | KAN,STR,TET | IncHI2 | aph, sph, strA, strB, tetC, tetD, ble |
| N20134 | AMP,GEN,STR,SUL | IncI1 | aac3-VI, aadA1, sul1, qacΔE |
| 29169 | Susceptible | None | None |
| 24391 | KAN,STR,SUL | IncI1 | aac3-VI, aadA1, sul1, qacΔE |
| 24359 | GEN,KAN,STR,SUL,TET | IncI1; IncHI2 | aac3-VI, aadA1, aph, sph, strA, strB, sul1, tetC, tetD, ble, qacΔE |
| 24388 | Susceptible | None | None |
| 20752 | KAN,STR,TET | IncHI2 | aph, sph, strA, strB, tetC, tetD, ble |
| N18413 | AMP,GEN,TET | IncI1 | aacC, aadA1, blaTEM1, tetA, tetR(A) |
| N18440 | AMP,GEN,STR,TET | IncI1 | aacC, aadA1, blaTEM1, tetA, tetR(A) |
| N18453 | AMP,GEN,STR,TET | IncI1 | aacC, aadA1, blaTEM1, tetA, tetR(A) |
Among the 36 S. Heidelberg isolates resistant to one or more antibiotics, 24 contained sequences indicative of presence of an IncI1 plasmid, 5 had an IncHI2 plasmid, and 1 carried an IncA/C plasmid. Three isolates contained two plasmids, one with IncI1 + IncHI2, and two isolates each having IncI1 + IncA/C replicons (table 4). Besides isolate 41565, which carries an IncHI2 plasmid, all Heidelberg isolates associated with 2011 outbreak carry an IncI1 plasmid, which was detected the most overall among our strains. The IncI1 plasmid carries several antibiotic resistance genes and components of the T4SS. The IncHI2 plasmid was found in eight isolates and carries several antibiotic resistance genes and genes conferring resistance to quaternary ammonium compounds, as well as genes encoding the conjugative T4SS. Furthermore, unique to the IncHI2 plasmid is the presence of genes encoding resistance to heavy metals, including tellurium, copper, cadmium, mercury, and silver.
The three isolates resistant to the most (ten or more) antibiotics carried the IncA/C multidrug resistance plasmid. In addition to carrying the genes encoding resistance to different antibiotics and quaternary ammonium compounds, this plasmid also carries the conjugative T4SS transfer system. All three IncA/C plasmids carried the resistance element blaCMY-2-blc-sugE2, which was only found in one other isolate, N189, on an IncI1 plasmid.
SARA 35, which was only resistant to ampicillin, did not carry a multidrug resistance plasmid but, instead, carried a smaller plasmid (∼8.0 kb) having only a single determinant that correlated with resistance to β-lactams (tnpA-blaTEM-1). This plasmid shared a high degree of sequence similarity to S. Typhimurium pAnkS (accession number: DQ916413), which also carried the transposon resistance element (tnpA-blaTEM-1). The Inc group for this plasmid could not be determined from the sequence.
Figure 8 shows a neighbor-joining tree generated from a presence/absence matrix of resistance genes for the 30 group 3 Heidelberg isolates having a common PFGE profile. As expected, resistance tended to segregate according to the particular Inc group plasmid they carried and not by clonality or evolutionary relationship. This observation indicated that the presence of specific resistance genes correlates with the particular Inc group of the plasmids. For example, the resistance genes (aphA1, blaCMY2, sul2, cmlA1, floR) were only detected in isolates that carry an IncA/C plasmid, whereas only isolates carrying an IncHI2 plasmid were found to contain the resistance genes aph, sph, tetC, tetD, and ble. Similarly, the aacC gene only appears to be present in those isolates identified as carrying an IncI1 plasmid. The association of different resistance genes with specific Inc groups is well known.
Fig. 8.—
Absence and presence tree of resistance genes among S. Heidelberg isolates associated with paraphyletic group 3. It is a similarity tree based on binary distances under neighbor-joining algorithm for tree construction. The resistance genes and incompatibility group of the resistance plasmid are mapped to the tree.
Mobilization Plasmids
Plasmids below 8 kb did not carry any antimicrobial resistance or virulence-associated genes. Instead, these smaller plasmids tended to carry genes capable of promoting plasmid mobilization, such as the mbeA, mbeB, mbeC, mbeD genes. S. Heidelberg isolates N189, N653, and N4403, which were originally isolated from chicken breast, carry a small, 3,772 bp plasmid that is identical to the S. Heidelberg SL476 plasmid pSL476_3 (GenBank accession number: CP001119) (Fricke et al. 2011). All group 3 isolates (fig. 3), except isolate 24393, have similar XbaI patterns (JF6X01.0032, JF6X01.0033, JF6X01.0034, JF6X01.0058), and carry a 4,473 bp plasmid that shares high sequence similarity with the S. Heidelberg plasmid pSH1148_4.8 (GenBank accession number: JX494965). SARA 35, which was isolated in Brazil, was found to carry a 6,647 bp ColE1 plasmid (pCFSAN000443_01) that is similar to E. coli plasmid ColE1 (GenBank accession number: J01566).
Discussion
Within the last several years, S. Heidelberg has been identified as one of the top serovars responsible for human illness, including a recent multistate outbreak of an antibiotic-resistant strain associated with consumption of contaminated ground turkey. Using whole-genome sequences, we characterized the genetic diversity of 46 S. Heidelberg isolated over ∼30 years. This study illustrates the novel ways in which WGS, combined with phylogenetic analysis, can be used to investigate and characterize the diversity of S. Heidelberg. Using this strategy with outbreak isolates, we were also able to comprehensively characterize and differentiate among highly clonal S. Heidelberg isolates. For example, SNP analysis in combination with phylogenetic analysis is able to determine the phylogenetic relationships among S. Heidelberg isolates, easily separating them from other S. Heidelberg strains, including isolates having the same XbaI and BlnI PFGE patterns and thought to be highly clonal.
We also found that the SNP analysis clustered isolates together that did not have the same PFGE profiles, such as the 2011 outbreak isolates, which were distinct on the PFGE two-enzyme dendrogram. SNP analysis tightly clustered the clinical and ground turkey outbreak isolates together and distinguished these from other apparently clonal isolates that shared the same PFGE and antibiotic resistance profiles. This result confirms the epidemiological data that the clinical and ground turkey isolates belong to the same outbreak, and are separate from other strains grouped with them by PFGE.
We found two nonsynonymous SNPs that were present only among the outbreak isolates. The first of these SNPs was found to be located on the gluconate transporter gntP gene. The biological role of GntP might be to permit the host to take up gluconate when only tiny amounts are present, thereby conferring an advantage to the host in a competitive environment (Klemm et al. 1996). The second of these SNPs was located within a gene encoding a putative hydrolase (phosphoserine phosphatase). This gene is involved in glycine and serine metabolism, and resulted in a proline to glutamine amino acid change that is known to change the secondary structure of this enzyme.
SNP-based phylogeny clearly has a distinct advantage in subtyping clonal isolates because the method offers nucleotide base resolution. We detected 4,053 SNPs within highly clonal S. Heidelberg isolates. Compared with the PFGE analyses that grouped isolates together having the same Xbal and BlnI patterns, the resolution of the SNP phylogeny conclusively demonstrated that genome sequencing distinguishes between outbreak and nonoutbreak isolates that shared the same XbaI and BlnI patterns. The results from the ML tree generated from the SNP matrix constructed with kSNP are concordant with those observed based on the SNP matrix developed with our custom pipeline. The study showed that SNP analysis is very consistent and reliable, confirming that even when different methods were used to identify SNPs, there are no appreciable changes in the overall phylogeny of the strains included in the analyses.
Only a few mutation “hotspot” genes were found to have a disproportional number of SNPs and that might serve as targets useful for rapidly subtyping serovar Heidelberg. Although these genes have several SNPs, the haplotype diversity is fairly low, indicating there are only few unique sequences among S. Heidelberg. This information supports the value of SNP analysis as a useful tool for subtyping this serovar.
WGS data and comparative genomics not only offer a useful subtyping method to differentiate closely related bacteria, but they also provide a better understanding of pathogenicity and evolution. Our results show substantial loss and gain of plasmids and phages between these S. Heidelberg isolates that were consistent with previously generated tree topologies. The phages found among these strains are known to carry cassette genes (morons) encoding factors that enhance the proliferation and dissemination of the prophage by improving the fitness and /or virulence of the lysogen (Boyd and Brussow 2002; Pelludat et al. 2003). This possibility has led to the assumption that phage-mediated distribution of virulence factors and fitness factors is a key driving force in the optimization of Salmonella–host interactions and the emergence of new epidemic clones (Brussow et al. 2004). For example, lambda-like phage Gifsy-2, which is present among all S. Heidelberg isolates examined in the study, carries the virulence factor periplasmic CuZn-superoxide dismutase (SodCI), an enzyme that has the potential to enhance the fitness of a Gifsy-2 lysogen (Brussow et al. 2004). Moreover, the prophage P4 and prophage P2-like were associated with isolates from group 3 which included the outbreak isolates. The P2-like phage carries the sopE1 gene that encodes part of the T3SS that translocates bacterial effector proteins directly into the cytosol of host cells. The phage P2-like has also been identified in S. Typhimurium strain DT49/DT204 that caused a major outbreaks in Europe in 1970s and 1980s. It has been speculated that lysogenic conversion with sopE1 was an important step in the emergence of this epidemic strain (Mirold et al. 2001). Interestingly, Mirold et al. reported that sopE1 is not restricted to a certain bacteriophage as a “vehicle,” identifying sopE1 in other serovars present on a Gifsy-like phage (Mirold et al. 2001). We also saw the same phenomenon among the S. Heidelberg of this study where the sopE1 gene, which was found to be present among all isolates, was carried by both the P2-like and the lambda-like Gifsy-2 phages.
The exchanges of genetic cassettes between unrelated phages further increase the degree of HGT noted among S. Heidelberg strains, clearly demonstrating the importance of phages in the emergence and evolution of Salmonella pathogens. To date, this is the first time that phages have been characterized by name among S. Heidelberg isolates and that a P2-like phage was identified in S. Heidelberg that carries the sopE1 gene. This phage and, subsequently, the sopE1 determinant, was present in the outbreak isolates, possibly serving as one of the virulence factors giving rise to these pathogens. Furthermore, we identified the presence of 74 virulence determinants in all S. Heidelberg isolates.
In the course of performing this study, both newly sequenced plasmids and plasmids sharing a high sequence similarity to previously described plasmids (Han et al. 2012) were identified. Similar to the phage presence and absence tree topology previously described, the mobilization plasmids absence/presence seemed to correlate with the tree topology according to the plasmid(s) they carried. For instance, the 3,772 bp mobilization plasmid was only identified in isolates belonging to paraphyletic group 2, whereas the 4,473 bp plasmid was found only in isolates which belong to the paraphyletic group 3 on the ML tree. The mobilization plasmids carry the genes necessary to encode the proteins involved with relaxosome formation and processing. Moreover, these plasmids are efficiently transferred by other plasmids, such as IncI1 and IncX (Francia et al. 2004), two plasmids frequently found among the S. Heidelberg in this and other studies (Folster et al. 2012; Han et al. 2012). The mobilization plasmids tend to be retained and are, continually carried by specific groups of S. Heidelberg isolates, suggesting that they must play an essential role in this serovar survival, although the precise role is unclear. Plasmid sequence analysis detected additional plasmids that encode several S. Heidelberg virulence determinants, including T4SS and antibiotic resistance genes. Unlike the smaller plasmids, the loss and gain of the larger plasmids (ranging from 33 to >200 kb) did not correlate with the tree topology. The plasmid analyses performed in this and previous studies (Folster et al. 2012; Han et al. 2012) demonstrated that S. Heidelberg isolates have at least four transmissible plasmids, including an IncA/C, IncI, and IncHI2 and IncX plasmid. The IncA/C, IncI, and IncHI2 plasmids carry genes that are important for virulence, colonization, and persistence, as well as those encoding resistance determinants to heavy metals, disinfection, and antimicrobial agents. We determined that most of the antimicrobial-resistance phenotypes could be accounted for by resistance genes carried on plasmids. We did not identify an instance of resistance occurring in the absence of a corresponding gene. This shows that genetic prediction of phenotypic resistance may be possible and may become necessary as clinical diagnostics moves toward culture-independent technologies. More studies are necessary to determine how well specific DNA sequences, and combinations of sequences, predict resistance or susceptibility to various antimicrobial agents.
Besides the IncI1 and the IncHI2 antibiotic resistance plasmids, an IncX plasmid that encodes the structural components of the VirB/D4 T4SS apparatus was detected in most of the outbreak isolates. The VirB/D4 T4SS apparatus is important in that it assists with “rapid dissemination of antibiotic resistance and virulence determinants” (Bhatty et al. 2013). Unlike other secretion systems, the T4SS is not only capable of acting in a conjugative fashion disseminating DNA to other bacteria, thereby contributing to genome plasticity and the evolution of infectious pathogens through dissemination of antibiotic resistance and virulence genes, but also capable of transferring protein effectors across both the bacterial and eukaryotic plasma membrane directly into the cytosol of the target cells, thereby contributing directly to bacterial pathogenicity (Juhas et al. 2008; Gokulan et al. 2013). The T4SS was first described for Agrobacterium tumefaciens and consists of 11 proteins encoded by the virB gene complex. Of particular interest was the fact that some plasmids carrying the T4SS, such as those of the IncX group, also carry systems that confer plasmid stability among the strains possessing the plasmid. For instance, the IncX plasmids carry the toxin–antitoxin system genes (hicA), (hicB), as well as the toxin/antitoxin stability genes (stbE), (stbD), both of which contribute to the plasmid stability to assure that the plasmid as well as its associated virulence determinants are inherited by all bacterial progeny (Yamaguchi and Inouye 2011; Unterholzner et al. 2013). As such, these genes serve to stabilize the “pathogenic potential” of these isolates.
Finding that the T4SS system was present in the majority of the pathogenic isolates and carried on plasmids also bearing determinants associated with assuring vertical transmission of the plasmid to progeny bacteria, we sought to assess the role of the T4SS in overall pathogenicity. Using the C. elegans virulence assay, we confirmed that S. Heidelberg isolates that carried components of the T4SS were more pathogenic in this model than isolates that did not carry the system, suggesting the T4SS plays a role in the pathogenicity of S. Heidelberg. Much more inclusive investigations must be performed to fully understand the role of plasmid-encoded genes in pathogenesis, with special emphasis on the role of virulence-associated VirB/D4 T4SS secretory mechanism.
In summary, the data presented in this study support the role of transmissible mobile genetic elements in the evolution of virulence and resistance among S. Heidelberg. Understanding the results and the role of these elements is very important since S. Heidelberg is the second most common serovar, following S. Typhimurium, responsible for Salmonella-related deaths in the United States (Kennedy et al. 2004; Patchanee et al. 2008). Although the mechanisms responsible for S. Typhimurium pathogenicity and colonization are well studied, fewer studies are available for S. Heidelberg (Han et al. 2011). Presumably, the transmissible plasmids identified in our strain set play a major role in the spread of virulence and resistance genes among S. Heidelberg and, most likely, other Salmonella serovars.
The results from this and other studies suggest that plasmids are being exchanged quickly. For example, it was noted that some strains associated with the 2011 outbreak isolated from the same state had different plasmid profiles. In addition to the ecological benefits conferred as a consequence of this rapid plasmid exchange, large plasmids can change PFGE patterns. We noted that, while most of the sequenced isolates associated with the 2011 outbreak had BlnI PFGE pattern JF6A26.0076, a single isolate, 41565, had a different, JF6A26.0017 BlnI pattern, most likely due to the presence of an IncHI2 plasmid rather than IncI1. Other closely related isolates not associated with the outbreak carrying the IncHI2 plasmid also exhibited BlnI PFGE pattern JF6A26.0017.
The results determined in this study have convincingly shown that WGS analysis could be a useful tool for identifying the source of contamination and studying the short-term evolution of these epidemic clones. In the context of the 2011 ground turkey-associated outbreak, SNP analysis provided an excellent tool for subtyping S. Heidelberg and clustering closely related isolates together. Using these data, we determined that the antibiotic-resistant outbreak isolates diverged from environmental isolates through the gain of several virulence factors, such as sopE1 gene carried on Phage P2 or the T4SS system carried on the IncX plasmid. Of particular interest, other isolates in paraphyletic group 3 on the ML tree carried the same virulence factors as the 2011 outbreak strain, suggesting that this strain has been seen earlier in the environment and that the outbreak occurred as a consequence of changes in outside influences such as the dose or the handling of the food.
CDC projected that for every case of Salmonella reported that 29.3 cases occur in the community and 28.3 cases goes undiagnosed or unreported (Scallan et al. 2011), suggesting that the 2011 outbreak actually may have sickened over 4,000 people in the United States. The World Health Organization has declared that most countries have documented a significant increase of microorganism-mediated foodborne illnesses (WHO 2013). Therefore, it is paramount to have a robust identification system that can distinguish closely related strains in an outbreak situation. To date, DNA “fingerprinting” facilitated by PFGE at PulseNet has often been used to establish the source of clinical isolates. However, several examples in the past have shown that PFGE may not be as useful for some serotypes that show less diversity by PFGE (CDC 2010a, 2010b). Therefore, it is essential to develop and evaluate other methods for their ability to effectively and accurately discriminate outbreak isolates from nonoutbreak isolates, like the combination of WGS and computationally efficient methods of analysis. Such a combination of approaches also will help to genetically link the implicated food sources of contamination with farm or factories. As more WGS data are gathered and analytical tools developed in combination with epidemiology investigations, we anticipate significant improvements in timeliness of outbreak investigations and attribution of human infections to specific sources.
Supplementary Material
Supplementary tables S1 and S2 and figures S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Ruth Timme for submission of the draft genomes to NCBI and the NCBI rapid annotation pipeline team, Bill Klimke, Dmitry Dernovoy, Stacy Ciufo, Kathleen O’Neill, Azat Badretdin, and Tatiana Tatusova, for key genome annotation service. Further they thank Yuansha Chen for sharing the CVM/DAFM database of antimicrobial resistance genes. No human subjects or animals were used in this study. All authors have read the manuscript and agreed to its contents, subject matter, and author line order. These data are novel and have not been previously published elsewhere. Disclosure forms provided by the authors will be available with the full text of this article. This work was supported by the Center for Veterinary Medicine and the Center for Food Safety and Applied Nutrition at the US Food and Drug Administration and by an appointment of MH by the Joint Institute for Food Safety and Applied Nutrition (JIFSAN), University of Maryland, College Park, MD.
Literature Cited
- Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol. 2000;10:1539–1542. doi: 10.1016/s0960-9822(00)00830-7. [DOI] [PubMed] [Google Scholar]
- Allard MW, et al. High resolution clustering of Salmonella enterica serovar Montevideo strains using a next-generation sequencing approach. BMC Genomics. 2012;13:32. doi: 10.1186/1471-2164-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard MW, et al. On the evolutionary history, population genetics and diversity among isolates of Salmonella enteritidis PFGE pattern JEGX01.0004. PLoS One. 2013;8:e55254. doi: 10.1371/journal.pone.0055254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazinet AL, Cummings MP. Computing the tree of life—leveraging the power of desktop and service grids. 2011 In: Proceedings of the Fifth Workshop on Desktop Grids and Volunteer Computing Systems PCGrid 2011. Available from: http://lattice.umiacs.umd.edu/latticefiles/publications/lattice/computing_tree_of_life.pdf. [Google Scholar]
- Bazinet AL, Myers DS, Fuetsch J, Cummings MP. Grid services base library: a high-level, procedural application program interface for writing Globus-based Grid services. Future Gener Comput Syst. 2007;23:517–522. [Google Scholar]
- Beltran P, et al. Reference collection of strains of the Salmonella typhimurium complex from natural populations. J Gen Microbiol. 1991;137:601–606. doi: 10.1099/00221287-137-3-601. [DOI] [PubMed] [Google Scholar]
- Bhatty M, Laverde Gomez JA, Christie PJ. The expanding bacterial type IV secretion lexicon. Res Microbiol. 2013;164(6):620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd EF, Brussow H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002;10:521–529. doi: 10.1016/s0966-842x(02)02459-9. [DOI] [PubMed] [Google Scholar]
- Brussow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Atlanta (GA): Centers for Disease Control and Prevention; 2010a. Centers for Disease Control and Prevention. 2010. Multistate outbreak of human Salmonella Montevideo infections (final ubdate) Available from: http://www.cdc.gov/salmonella/montevideo. [Google Scholar]
- CDC. Atlanta (GA): Centers for Disease Control and Prevention; 2010b. Centers for Disease Control and Prevention. 2011. Multistate outbreak of human Salmonella Enteritidis infections associated with shell eggs (final ubdate) Available from: http://www.cdc.gov/salmonella/enteritidis. [Google Scholar]
- CDC. Atlanta (GA): Centers for Disease Control and Prevention; 2011. Centers for Disease Control and Prevention. 2011. Multistate outbreak of human Salmonella Heidelberg infections linked to ground turkey. Available from: http://www.cdc.gov/salmonella/heidelberg/ [Google Scholar]
- CDC. Atlanta (GA): Centers for Disease Control and Prevention; 2013. Centers for Disease Control and Prevention. 2013. National enteric disease surveillance: Salmonella annual report, 2011. Available from: http://www.cdc.gov/ncezid/dfwed/PDFs/salmonella-annual-report-2011-508c.pdf. [Google Scholar]
- Chittick P, Sulka A, Tauxe RV, Fry AM. A summary of national reports of foodborne outbreaks of Salmonella Heidelberg infections in the United States: clues for disease prevention. J Food Prot. 2006;69:1150–1153. doi: 10.4315/0362-028x-69.5.1150. [DOI] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump JA, et al. Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob Agents Chemother. 2011;55:1148–1154. doi: 10.1128/AAC.01333-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley SL, et al. Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl Environ Microbiol. 2011;77:4273–4279. doi: 10.1128/AEM.00598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folster JP, et al. Characterization of multidrug-resistant Salmonella enterica serovar Heidelberg from a ground turkey-associated outbreak in the United States in 2011. Antimicrob Agents Chemother. 2012;56:3465–3466. doi: 10.1128/AAC.00201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia MV, et al. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 2004;28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Fricke WF, et al. Comparative genomics of 28 Salmonella enterica isolates: evidence for CRISPR-mediated adaptive sublineage evolution. J Bacteriol. 2011;193:3556–3568. doi: 10.1128/JB.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SN, Slezak T. Scalable SNP analyses of 100+ bacterial or viral genomes. J Forensic Res. 2010;01:107. [Google Scholar]
- Gokulan K, et al. Impact of plasmids, including those encodingVirB4/D4 type IV secretion systems, on Salmonella enterica serovar Heidelberg virulence in macrophages and epithelial cells. PLoS One. 2013;8:e77866. doi: 10.1371/journal.pone.0077866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn JS, Alpuche-Aranda CM, Loomis WP, Belden WJ, Miller SI. Characterization of the Salmonella typhimurium pagC/pagD chromosomal region. J Bacteriol. 1995;177:5040–5047. doi: 10.1128/jb.177.17.5040-5047.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, et al. Comparison of Salmonella enterica serovar Heidelberg isolates from human patients with those from animal and food sources. J Clin Microbiol. 2011;49:1130–1133. doi: 10.1128/JCM.01931-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, et al. DNA sequence analysis of plasmids from multidrug resistant Salmonella enterica serotype Heidelberg isolates. PLoS One. 2012;7:e51160. doi: 10.1371/journal.pone.0051160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, et al. Genome sequences of five Salmonella enterica serovar Heidelberg isolates associated with a 2011 multistate outbreak in the United States. J Bacteriol. 2012;194:3274–3275. doi: 10.1128/JB.00419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huehn S, Bunge C, Junker E, Helmuth R, Malorny B. Poultry-associated Salmonella enterica subsp. enterica serovar 4,12:d:- reveals high clonality and a distinct pathogenicity gene repertoire. Appl Environ Microbiol. 2009;75:1011–1020. doi: 10.1128/AEM.02187-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huehn S, et al. Virulotyping and antimicrobial resistance typing of Salmonella enterica serovars relevant to human health in Europe. Foodborne Pathog Dis. 2010;7:523–535. doi: 10.1089/fpd.2009.0447. [DOI] [PubMed] [Google Scholar]
- Johnson TJ, et al. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Johnson TJ, Nolan LK. Plasmid replicon typing. Methods Mol Biol. 2009;551:27–35. doi: 10.1007/978-1-60327-999-4_3. [DOI] [PubMed] [Google Scholar]
- Juhas M, Crook DW, Hood DW. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol. 2008;10:2377–2386. doi: 10.1111/j.1462-5822.2008.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M, et al. Hospitalizations and deaths due to Salmonella infections, FoodNet, 1996–1999. Clin Infect Dis. 2004;38(Suppl. 3):S142–S148. doi: 10.1086/381580. [DOI] [PubMed] [Google Scholar]
- Klemm P, Tong S, Nielsen H, Conway T. The gntP gene of Escherichia coli involved in gluconate uptake. J Bacteriol. 1996;178:61–67. doi: 10.1128/jb.178.1.61-67.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimke W, et al. The National Center for Biotechnology Information’s Protein Clusters Database. Nucleic Acids Res. 2009;37:D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, et al. Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major serovars of Salmonella enterica subsp. enterica. Appl Environ Microbiol. 2011;77:1946–1956. doi: 10.1128/AEM.02625-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirold S, et al. Salmonella host cell invasion emerged by acquisition of a mosaic of separate genetic elements, including Salmonella pathogenicity island 1 (SPI1), SPI5, and sopE2. J Bacteriol. 2001;183:2348–2358. doi: 10.1128/JB.183.7.2348-2358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NARMS. Bethesda (MD): Executive Report FDA; 2010. National Antimicrobial Resistance Monitoring System-Enteric Bacteria (NARMS) [Google Scholar]
- Norman A, Hansen LH, She Q, Sorensen SJ. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid. 2008;60:59–74. doi: 10.1016/j.plasmid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Patchanee P, Zewde BM, Tadesse DA, Hoet A, Gebreyes WA. Characterization of multidrug-resistant Salmonella enterica serovar Heidelberg isolated from humans and animals. Foodborne Pathog Dis. 2008;5:839–851. doi: 10.1089/fpd.2008.0149. [DOI] [PubMed] [Google Scholar]
- Pelludat C, Mirold S, Hardt WD. The SopEPhi phage integrates into the ssrA gene of Salmonella enterica serovar Typhimurium A36 and is closely related to the Fels-2 prophage. J Bacteriol. 2003;185:5182–5191. doi: 10.1128/JB.185.17.5182-5191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol Ecol. 2004;4:137–138. [Google Scholar]
- Sahu SN, et al. Genomic analysis of immune response against Vibrio cholerae hemolysin in Caenorhabditis elegans. PLoS One. 2012;7:e38200. doi: 10.1371/journal.pone.0038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E, et al. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprando RL, Olejnik N, Cinar HN, Ferguson M. A method to rank order water soluble compounds according to their toxicity using Caenorhabditis elegans, a complex object parametric analyzer and sorter, and axenic liquid media. Food Chem Toxicol. 2009;47:722–728. doi: 10.1016/j.fct.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Sunde M, Nortström M. The genetic backround for streptomycin resistance in Escherichia coli infuences the distribution of MICs. Antimicrob Agents Chemother. 2005;56:87–90. doi: 10.1093/jac/dki150. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Timme RE, et al. Phylogenetic diversity of enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol Evol. 2013;5(11):2109–2123. doi: 10.1093/gbe/evt159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner SJ, Hailer B, Poppenberger B, Rozhon W. Characterisation of the stbD/E toxin-antitoxin system of pEP36, a plasmid of the plant pathogen Erwinia pyrifoliae. Plasmid. 2013;70:216–225. doi: 10.1016/j.plasmid.2013.04.002. [DOI] [PubMed] [Google Scholar]
- WHO. Food safety: general information related to microbiological risks in food. 2013 Available from: http://www.who.int/foodsafety/en/ [Google Scholar]
- Wilmshurst P, Sutcliffe H. Splenic abscess due to Salmonella Heidelberg. Clin Infect Dis. 1995;21:1065. doi: 10.1093/clinids/21.4.1065. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Inouye M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat Rev Microbiol. 2011;9:779–790. doi: 10.1038/nrmicro2651. [DOI] [PubMed] [Google Scholar]
- Zhao S, et al. Antimicrobial resistance in Salmonella enterica serovar Heidelberg isolates from retail meats, including poultry, from 2002 to 2006. Appl Environ Microbiol. 2008;74:6656–6662. doi: 10.1128/AEM.01249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl D. Austin (TX): University of Texas; 2006a. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence dataset under the maximum likelihood criterion. [Google Scholar]
- Zwickl DJ. [Austin (TX)]: The University of Texas; 2006b. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion [PhD dissertation] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.