Abstract
Acquisition of Elizabethkingia infections in intensive care units (ICUs) has risen in the past decade. Treatment of Elizabethkingia infections is challenging due to the lack of effective therapeutic regimens, leading to a high mortality rate. Elizabethkingia infections have long been attributed to Elizabethkingia meningoseptica. Recently, we used whole-genome sequencing to reveal that E. anophelis is the pathogenic agent for an Elizabethkingia outbreak at two ICUs. We performed comparative genomic analysis of seven hospital-isolated E. anophelis strains with five available Elizabethkingia spp. genomes deposited in the National Center for Biotechnology Information Database. A pan-genomic approach was applied to identify the core- and pan-genome for the Elizabethkingia genus. We showed that unlike the hospital-isolated pathogen E. meningoseptica ATCC 12535 strain, the hospital-isolated E. anophelis strains have genome content and organization similar to the E. anophelis Ag1 and R26 strains isolated from the midgut microbiota of the malaria mosquito vector Anopheles gambiae. Both the core- and accessory genomes of Elizabethkingia spp. possess genes conferring antibiotic resistance and virulence. Our study highlights that E. anophelis is an emerging bacterial pathogen for hospital environments.
Keywords: Elizabethkingia, comparative genomics, pan/core-genomes
Introduction
Elizabethkingia is a genus of aerobic, nonmotile, Gram-negative rods that is ubiquitous in nature. Members of this genus thrive in wet habitats and hospital settings, in particular water supplies and saline flushing solutions. Among the Elizabethkingia spp., the species Elizabethkingia meningoseptica is well established as a serious causative agent of neonatal meningitis and sepsis (Dooley et al. 1980), and a notable rise in E. meningoseptica nosocomial infections has been recorded in recent years. Treatment of E. meningoseptica infections is notoriously difficult, and there is a lack of effective specific therapeutic regimens (Hsu et al. 2011). The mortality rate of nosocomial infections caused by E. meningoseptica can reach as high as 52% in neonates (Bloch et al. 1997) and ranges from 23% (Teres 1974) to 33% (Bloch et al. 1997) in nonneonates. Hence, the acquisition of E. meningoseptica in intensive care units (ICUs) is used as a significant predictor of mortality (Teres 1974). Except for E. meningoseptica, it is rarely reported that other Elizabethkingia species can cause infections although E. miricola has been reported to be associated with sepsis (Green et al. 2008).
The limited genomic information available for Elizabethkingia hinders our understanding of the virulence mechanisms and molecular epidemiology of its member species. Recently, we used whole-genome sequencing to investigate an Elizabethkingia outbreak in two ICUs at the National University Hospital, Singapore (Balm et al. 2013). All the patient-associated Elizabethkingia strains and hand hygiene sink aerator-associated Elizabethkingia strains were isolated within a 1-month period at the ICUs. We found the outbreak agent to be a novel species—E. anophelis (Teo et al. 2013), usually found in the midgut microbiota of the malaria mosquito vector Anopheles gambiae. Here, we report comparative genomic analysis on the Elizabethkingia spp. to investigate their mechanism for virulence, stress response, and niche adaptation.
General Characteristics of the Outbreak E. anophelis Strains
The general genomic characteristics of the seven Elizabethkingia spp. strains derived from this study, three patient (NUHP1, NUHP2, and NUHP3) and four sink isolates (NUH1, NUH4, NUH6, and NUH11), obtained from the RAST server (Aziz et al. 2008) are presented in supplementary table S1, Supplementary Material online.
The Whole-Genome Shotgun (WGS) sequences of five previously sequenced E. anophelis strains Ag1 (Bioproject accession number, PRJNA80705) and R26 (Bioproject accession number, PRJNA178189), E. meningoseptica ATCC 12535 (NITE) (Bioproject accession number, PRJNA199489), E. meningoseptica ATCC 12535 (OSU) (Bioproject accession number, PRJNA198814), and E. meningoseptica 502 (Bioproject accession number, PRJNA176121) were also submitted to the RAST server; supplementary table S2, Supplementary Material online, shows the general characteristics of these strains. The genome sizes and GC content of the hospital-isolated strains are similar to the other E. anophelis strains and one of the E. meningoseptica strains (502) (supplementary tables S1 and S2, Supplementary Material online).
Comparative Genomic Analysis
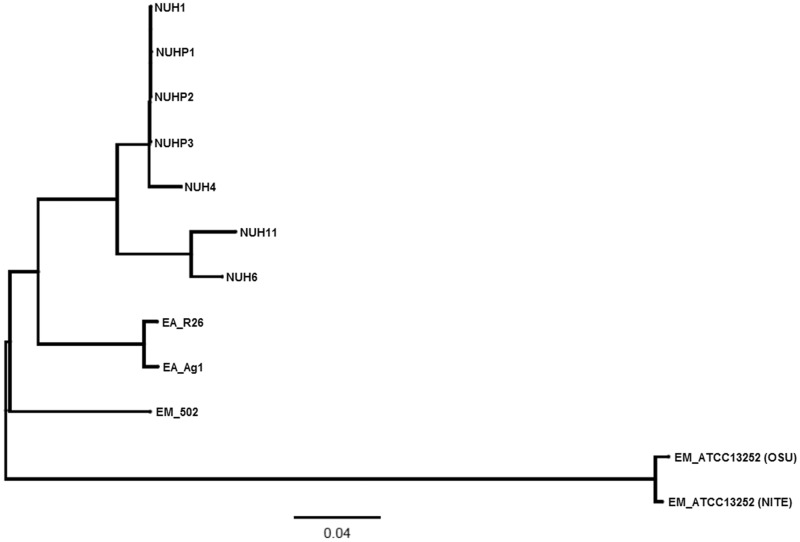
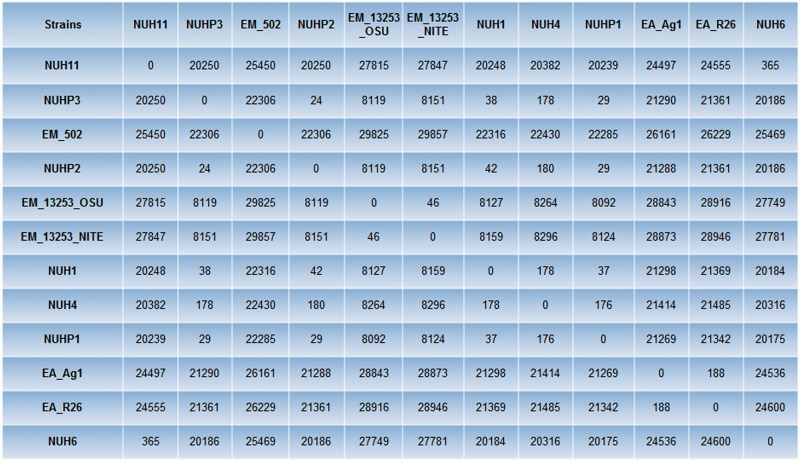
Multiple genome alignment was performed by using Progressive Mauve (Darling et al. 2010) to compare all the genomes of the Elizabethkingia spp. The phylogenetic tree based on the multiple genome alignment showed that E. anophelis and E. meningoseptica genomes belong to distinct groups (fig. 1). Single-nucleotide polymorphism (SNP) differences between each pair of Elizabethkingia spp. was calculated using the snpTree web server (Leekitcharoenphon et al. 2012) and shown in figure 2. These results showed that the genomes of the patient isolates from the current outbreak (NUHP1, NUHP2, and NUHP3) were very similar to each other, NUH1 and NUH4, which suggests that the E. anophelis strains NUHP1, NUHP2, NUHP3, NUH1, and NUH4 may be clonal. The NUH6 and NUH11 strains may exist as a separate clone as they were found to have high-genomic similarity to each other but not to the other hospital isolates. The E. anophelis Ag1 and R26 strains formed a distinct group in the phylogenetic tree. Also, the E. meningoseptica ATCC 12535 (NITE) and ATCC 12535 (OSE) strains formed a distinct group in the phylogenetic tree, whereas the E. meningoseptica 502 formed yet another distinct group.
Fig. 1.—
A phylogenetic tree showing the 12 Elizabethkingia spp. This phylogenetic tree was produced by pair-wise genome comparisons by Progressive Mauve.
Fig. 2.—
SNP distance matrix among the 12 Elizabethkingia spp. SNP difference between each pair of Elizabethkingia spp. was calculated by using the snpTree web server.
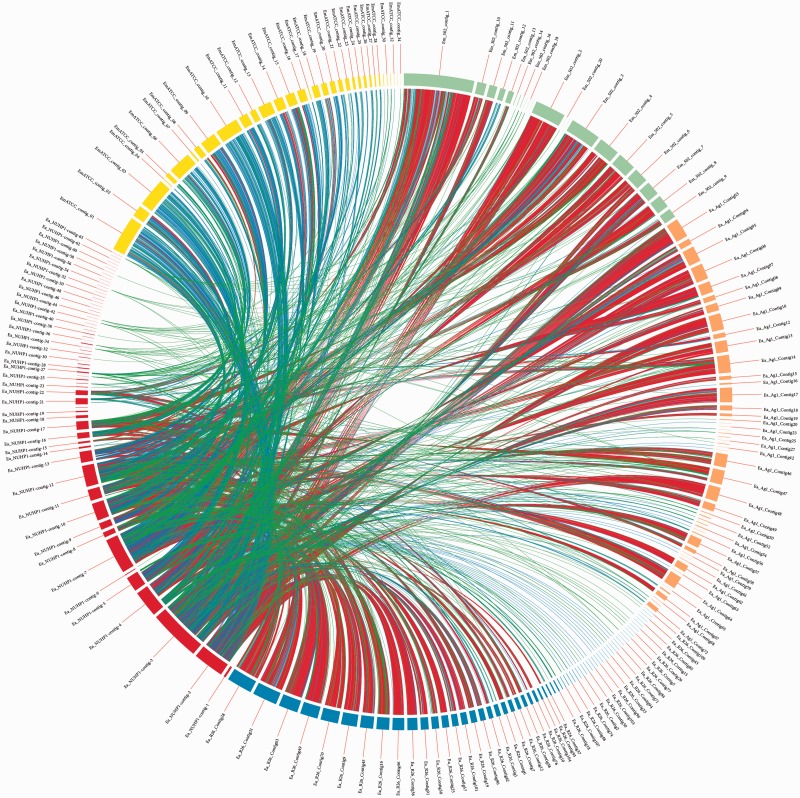
To reach a high-resolution comparison, multiple whole-genome sequence alignments were performed with CIRCOS v0.64 (Krzywinski et al. 2009), using the E. anophelis NUHP1 strain, E. anophelis Ag1 and R26 strains, and the E. meningoseptica ATCC 12535 (NITE) and 502 strains (fig. 3). The red links show a high number of large homologous regions (longer than 10,000 nucleotides) between E. anophelis NUHP1 strain and the E. anophelis Ag1 strain, E. anophelis R26 strain, and the E. meningoseptica 502 strain. The genome of the E. meningoseptica ATCC 12535 (NITE) is less close to the E. anophelis NUHP1 (fig. 3). This result correlates well with the phylogenetic tree result (fig. 1) and GC content (supplementary tables S1 and S2, Supplementary Material online) of the genomes of these Elizabethkingia spp.
Fig. 3.—
Sequence comparison by alignment. The Elizabethkingia anophelis NUHP1 strain (red arc) was aligned against: E. anophelis R26 strain (blue arc), E. anophelis Ag1 (orange arc), E. meningoseptica ATCC 12535 (NITE) (yellow arc), and E. meningoseptica 502 (light green arc). Colored links between contigs represent homologous regions spanning: 102–103 bp (green), 103–104 bp (blue), and above 104 bp (red). The presence of red links between the NUHP1 strain and E. anophelis R26, Ag1 contigs, and E. meningoseptica 502 contigs indicate a high degree of similarity between these genomes.
Core/Pan-Genomic Analysis of Elizabethkingia spp.
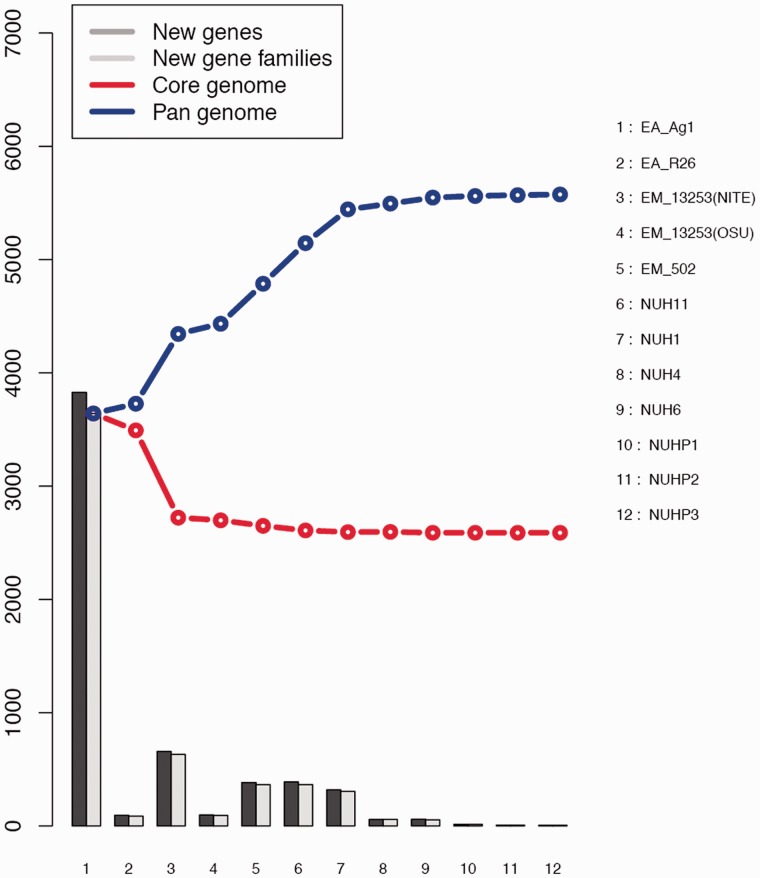
We performed core/pan-genomic analysis of the Elizabethkingia spp. The results of all permutations of the order of addition for each of the 12 genomes are presented in figure 4. As expected, the number of genes in the core-genome initially decreases and that of the pan-genome initially increases with addition of each new genome sequence. Extrapolation of the curve indicates that the core-genome reaches a minimum of 2,589 “core" genes (fig. 4). In accordance with the core-genome size, the pan-genome reaches a maximum of 5,575 genes in total across 12 genomes (fig. 4).
Fig. 4.—
Curves for the core-genomes, pan-genomes, and numbers of new genes of the 12 Elizabethkingia spp.
Resistance and Virulence Profile of the Elizabethkingia spp.
The core/accessory genomes were searched against the Comprehensive Antibiotic Resistance Database (McArthur et al. 2013) and Virulence Factors of Pathogenic bacteria Database (VFDB) (Chen et al. 2012) to identify antibiotic resistant genes and virulence genes. Thirty percentage identity and expectation value < 1e-5 were used as a threshold when performing the BLASTP searches because the genomes of Elizabethkingia spp. are very new and highly likely to not have been included in any of these databases before. Sixteen and 19 antibiotic-resistant genes were identified from the core- and accessory genomes of Elizabethkingia spp., respectively, which cover genes conferring resistance to aminoglycosides, beta-lactamases, fluoroquinolones, glycopeptides, macrolide-lincosamide-streptogramin, tetracyclines, trimethoprim, and rifampicin (table 1). These genes correlate with the reported antibiotic resistant profiles of Elizabethkingia spp. (Hsu et al. 2011). The patient-isolated NUHP1, NUHP2, and NUHP3 strains are resistant to tetracycline (minimum inhibitory concentration, MIC = 64 µg/ml), ciprofloxacin (MIC = 16 µg/ml), erythromycin (MIC > 512 µg/ml), ceftazidime (MIC = 64 µg/ml), tobramycin (MIC > 512 µg/ml), and vancomycin (MIC = 32 µg/ml). Some of the antibiotic resistance genes might be nonfunctional. A complete genome sequence is required for intact analysis of these resistance gene operons.
Table 1.
Proteins Involved in Antibiotic Resistance Encoded from Core- and Accessory Genomes of the Elizabethkingia spp. Identified by BLAST Search against the Comprehensive Antibiotic Resistance Database
| Database ID | % Identity | Antibiotic Resistance | Annotation |
|---|---|---|---|
| Core genome | |||
| (AGly)ApmA:FN806789:2858–3682:822 | 44.71 | Aminoglycosides | Aminocyclitol acetyltransferase. Confers apramycin resistance. |
| (Bla)AIM-1:AM998375:1173–2084:912 | 30.88 | Beta-lactamases | Metallo-beta-lactamase AIM-1. Imipenemase. |
| (Bla)B-1:AF189298:1–750:750 | 94.35 | Beta-lactamases | Chryseobacterium meningosepticum PINT class B carbapenemase BlaB-1 gene. Elizabethkingia meningoseptica class B carbapenemase BlaB-1 |
| (Bla)Beta-lactamase_class-A:NC_010410:1803480–1804499:1020 | 35.09 | Beta-lactamases | Beta-lactamase_class-A |
| (Bla)SFO-1:FJ848785:4719–5594:876 | 32.5 | Beta-lactamases | AmpR of SFO-1; activator of ampA. |
| (Flq)OqxA:EU370913:46652–47827:1176 | 30.71 | Fluoroquinolones | OqxA membrane-fusion protein. component of RND-type multidrug efflux pump that confers resistance to olaquindox |
| (Flq)OqxBgb:EU370913:47851–51003:3153 | 39.96 | Fluoroquinolones | OqxB integral membrane protein. component of RND-type multidrug efflux pump that confers resistance to olaquindox |
| (Gly)VanH:DQ246438:124–1179:1056 | 33.65 | Glycopeptides | VanH, D-lactate dehydrogenase. |
| (Gly)VanR-A:M97297:3976–4671:696 | 32.33 | Glycopeptides | VanR. |
| (Gly)VanR-B:AY655721:69–728:660 | 34.15 | Glycopeptides | VanRB, regulator protein. |
| (Gly)VanT:AF162694:3008–5104:2097 | 32.25 | Glycopeptides | Serine racemase VanT. converts L-serine to D-serine; involved in vancomycin resistance |
| (Gly)VanW-B:AY655721:3069–3896:828 | 33.57 | Glycopeptides | VanW. |
| (MLS)CarA:M80346:411–2066:1656 | 41.54 | Macrolide-lincosamide-streptogramin | CarA, carbomycin resistance protein. |
| (MLS)CfrA:AM408573:10028–11077:1050 | 31.9 | Macrolide-lincosamide-streptogramin | Cfr, rRNA methylase, mediates the PhLOPSA resistance phenotype |
| (Tet)otrA:X53401:349–2341:1992 | 43.66 | Tetracyclines | OtrA, oxytetracycline resistance |
| (Tmt)Dfr16:AF077008:115–558:474 | 34.03 | Trimethoprim | DHFRXVI, trimethoprim resistant dihydrofolate reductase |
| Accessory genome | |||
| (AGly)Aac3-Ig:CP000282:2333620–2334096:477 | 33.56 | Aminoglycosides | Sde_1840,Gentamicin 3'-N-acetyltransferase |
| (AGly)Aac6:DQ302723:81–482:402 | 31.09 | Aminoglycosides | Aac(6'), aminoglycoside-6'-N-acetyltransferase |
| (Bla)ACC-3:AF180957:1–1131:1131 | 30.56 | Beta-lactamases | ACC-3, AMPC cephalosporinase precursor protein ACC-3 |
| (Bla)IND-7:AB529520:1–720:720 | 72.73 | Beta-lactamases | BlaIND-7, metallo-beta-lactamase IND-7 |
| (Bla)OCH-7:AJ295345:1–1173:1173 | 30.15 | Beta-lactamases | Bla OCH-7, beta-lactams hydrolysis |
| (Flq)OqxBgb:EU370913:47851–51003:3153 | 31.64 | Fluoroquinolones | pOLA52_67, OqxB integral membrane protein, component of RND-type multidrug efflux pump that confers resistance to olaquindox |
| (Gly)VanRc3:AY033764:3846–4541:696 | 30.34 | Glycopeptides | VanRc3 |
| (Gly)VanR-M:FJ349556:982–1680:699 | 32.35 | Glycopeptides | Van operon, response regulator |
| (MLS)CarA:M80346:411–2066:1656 | 34.88 | Macrolide-lincosamide-streptogramin | Carbomycin resistance protein (carA) |
| (MLS)LmrA:X59926:318–1763:1446 | 30.43 | Macrolide-lincosamide-streptogramin | LmrA gene for lincomycin resistance protein |
| (MLS)MefB:FJ196385:11084–12313:1230 | 36.18 | Macrolide-lincosamide-streptogramin | Macrolide efflux pump. |
| (MLS)MsrE:JF769133:7246–8721:1476 | 36.96 | Macrolide-lincosamide-streptogramin | Macrolide efflux protein. |
| (MLS)OleB:L36601:1421–3130:1710 | 37.04 | Macrolide-lincosamide-streptogramin | ATP-binding protein. oleandomycin resistance and secretion |
| (MLS)TlrC:M57437:277–1923:1647 | 30.77 | Macrolide-lincosamide-streptogramin | Tylosin resistance protein (tlrC) gene |
| (MLS)VatF:AF170730:70–735:666 | 31.21 | Macrolide-lincosamide-streptogramin | Streptogramin A acetyl transferase (sat) gene. Confers resistance to class A streptogramins |
| (MLS)vgaa-LC:DQ823382:1–1569:1569 | 31.11 | Macrolide-lincosamide-streptogramin | Lincosamide-streptogramin A resistance protein (vga(A)LC) gene |
| (Rif)Arr7:FN397623:1189–1641:453 | 41.73 | Rifampicin | ADP-ribosyltransferase. Resistance to rifampin. |
| (Tet)OtrB:AF079900:40–1733:1692 | 30.27 | Tetracyclines | Tetracycline efflux protein (otrB) gene |
| (Tet)TetX:M37699:586–1752:1167 | 59.36 | Tetracyclines | Transposon Tn4351 tetracycline resistance protein (tetX) gene |
In our study, 146 and 70 virulence genes were identified from the core- and accessory genomes of Elizabethkingia spp., respectively, which include genes involved in lipopolysaccharide biosynthesis, iron siderophore synthesis, heme uptake, transposase synthesis, alginate synthesis, and so on (supplementary table S3, Supplementary Material online).
Functional Classification of Genes Only Belongs to E. meningoseptica or E. anophelis
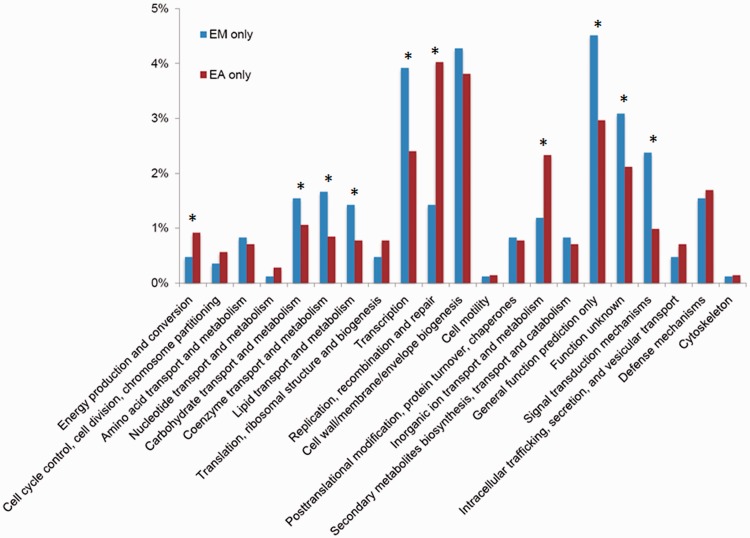
To gain knowledge about the difference in metabolic capacity between E. meningoseptica and E. anophelis, we enriched genes that exist only in E. meningoseptica but not in E. anophelis (referred to as EM only) and genes that exist only in E. anophelis but not in E. meningoseptica (EA only) based on BLAST analysis. There were 842 genes unique to the five E. meningoseptica genomes and 1,416 genes unique to the seven E. anophelis genomes. These unique genes were then classified according to their predicted functional role (fig. 5). When compared with the EA-only genes, the EM-only genes are enriched in predicted proteins belonging to Clusters of Orthologous Group (COG) category G (carbohydrate transport and metabolism), H (coenzyme transport and metabolism), I (lipid transport and metabolism), K (transcription), R (general function prediction only), S (function unknown), and T (signal transduction mechanisms) (fig. 5). Conversely, the EA-only genes are enriched in C (energy production and conversion), L (replication, recombination, and repair), and P (inorganic ion transport and metabolism) (fig. 5).
Fig. 5.—
The identified EM-only and EA-only proteins were assigned to Clusters of Orthologous Groups (COGs). The y axis indicates the percentage of genes in a specific function cluster out of the total numbers of EM-only and EA-only proteins, respectively. *The abundances of specific function clusters were compared statistically as described in Rodriguez-Brito et al. (2006) and Allen et al. (2009) using a subsample size of 500 and 1,000 bootstrap replicates at a statistical confidence of 99%.
In conclusion, our study revealed the emergence of a novel pathogen E. anophelis in the hospital environment. Elizabethkingia anophelis is well known to be a dominant species in the gut microbiota of the malaria mosquito vector A. gambiae (Dong et al. 2009; Boissiere et al. 2012; Osei-Poku et al. 2012). Elizabethkingia species in the midgut of the malaria vector may modulate the anti-Plasmodium effects of the host's immune genes, thus prolonging the life span of the Plasmodium-infected malaria mosquito vector (Dong et al. 2009). The genome content and organization of E. anophelis is similar to, yet distinct from the well-known E. meningoseptica, which often causes high mortality among hospital acquired infections. Our study suggests that the mosquito vector might be a potential mobile reservoir of antibiotic and virulence genes for emerging bacterial pathogens. However, we should notice that the core- and pan-genome analysis is based on only a small group of genomes. More genomes of the Elizabethkingia spp. are required for sophisticated comparative genomic analysis. Further studies will be carried out to comparatively investigate the pathogenesis mechanisms employed by E. anophelis and E. meningoseptica in causing human infections.
Materials and Methods
Ethics Statement
Ethical approval was not required for the study because it was done as part of surveillance and management of healthcare-associated infection.
Genome Sequencing, Assembly, and Comparative Genomic Analyses
The genomes of seven Elizabethkingia spp. strains: Three patient isolates (NUHP1, NUHP2, and NUHP3) and four environmental isolates (NUH1, NUH4, NUH6, and NUH11) were sequenced in this study. Whole-genome DNA of these E. anophelis strains were purified using QIAamp DNA Mini Kit (QIAgen) and sequenced on an Illumina MiSeq platform generating 150-bp-long paired-end reads. Reads were assembled into contigs using de novo assembly in CLCBio's Genomics Workbench NGS suite (CLCBio, version 6.0.3) with default settings. Average genomic coverages across the strains ranged from 111 to 160-fold. The assembled genomes were compared with the five available Elizabethkingia spp. genomes deposited in the National Center for Biotechnology Information (NCBI) Database (supplementary table S2, Supplementary Material online).
Multiple genome alignment was used to compare the genomes of the Elizabethkingia spp. by using Progressive Mauve with match seed weight 15, min Locally Collinear Block weight 45, minimum island size 50, maximum backbone gap size 50, and minimum backbone size 50 (Darling et al. 2010). Phylogenetic tree diagrams were prepared using the software FigTree ver 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/, last accessed May 9, 2014). SNP differences between each pair of Elizabethkingia spp. was calculated using the snpTree web server for assembled genomes with minimum coverage 10 and minimum distance between SNPs 10 (Leekitcharoenphon et al. 2012). Contigs of the NUHP1 genome were aligned with the three other Elizabethkingia species to identify homologous sequences (above 100 bp in length and at least 80% homology), using BLASTn as implemented in NCBI BLAST+ v. 2.2.8 (Camacho et al. 2009). A visual representation linking homologous regions between NUHP1 and the three strains was constructed with CIRCOS v. 0.64 (Krzywinski et al. 2009).
Core/Pan-Genomic Analysis
Core- and pan-genomes were calculated, and curves for pan-genome and core-genome were generated by the CMG-biotools package with BLAST cutoff of 50% identity and 50% coverage of the longest gene (Vesth et al. 2013). The identified core- and accessory genomes were BLAST searched against the Comprehensive Antibiotic Resistance Database (McArthur et al. 2013) and VFDB (Chen et al. 2012) to identify antibiotic resistant genes and virulence genes by using Bio-Edit (Ibis Biosciences, Carlsbad, CA, http://www.mbio.ncsu.edu/bioedit/bioedit.html, last accessed May 9, 2014) (minimum 30% identity with E value < 1e-5).
EM-only proteins and EA-only proteins were enriched by the CMG-biotools package (Vesth et al. 2013). The identified EM-only proteins and EA-only proteins were assigned to COGs, and their abundances compared statistically as described in Rodriguez-Brito et al. (2006) and Allen et al. (2009) using a subsample size of 500 and 1,000 bootstrap replicates at a statistical confidence of 99%.
Nucleotide Sequence Accession Numbers
Each of the seven outbreak E. anophelis genomes were deposited at DDBJ/EMBL/GenBank as individual WGS bioprojects. The respective accession numbers of the seven E. anophelis genomes are NUHP1 (ASYE00000000), NUHP2 (ASYF00000000), NUHP3 (ASYG00000000), NUH1 (ASYH00000000), NUH4 (ASYI00000000), NUH6 (ASYJ00000000), NUH11 (ASYK00000000), respectively.
Supplementary Material
Supplementary tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Dr. Daniela Drautz, Mr. Yap Zhei Hwee and Rikky Wenang Purbojati for helping with genome sequencing experiment. This research is supported by the National Research Foundation and Ministry of Education Singapore under its Research Centre of Excellence Programme and the Start-up Grants (M4330002.C70) from Nanyang Technological University, Singapore. This work was supported by grants from the Danish Council for Strategic Research (M.G.)
Literature Cited
- Allen MA, et al. The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: the role of genome evolution in cold adaptation. ISME J. 2009;3:1012–1035. doi: 10.1038/ismej.2009.45. [DOI] [PubMed] [Google Scholar]
- Aziz R, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balm MND, et al. Bad design, bad practices, bad bugs—frustrations in controlling an outbreak of Elizabethkingia meningoseptica in intensive care units. J Hosp Infect. 2013;85:134–140. doi: 10.1016/j.jhin.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Bloch KC, Nadarajah R, Jacobs R. Chryseobacterium meningosepticum: an emerging pathogen among immunocompromised adults. Report of 6 cases and literature review. Medicine. 1997;76:30–41. doi: 10.1097/00005792-199701000-00003. [DOI] [PubMed] [Google Scholar]
- Boissiere A, et al. Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 2012;8:e1002742. doi: 10.1371/journal.ppat.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Xiong Z, Sun L, Yang J, Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2012;40:D641–D645. doi: 10.1093/nar/gkr989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley JR, Nims LJ, Lipp VH, Beard A, Delaney LT. Meningitis of infants caused by Flavobacterium meningosepticum: report of a patient and analysis of 63 infections. J Trop Pediatr. 1980;26:24–30. doi: 10.1093/tropej/26.1.24. [DOI] [PubMed] [Google Scholar]
- Green O, Murray P, Gea-Banacloche JC. Sepsis caused by Elizabethkingia miricola successfully treated with tigecycline and levofloxacin. Diagn Microbiol Infect Dis. 2008;62:430–432. doi: 10.1016/j.diagmicrobio.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MS, et al. Clinical features, antimicrobial susceptibilities, and outcomes of Elizabethkingia meningoseptica (Chryseobacterium meningosepticum) bacteremia at a medical center in Taiwan, 1999-2006. Eur J Clin Microbiol Infect Dis. 2011;30:1271–1278. doi: 10.1007/s10096-011-1223-0. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leekitcharoenphon P, et al. snpTree—a web-server to identify and construct SNP trees from whole genome sequence data. BMC Genomics. 2012;13(Suppl 7):S6. doi: 10.1186/1471-2164-13-S7-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur AG, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osei-Poku J, Mbogo CM, Palmer WJ, Jiggins FM. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol Ecol. 2012;21:5138–5150. doi: 10.1111/j.1365-294X.2012.05759.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Rohwer F, Edwards RA. An application of statistics to comparative metagenomics. BMC Bioinformatics. 2006;7:162. doi: 10.1186/1471-2105-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo J, et al. The first case of intensive care unit outbreak by the novel agent Elizabethkingia anophelis. Lancet. 2013;382:855–856. doi: 10.1016/S0140-6736(13)61858-9. [DOI] [PubMed] [Google Scholar]
- Teres D. ICU-acquired pneumonia due to Flavobacterium meningosepticum. J Am Med Assoc. 1974;228:732. [PubMed] [Google Scholar]
- Vesth T, Lagesen K, Acar O, Ussery D. CMG-biotools, a free workbench for basic comparative microbial genomics. PLoS One. 2013;8:e60120. doi: 10.1371/journal.pone.0060120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.