Abstract
To maintain linear DNA genomes, organisms have evolved numerous means of solving problems associated with DNA ends (telomeres), including telomere-associated retrotransposons, palindromes, hairpins, covalently bound proteins and the addition of arrays of simple DNA repeats. Telomeric arrays can be maintained through various mechanisms such as telomerase activity or recombination. The recombination-dependent maintenance pathways may include telomeric loops (t-loops) and telomeric circles (t-circles). The potential involvement of t-circles in telomere maintenance was first proposed for linear mitochondrial genomes. The occurrence of t-circles in a wide range of organisms, spanning yeasts, plants and animals, suggests the involvement of t-circles in many phenomena including the alternative-lengthening of telomeres (ALT) pathway and telomere rapid deletion (TRD). In this Perspective, we summarize these findings and discuss how t-circles may be related to t-loops and how t-circles may have initiated the evolution of telomeres.
The linearity of DNA generates numerous difficulties, including the need to replicate the very ends of the DNA and the need to protect the ends against nucleolytic attacks and inappropriate DNA repair1–3. Although the term ‘telomere’ is primarily used for a specific nucleoprotein complex at the ends of eukaryotic nuclear chromosomes, any linear DNA genome, regardless of the phylogenetic origin of its host (eukaryotic, prokaryotic or viral) or its localization within the cell (for example, nuclear or mitochondrial), possesses at least two telomeres that must cope with the problems associated with chromosomal ends.
There are many types of telomeric structures that represent diverse solutions to these obstacles (Table 1). Elongation of telomeric repeats by the reverse transcriptase telomerase is the most common solution in eukaryotic nuclei2, where telomeres consist of t-arrays (Table 1)—double-stranded DNA tracts of short repeat motifs (for example, 5′-TTAGGG-3′ in mammals) that terminate in a G-rich, single-stranded 3′ overhang. There are other means of maintaining telomeres in eukaryotic (both nuclear and organellar), prokaryotic and viral genomes4. For example, the nuclear chromosomes of Drosophila melanogaster lack short DNA arrays and instead use retrotransposons (t-posons, Table 1) to maintain the chromosomal ends5, whereas in Alliaceae (a group of monocots in the plant order Asparagales that includes the onions) ribosomal DNA repeats cap the ends6. Additional solutions for protecting linear DNA ends are illustrated by some bacteriophages, animal viruses and plasmids, with various types of terminal arrays of tandem repeats, terminal palindromes or hairpins, and terminal proteins covalently attached to the 5′ ends4. Importantly, even though the architecture of telomeres in these cases may differ from that found in a typical eukaryotic nucleus, their experimental analyses have not only revealed the molecular mechanisms involved in the telomerase-independent maintenance of nuclear telomeres, but they have also provided clues about the origin and evolution of telomeres in general4,7,8 (see also below).
Table 1.
Glossary of t-elements
| t-array |
|
Telomeric array. A tract of tandemly repeated sequences at the end of a linear chromosome, usually ending with a single-stranded protrusion (t-overhang). The number of repeats in an array, as well as their length, can vary; they are a prerequisite for the formation of higher-order structures such as t-loops. |
| t-circle |
|
Telomeric circle. An extrachromosomal duplex or single-stranded circular DNA molecule composed of t-arrays. |
| t-hairpin |
|
Telomeric hairpin. A covalently closed single-stranded hairpin present at the ends of linear DNA (for example, the poxviral genome, chromosomes in Borrelia and S. cerevisiae mutants, mitochondrial DNA in Williopsis and Pichia). It represents a resolved (monomeric) form of a t-palindrome. |
| t-palindrome |
|
Telomeric palindrome. A DNA element (within a chromosome or a genome) that consists of inverted repeats, which can be processed into a t-hairpin. Resolution of t-palindromes into t-hairpins is associated with a change in the chromosome or genome topology (that is, a circular-to-linear change or multiple linear chromosomes). |
| t-loop |
|
Telomeric loop. A higher-order structure that is generated by invasion of a t-overhang into the duplex region of a t-array. The occurence of t-loops in a wide variety of organisms indicates that they represent a universal capping structure. However, owing to technical reasons it is difficult to directly quantify the fraction of telomeres present as t-loops in vivo. |
| t-overhang |
|
Telomeric overhang. A single-stranded stretch of DNA extending from the duplex region of the telomere. Depending on the system, the extension may be either of the 3′ (typical nuclear telomeres) or the 5′ strand (for example, mitochondrial telomeres). |
| t-poson |
|
Telomeric retrotransposon. A transposable or mobile genetic element (for example, HeT-A, TART and Tahre in D. melanogaster) that is reverse-transcribed directly onto the end of the chromosome, extending the end by successive retrotransposition events. |
| t-protein |
|
Telomeric protein. A protein that is covalently attached to the 5′ end of linear DNA chromosomes (for example, adenoviral genomes, chromosomes in Streptomyces), possessing a serine or threonine residue whose hydroxyl group serves as a primer for DNA synthesis. |
The architecture of various types of telomeres reflects their ability to acquire different states. In general terms, telomeres may adopt various types of open or closed (capped) conformations, which differ in their response to the protein machineries involved in their maintenance and dynamics (for example, telomerase or other proteins involved in telomere replication, repair, recombination or attachment to a defined site in a cellular compartment9). The formation of a particular state is governed by a number of proteins that are directly or indirectly associated with the telomere, and access to the chromosome end is regulated by transcriptional control, protein-protein interactions or post-translational modifications10,11.
The different states of a telomere vary not only in their accessibility to telomerase, but also in their susceptibility to recombination. In the absence of telomerase, telomeres may be maintained by recombinational mechanisms. The ALT pathway is found in telomerase-negative tumors12,13 and in clones of budding yeast Saccharomyces cerevisiae surviving a senescence crisis caused by deletion of genes encoding components of telomerase14–16. In numerous instances, this seems to involve the generation of duplex or single-stranded DNA circles formed from telomeric repeat sequences (t-circles; Table 1). As t-circles have been observed in phylogenetically distant species, including vertebrates, yeasts, plants, and even in yeast mitochondria containing a linear DNA genome, they seem to represent an evolutionarily conserved feature and may have an important role in the biology of telomeres4,7,8. Here we review the discovery of t-circles and their relationship to t-loops and discuss possible pathways that could generate abundant t-circles.
T-circles: historical background
The path toward the discovery of t-circles and their potential role in telomere maintenance began in the mid-1980s with work on the replication of the linear mitochondrial genome of yeast Candida parapsilosis17. Analogous to a canonical nuclear telomere, the mitochondrial DNA (mtDNA) of this opportunistic pathogen terminates with an array (up to 12) of 738-bp tandem repeat units and a 5′ single-stranded overhang of about 110 nucleotides18. The overhang is protected by a mitochondrial telomere-binding protein (mtTBP)19, which was later shown to have a dual function in C. parapsilosis mitochondria: specific telomere binding and general single-stranded DNA binding20–22. However, neither a detailed description of the molecular architecture of C. parapsilosis telomeres, nor the identification of mtTBP, revealed any molecular mechanism(s) involved in the maintenance of this type of mitochondrial telomere. It has not been possible to identify a mitochondrial equivalent of telomerase by sequence homology searches, nor to detect its activity.
Investigation of in vivo nucleoprotein structure and replication intermediates of the C. parapsilosis mtDNA resulted in identification of extragenomic DNA fragments derived exclusively from the terminal regions of the mitochondrial genome23. The migration of the fragments on two-dimensional agarose gels indicated that they might be represented by circular DNA molecules. To explore this possibility, we carried out DNA analysis by EM, taking advantage of the fact that the fragments could be highly enriched by alkaline lysis of the purified mitochondria. The resulting DNAs appeared as tightly supertwisted DNA molecules (rods). Mild treatment with DNase I produced fields of open DNA circles whose sizes corresponded to integral multiples of 738 bp (Fig. 1a). Similar results were obtained with other yeast species (Candida salmanticensis, Candida metapsilosis and Pichia philodendri) that also harbor linear mtDNA terminating in an array of tandemly repeated sequences23,24. Subsequent studies demonstrated that mitochondrial t-circles are not just by-products of recombination between tandemly repeated sequences, but they also provide substrates for rolling circle– dependent amplification, which in turn could provide substrates for recombinational telomere elongation (RTE)24 (Fig. 1a). Importantly, a search for yeast mutants lacking mitochondrial t-circles revealed that they contain circularized derivatives of the original linear mitochondrial genome formed by end-to-end fusions, suggesting that the t-circle–dependent mechanism may represent the main, or perhaps the only, telomere maintenance pathway operating in the mitochondria of these yeasts25,26.
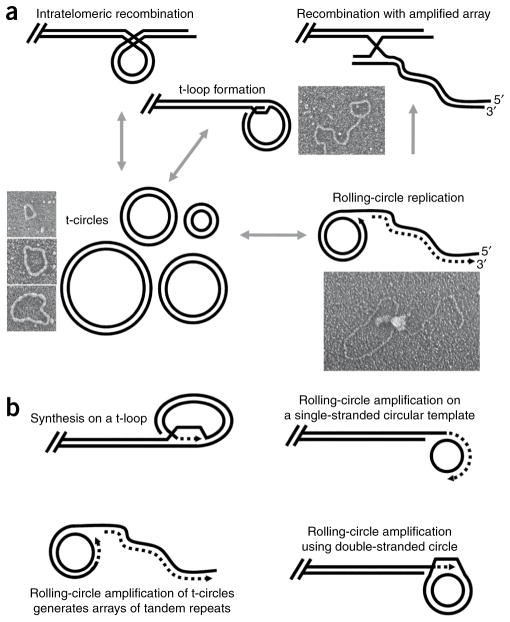
Figure 1.
t-circles and t-loops seem to be common players in telomere maintenance. (a) Proposed pathway in the maintenance of mitochondrial telomeres in the yeast C. parapsilosis. The mitochondrial telomeres consist of t-arrays (n × 738 bp) with a 5′ single-stranded overhang of about 110 nt. Invasion of the overhang into the double-stranded region of t-arrays can form t-loop structures. The t-circles can be formed either by intramolecular recombination between repeats within t-arrays or by excision from t-loops. The t-circles amplify autonomously via a rolling-circle mechanism (forming σ-form ‘tailed circles’), thus generating long extrachromosomal t-arrays, which can recombine with the main genome and extend the termini. EM revealed the key players in the proposed pathway of the mitochondrial telomere maintenance18,23,24,67,68. (b) Proposed role for the rolling-circle synthesis in nuclear telomere maintenance. Rolling-circle synthesis can extend the termini in situ (on a t-loop or single-stranded or double-stranded t-circles), or extrachromosomal t-circles may replicate autonomously.
Mitochondrial versus nuclear telomeres: common themes
Although a few reports demonstrated the presence of various types of extragenomic telomeric fragments, especially in ALT cells27–30, the field truly blossomed after the identification of t-circles in the mitochondria of budding yeast with linear mitochondrial genomes. t-circles have been subsequently observed in the yeast Kluyveromyces lactis31,32, the nematode Caenorhabditis elegans33, the plant Arabidopsis thaliana34, Xenopus laevis embryonic cells35 and mammalian ALT cells36–38. In mammals, various forms of t-circles were shown to constitute a substantial part of the extrachromosomal telomeric DNA37. t-circles arising from cleavage of t-loops could serve as a template for generation of rolling-circle replication intermediates, including the σ-form ‘tailed circles’ (Fig. 1a) and single-stranded DNA circles with double-stranded portions. As seen in linear mtDNA of yeast23,24, t-circle–mediated rolling-circle replication could provide an efficient means of telomere elongation, including synthesis on a t-loop, as well as using single- and/or double-stranded circular DNA as templates (Fig. 1b).
Indeed, in ALT cells, the t-circle–mediated telomere maintenance pathway represents an alternative form of homologous recombination– driven telomere elongation. Recent reports have identified t-circles in human somatic cells deficient in Ku86 (ref. 39) and in telomerase- positive cancer cells40. The latter report also demonstrated an association between shortening of overlengthened telomeres and t-circle generation, indicating that t-circle formation is part of the mechanism of preventing overlengthening of telomeres (by telomerase and/or ALT). Interestingly, t-circle formation is developmentally regulated in X. laevis. During early development (before mid- blastula transition; pre-MBT stage), the embryo contains a substantial amount (up to 10% of the total cellular telomere content) of t-circles, but they are dramatically reduced in germline and adult tissues35. This may parallel the recombination-dependent lengthening of telomeres (via extensive telomere sister-chromatid exchange; T-SCE) in the early mammalian embryo, which is replaced by a telomerase maintenance mode in later embryonic stages41. This suggests that the ALT pathways (including the t-circle–dependent ones) do not solely represent evolutionarily retained back-ups for the loss of telomerase but may have been selected for their specific role(s) in the development of multicellular organisms.
It is possible that the ubiquitous occurrence of t-circles in phylogenetically distant groups reflects the fact that they are a common by-product of telomere maintenance processes and either just prevent telomere overlengthening or have no direct role in telomere length maintenance at all. However, studies on the natural t-circle–dependent system in yeast mitochondria and using synthetic t-circles in yeast nuclei23,32,42 point toward their general importance in telomere maintenance and a potential role as an ancient mechanism for replication and stabilization of the ends of linear DNA molecules4.
The strongest evidence that t-circles have direct roles in telomere maintenance comes from studies on K. lactis. RTE was invoked by the introduction of synthetic t-circles into K. lactis ter1-16T mutant cells with altered telomerase RNA, whose telomeres are uncapped owing to a base change within the telomeric repeat that changes the binding site for the telomere-associated protein Rap1 (refs. 43,44). Moreover, it was shown that telomere uncapping caused by the ter1-16T mutation increases homologous recombination leading to t-loop and t-circle formation31. Resulting t-circles have been implicated in a roll-and-spread telomere-lengthening pathway. Here, t-circle amplification generates a substrate for homologous recombination–dependent and telomerase-independent Rad52-dependent telomere maintenance mechanisms, which recombine the amplified telomeric sequence into the telomeric regions, thus leading to a rapid homogenization of chromosomal ends36,44.
Several lines of evidence indicate that t-circles have important roles in telomere dynamics and contribute to the stochastic changes observed in telomere lengths45. Intrachromosomal recombination within telomeric arrays or excision from a t-loop may result in t-circle formation and, at the same time, lead to shortening of the chromosome ends, known as telomere rapid deletion (TRD)46. Conversely, RTE mediated by amplification of t-circles may counterbalance the loss of telomeric sequences caused by TRD. Importantly, both RTE and telomerase-dependent replication may coexist within the same cells47, and telomerase activity does not necessarily interfere with t-loop and t-circle formation36.
The situation in mammalian cells is much more complex than in simpler models. This is due to the involvement of a wide repertoire of proteins, including the major telomere-capping protein TRF2, which inhibits the formation of t-circles through recruitment of the RecA- and Rad51-related protein XRCC3 (ref. 38), the origin recognition complex subunit ORC2 (ref. 48) or the Werner’s syndrome-associated WRN helicase49 to telomeres. This implies that the formation of t-circles is repressed under normal circumstances. This repression can be alleviated either by treatment with chemicals (for example, the alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) or the ribonucleotide reductase inhibitor hydroxyurea29,48), or genetically (for example, TRF2ΔB mutant cells lacking the N-terminal basic domain of TRF2 (involved in regulation of recombination at telomeres)38,48 or through deletion of Ku86 (refs. 38,39,48)). The involvement of Ku70–Ku80 in regulation of t-circle formation has also been demonstrated in plants. Experiments with A. thaliana telomerase-negative cells have revealed that, whereas Ku70–Ku80 inhibits ALT, inactivation of this complex promotes t-circle generation34.
The formation of t-circles is controlled by additional proteins. For example, depletion of ORC2 increases the levels of t-circles, indicating that TRF2 may inhibit generation of t-circles via recruitment of ORC subunits and other proteins to telomeres48. In addition, studies in Werner’s syndrome cells showed that the TRF2ΔB-induced homolous recombination t-loop pathway requires WRN helicase, and both the helicase and endonuclease activities of WRN are essential for suppressing spontaneous formation of t-circles. In contrast to other cell lines38, t-circle formation in Werner’s syndrome cells does not seem to involve the recombination factor XRCC3, indicating that an alternative mechanism to homolous recombination takes place in these cells. As in the previous example, TRF2 seems to recruit the helicase, which represses the excision of t-loops into t-circles49.
Another important player in t-circle dynamics in mammalian cells is the MRN complex (MRE11–RAD50–NBS1). The NBS1 protein (but not the other two components of the complex) is required for both the ALT phenotype and the formation of t-circles50,51. However, although cells depleted for NBS1 show greatly diminished t-circle levels, they can still proliferate50. It is possible that the residual amount of ALT activity remaining after the incomplete knockdown of NBS1 activity becomes more efficient as the telomeres get shorter; however, these results suggest that the cells use t-circle–independent telomere maintenance. This is in line with the idea that more than one ALT mechanism may operate in mammalian cells52–54. Alternatively, only a few t-circles may be required to maintain telomeres once the mechanism is established.
Two recent studies have shown that t-circles can exist in the presence of telomerase and do not necessarily reflect a cellular response to the loss of telomerase. Pickett et al.40 showed that increased expression of telomerase in an already telomerase-positive human cancer cell line led to telomere elongation, followed by telomere trimming by a mechanism involving the generation of t-circles. It is also interesting to note that C. elegans chromosomes contain both 3′ and 5′ single-stranded telomeric overhangs and also possess t-circles in the presence of telomerase33. We speculate that the simultaneous occurrence of these two mechanisms of telomere amplification is justified by the unusual presence of both 3′ and 5′ single-stranded overhangs, which are, respectively, the specific substrates for telomerase (3′ single-stranded overhangs serve both as a substrate for annealing of telomerase RNA and as a primer for DNA synthesis) and products of rolling-circle synthesis-dependent telomere elongation (recombination of telomeres with the linear DNA product of rolling-circle replication containing a single-stranded DNA at the 5′ end would generate 5′ single-stranded telomeric overhangs (Fig. 1a)).
The discovery of t-circles provides further evidence that normal chromosomes are capped by t-loops. The close correlation between the distribution of sizes of the circular portion of the t-loops observed in human and K. lactis cells, and the sizes of the t-circles isolated from the same cells, argue that t-circles arise from cleavage of normal t-loops. It is not fully known which factors facilitate or protect the t-loop from cleavage, but they should include proteins that bind DNA junctions. Candidates include the WRN helicase, which is known to bind four-way junctions55 and whose loss in human cells results in the appearance of t-circles49,56. Full-length TRF2 binds tightly to three-way and four-way junctions, but TRF2ΔB does not57, and overexpression of TRF2ΔB in human cells results in the appearance of t-circles38. This result argues that, when TRF2 is not specifically targeted to the t-loop junction (TRF2ΔB protein still binds duplex telomeric DNA), then the junction becomes sensitive to cleavage. Recently it was observed that the induced loss of Ku86 in a human somatic cell line with a conditionally null allele of Ku86 resulted in the dramatic loss of telomeres in these cells and the appearance of abundant t-circles39. These results support the presence of a multiprotein complex bound at the t-loop junction, which forms, stabilizes and protects the t-loop until cell cycle–specific signals (for example, during DNA replication) dictate opening of the loop. The most likely candidate is TRF2–shelterin, a protein complex that has DNA remodeling activity and that, together with telomere-associated DNA repair factors, mediates structural changes of the telomeric DNA58.
t-circles in an evolutionary perspective
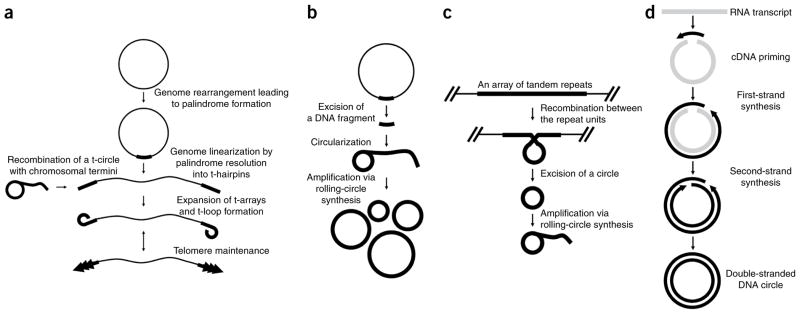
The wide occurrence of t-circles strongly suggests that they represent a primordial means of telomere maintenance. According to one view, t-circles may have a role in the origin of linear DNA genophores (chromosomal DNA equivalents) (Fig. 2a). As we proposed recently4, telomeres might have evolved from a selfish element (functionally related to the t-circles) that integrated into a primitive eukaryotic genome (presumably circular), forced its conversion toward a linear form and produced amplified tandem arrays at its termini. Expanded telomeric arrays subsequently allowed formation of the t-loop structures, as proposed by de Lange59. In other words, t-circles would have generated problems associated with DNA linearity and at the same time made themselves essential by providing their solutions4. If so, this raises questions about their evolutionary origin and how have they participated in the generation of linear chromosomes. We have recently addressed these questions and postulated several scenarios with general implications for the origin of telomeres7.
Figure 2.
Evolutionary scenario for the emergence of linear chromosomes terminating with t-arrays and hypotheses on the emergence of t-circles. (a) Genome rearrangements within a circular genome may result in formation of a palindrome. Alternatively, a selfish genetic element may ‘infect’ a circular genome. Subsequent resolution of the palindrome eventually generates a linear molecule with t-hairpins at the ends. The termini represent a substrate for recombination with t-circles and/or elongation by telomerase. Expanded t-arrays allow the formation of higher-order structures such as t-loops and the emergence of telomere maintenance mechanism(s)4. We have proposed three mechanisms that may explain the emergence of t-circles7. (b) Similarities between mitochondrial t-circles and hypersuppresive (ρ −) petite genomes indicate that they could have emerged by a common mechanism. Hypersuppressive genomes represent short fragments of a mitochondrial genome, which replicates via rolling-circle synthesis. In baker’s yeast, hypersuppressive genomes can outcompete the main genome. However, in strictly aerobic petite-negative organisms, the two replicons (mtDNA and hypersuppressive-like t-circles) can coexist, and eventually the t-circles can recombine with mtDNA, which may result in the formation of a linear DNA genome with t-arrays at the ends. Rolling-circle replicating t-circles may escape from the mitochondria to the nucleus and analogously affect the nuclear genome. (c) Alternatively, t-circles may emerge from intra-strand recombination of units within a preexisting intrachromosomal array of tandem repeats. (d) T-circles may also emerge by reverse transcription of an RNA molecule (or an RNA fragment, shown in gray), which serves as a template for the first-strand cDNA synthesis, followed by template degradation, synthesis of the second cDNA strand and ligation of nicks. The double-stranded DNA circle is either discarded (most cases) because of its inability to replicate, or if it is able to replicate it can be preserved and may invade into the main genome or chromosome, as shown in a.
The first scenario stems from comparison with respiratory-deficient mutants of S. cerevisiae, dubbed cytoplasmic petites, which have alterations in their mitochondrial genome. In some petite strains, mtDNAs are deleted for most of the standard (ρ+) genome, with the remaining sequences amplified and organized as oligomeric repeats in a form of a ρ− genome. In case of some ρ+ × ρ − crosses, >95% of the progeny contain exclusively the ρ− genome. The types of ρ − mtDNA with this property are called hypersuppressive ρ − (ref. 60). Interestingly, similarly to t-circles in mitochondria of species with linear mitochondrial genome, hypersuppressive ρ − mtDNA also uses DNA circles as replication intermediates61. This indicates that t-circles and hypersuppressive ρ − derivatives of mitochondrial DNA might have a common origin (Fig. 2b). Hypersuppressive cells harbor amplified subgenomic mtDNA fragments that can out-compete the wild-type mitochondrial genome for replication and/or segregation machineries without any apparent advantage for its host. Hence, hypersuppressive genomes can be classified as selfish genetic elements. One can hypothesize that mitochondrial t-circles in C. parapsilosis originated from an hypersuppressive derivative that integrated into an ancestral (presumably circular) mitochondrial genome and subsequently became essential for maintenance of its linearized form. In contrast to baker’s yeast, C. parapsilosis is a strictly aerobic organism that does not tolerate the loss of mtDNA. Therefore, hypersuppressive-like mtDNA derivatives would be unable to eliminate the wild-type genome from this yeast and might have been forced to coexist with the main mitochondrial genome.
Second, extrachromosomal circular DNA molecules result from intramolecular recombination between two or more head-to-tail repeats present within the genome (Fig. 2c). The ability of a small subset of molecules to replicate via rolling-circle synthesis would lead to their stable maintenance and further amplification. Eventually, the resulting linear tandem arrays would reintegrate into the circular genome, leading to the formation of a linearized chromosome.
Finally, several reverse transcriptases have the ability to initiate end-to-end intramolecular reverse transcription that might result in extra-chromosomal circular DNA molecules62. There are numerous reverse transcriptases (including the mitochondrial Mauriceville retroplasmid of Neurospora crassa63,64) that effectively use endogenous RNA molecules as substrates. The RNA molecules do not necessarily have to be full-length, but can also be degradation products. Furthermore, some reverse transcriptases are capable of non-templated extension of existing RNA molecules and thus generate templates of high sequence variability. These templates then may lead to the generation of circular DNA molecules. Although most of these pieces of circular DNA will be lost owing to their inability to replicate, a DNA circle with appropriate structural features that enable it to amplify might appear. For example, short palindromes can form a cruciform structure that is nicked to generate a free 3′ OH end for leading-strand DNA synthesis in the course of a rolling-circle replication. Such a circle could invade the main chromosome (for example, mtDNA) via homologous recombination (as it is derived from an RNA molecule transcribed from endogenous DNA) and thus lead to linearization of the chromosome (Fig. 2d). This scenario explains (i) how the t-circles appear, (ii) why their sequences are very different and (iii) how they can integrate into the main genome.
The reverse transcriptase–assisted emergence of t-circles may not be limited to mitochondrial telomeres, but may be at the heart of the origin of the first nuclear telomere. An intriguing possibility is that t-circles generated in the mitochondrial compartment escaped from this organelle and entered the nucleus, where they linearized the genome and established a t-circle–dependent mechanism of telomere maintenance. Alternatively, nuclear t-circles may have evolved independently, so that telomerase is the remnant of the event: as a reverse transcriptase, it produced the first t-circle by end-to-end reverse transcriptase activity and then adopted the role of maintaining nuclear telomeres through extension of the 3′ overhang. It would be interesting to see whether the G-rich or C-rich telomeric transcripts found in mammalian nuclei65,66 are substrates for reverse transcriptases, thus leading to generation of telomeric tracts of DNAs that can circularize. Another question is why the G-rich telomeric repeat motif (such as TTAGGG in vertebrates) has been selected in most eukaryotic species. Perhaps one of the (presumably several) factors favoring telomeric sequences containing this repeat motif was their inherent ability to undergo transactions involved in generation and propagation of t-circles, thus providing one of the robust means for telomere maintenance. In any case, the origin of nuclear telomeres seems to have required both reverse transcriptase and t-circles, whose emergence might have been dependent on reverse transcriptase activity. The question of which was first, telomerase or t-circles, would then become meaningless7.
Acknowledgments
We wish to thank L. Kovac (Comenius University) for inspiration and continuous support and members of our laboratories for discussions. We also thank two anonymous reviewers for valuable comments and suggestions. Our work related to telomere biology is supported by grants from a Fogarty International Research Collaboration Award (2-R03-TW005654-04A1), the Howard Hughes Medical Institute (55005622), the Slovak grant agencies APVT (20-001604 and 0024-07) and VEGA (1/0132/09 and 1/0219/08), and grants to J.D.G. (US National Institutes of Health grants GM31819 and ES13773 and awards from the Ellison and Glenn foundations).
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Gilson E, Géli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 2.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 3.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 4.Nosek J, Kosa P, Tomaska L. On the origin of telomeres: a glimpse at the pre-telomerase world. Bioessays. 2006;28:182–190. doi: 10.1002/bies.20355. [DOI] [PubMed] [Google Scholar]
- 5.Pardue ML, DeBaryshe PG. Drosophila telomeres: a variation on the telomerase theme. In: Nosek J, Tomaska L, editors. Origin and Evolution of Telomeres. Landes Bioscience; Austin, Texas: 2008. pp. 27–44. [DOI] [PubMed] [Google Scholar]
- 6.Pich U, Fuchs J, Schubert I. How do Alliaceae stabilize their chromosome ends in the absence of TTTAGGG sequences? Chromosome Res. 1996;4:207–213. doi: 10.1007/BF02254961. [DOI] [PubMed] [Google Scholar]
- 7.Nosek J, Tomaska L. Mitochondrial telomeres: an evolutionary paradigm for the emergence of telomeric structures and their replication strategies. In: Nosek J, Tomaska L, editors. Origin and Evolution of Telomeres. Landes Bioscience; Austin, Texas: 2008. pp. 163–171. [Google Scholar]
- 8.Tomaska L, McEachern MJ, Nosek J. Alternatives to telomerase: keeping linear chromosomes via telomeric circles. FEBS Lett. 2004;567:142–146. doi: 10.1016/j.febslet.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 9.Blackburn EH. Telomere states and cell fates. Nature. 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- 10.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 11.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 12.Cesare AJ, Reddel RR. Telomere uncapping and alternative lengthening of telomeres. Mech Ageing Dev. 2008;129:99–108. doi: 10.1016/j.mad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Cesare AJ, Reddel RR. Alternative lengthening of telomeres in mammalian cells. In: Nosek J, Tomaska L, editors. Origin and Evolution of Telomeres. Landes Bioscience; Austin, Texas: 2008. pp. 45–57. [Google Scholar]
- 14.Lundblad V. Telomere maintenance without telomerase. Oncogene. 2002;21:522–531. doi: 10.1038/sj.onc.1205079. [DOI] [PubMed] [Google Scholar]
- 15.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 16.Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovac L, Lazowska J, Slonimski PP. A yeast with linear molecules of mitochondrial DNA. Mol Gen Genet. 1984;197:420–424. doi: 10.1007/BF00329938. [DOI] [PubMed] [Google Scholar]
- 18.Nosek J, Dinouel N, Kovac L, Fukuhara H. Linear mitochondrial DNAs from yeasts: telomeres with large tandem repetitions. Mol Gen Genet. 1995;247:61–72. doi: 10.1007/BF00425822. [DOI] [PubMed] [Google Scholar]
- 19.Tomaska L, Nosek J, Fukuhara H. Identification of a putative mitochondrial telomere-binding protein of the yeast Candida parapsilosis. J Biol Chem. 1997;272:3049–3056. doi: 10.1074/jbc.272.5.3049. [DOI] [PubMed] [Google Scholar]
- 20.Nosek J, Tomaska L, Pagacova B, Fukuhara H. Mitochondrial telomere-binding protein from Candida parapsilosis suggests an evolutionary adaptation of a nonspecific single-stranded DNA-binding protein. J Biol Chem. 1999;274:8850–8857. doi: 10.1074/jbc.274.13.8850. [DOI] [PubMed] [Google Scholar]
- 21.Tomaska L, Makhov AM, Nosek J, Kucejova B, Griffith JD. Electron microscopic analysis supports a dual role for the mitochondrial telomere-binding protein of Candida parapsilosis. J Mol Biol. 2001;305:61–69. doi: 10.1006/jmbi.2000.4254. [DOI] [PubMed] [Google Scholar]
- 22.Tomaska L, Nosek J, Kucejova B. Mitochondrial single-stranded DNA-binding proteins: in search for new functions. Biol Chem. 2001;382:179–186. doi: 10.1515/BC.2001.025. [DOI] [PubMed] [Google Scholar]
- 23.Tomaska L, Nosek J, Makhov AM, Pastorakova A, Griffith JD. Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res. 2000;28:4479–4487. doi: 10.1093/nar/28.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nosek J, Rycovska A, Makhov AM, Griffith JD, Tomaska L. Amplification of telomeric arrays via rolling-circle mechanism. J Biol Chem. 2005;280:10840–10845. doi: 10.1074/jbc.M409295200. [DOI] [PubMed] [Google Scholar]
- 25.Kosa P, Valach M, Tomaska L, Wolfe KH, Nosek J. Complete DNA sequences of the mitochondrial genomes of the pathogenic yeasts Candida orthopsilosis and Candida metapsilosis: insight into the evolution of linear DNA genomes from mitochondrial telomere mutants. Nucleic Acids Res. 2006;34:2472–2481. doi: 10.1093/nar/gkl327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rycovska A, Valach M, Tomaska L, Bolotin-Fukuhara M, Nosek J. Linear versus circular mitochondrial genomes: intraspecies variability of mitochondrial genome architecture in Candida parapsilosis. Microbiology. 2004;150:1571–1580. doi: 10.1099/mic.0.26988-0. [DOI] [PubMed] [Google Scholar]
- 27.Cohen S, Regev A, Lavi S. Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability. Oncogene. 1997;14:977–985. doi: 10.1038/sj.onc.1200917. [DOI] [PubMed] [Google Scholar]
- 28.Ogino H, et al. Release of telomeric DNA from chromosomes in immortal human cells lacking telomerase activity. Biochem Biophys Res Commun. 1998;248:223–227. doi: 10.1006/bbrc.1998.8875. [DOI] [PubMed] [Google Scholar]
- 29.Regev A, Cohen S, Cohen E, Bar-Am I, Lavi S. Telomeric repeats on small polydisperse circular DNA (spcDNA) and genomic instability. Oncogene. 1998;17:3455–3461. doi: 10.1038/sj.onc.1202250. [DOI] [PubMed] [Google Scholar]
- 30.Tokutake Y, et al. Extra-chromosomal telomere repeat DNA in telomerase-negative immortalized cell lines. Biochem Biophys Res Commun. 1998;247:765–772. doi: 10.1006/bbrc.1998.8876. [DOI] [PubMed] [Google Scholar]
- 31.Cesare AJ, Groff-Vindman C, Compton SA, McEachern MJ, Griffith JD. Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol Cell Biol. 2008;28:20–29. doi: 10.1128/MCB.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groff-Vindman C, Cesare AJ, Natarajan S, Griffith JD, McEachern MJ. Recombination at long mutant telomeres produces tiny single- and double-stranded telomeric circles. Mol Cell Biol. 2005;25:4406–4412. doi: 10.1128/MCB.25.11.4406-4412.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raices M, et al. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell. 2008;132:745–757. doi: 10.1016/j.cell.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 34.Zellinger B, Akimcheva S, Puizina J, Schirato M, Riha K. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol Cell. 2007;27:163–169. doi: 10.1016/j.molcel.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 35.Cohen S, Méchali M. Formation of extrachromosomal circles from telomeric DNA in Xenopus laevis. EMBO Rep. 2002;3:1168–1174. doi: 10.1093/embo-reports/kvf240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nabetani A, Ishikawa F. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol Cell Biol. 2009;29:703–713. doi: 10.1128/MCB.00603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates t-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci USA. 2009;106:12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009;28:799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L, et al. Telomere lengthening early in development. Nat Cell Biol. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- 42.Topcu Z, Nickles K, Davis C, McEachern MJ. Abrupt disruption of capping and a single source for recombinationally elongated telomeres in Kluyveromyces lactis. Proc Natl Acad Sci USA. 2005;102:3348–3353. doi: 10.1073/pnas.0408770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Natarajan S, Groff-Vindman C, McEachern MJ. Factors influencing the recombinational expansion and spread of telomeric tandem arrays in Kluyveromyces lactis. Eukaryot Cell. 2003;2:1115–1127. doi: 10.1128/EC.2.5.1115-1127.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natarajan S, McEachern MJ. Recombinational telomere elongation promoted by DNA circles. Mol Cell Biol. 2002;22:4512–4521. doi: 10.1128/MCB.22.13.4512-4521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomaska L, Nosek J. Telomere heterogeneity: taking advantage of stochastic events. FEBS Lett. 2009;583:1067–1071. doi: 10.1016/j.febslet.2009.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lustig AJ. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev Genet. 2003;4:916–923. doi: 10.1038/nrg1207. [DOI] [PubMed] [Google Scholar]
- 47.Cerone MA, Londono-Vallejo JA, Bacchetti S. Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum Mol Genet. 2001;10:1945–1952. doi: 10.1093/hmg/10.18.1945. [DOI] [PubMed] [Google Scholar]
- 48.Deng Z, Dheekollu J, Broccoli D, Dutta A, Lieberman PM. The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr Biol. 2007;17:1989–1995. doi: 10.1016/j.cub.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 49.Li B, Jog SP, Reddy S, Comai L. WRN controls formation of extrachromosomal telomeric circles and is required for TRF2ΔB-mediated telomere shortening. Mol Cell Biol. 2008;28:1892–1904. doi: 10.1128/MCB.01364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Compton SA, Choi JH, Cesare AJ, Ozgur S, Griffith JD. Xrcc3 and Nbs1 are required for the production of extrachromosomal telomeric circles in human alternative lengthening of telomere cells. Cancer Res. 2007;67:1513–1519. doi: 10.1158/0008-5472.CAN-06-3672. [DOI] [PubMed] [Google Scholar]
- 51.Zhong ZH, et al. Disruption of telomere maintenance by depletion of the MRE11/ RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J Biol Chem. 2007;282:29314–29322. doi: 10.1074/jbc.M701413200. [DOI] [PubMed] [Google Scholar]
- 52.Cerone MA, Autexier C, Londono-Vallejo JA, Bacchetti S. A human cell line that maintains telomeres in the absence of telomerase and of key markers of ALT. Oncogene. 2005;24:7893–7901. doi: 10.1038/sj.onc.1208934. [DOI] [PubMed] [Google Scholar]
- 53.Jiang WQ, et al. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol Cell Biol. 2005;25:2708–2721. doi: 10.1128/MCB.25.7.2708-2721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muntoni A, Reddel RR. The first molecular details of ALT in human tumor cells. Hum Mol Genet. 2005;14:R191–R196. doi: 10.1093/hmg/ddi266. [DOI] [PubMed] [Google Scholar]
- 55.Compton SA, Tolun G, Kamath-Loeb AS, Loeb LA, Griffith JD. The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J Biol Chem. 2008;283:24478–24483. doi: 10.1074/jbc.M803370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poulet A, et al. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009;28:641–651. doi: 10.1038/emboj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fouché N, et al. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J Biol Chem. 2006;281:37486–37495. doi: 10.1074/jbc.M608778200. [DOI] [PubMed] [Google Scholar]
- 58.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 59.de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 60.Blanc H, Dujon B. Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc Natl Acad Sci USA. 1980;77:3942–3946. doi: 10.1073/pnas.77.7.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacAlpine DM, Kolesar J, Okamoto K, Butow RA, Perlman PS. Replication and preferential inheritance of hypersuppressive petite mitochondrial DNA. EMBO J. 2001;20:1807–1817. doi: 10.1093/emboj/20.7.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bibillo A, Eickbush TH. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J Biol Chem. 2004;279:14945–14953. doi: 10.1074/jbc.M310450200. [DOI] [PubMed] [Google Scholar]
- 63.Chen B, Lambowitz AM. De novo and DNA primer-mediated initiation of cDNA synthesis by the mauriceville retroplasmid reverse transcriptase involve recognition of a 3′ CCA sequence. J Mol Biol. 1997;271:311–332. doi: 10.1006/jmbi.1997.1185. [DOI] [PubMed] [Google Scholar]
- 64.Kennell JC, Wang H, Lambowitz AM. The Mauriceville plasmid of Neurospora spp. uses novel mechanisms for initiating reverse transcription in vivo. Mol Cell Biol. 1994;14:3094–3107. doi: 10.1128/mcb.14.5.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 66.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 67.Nosek J, et al. Complete DNA sequence of the linear mitochondrial genome of the pathogenic yeast Candida parapsilosis. Mol Genet Genomics. 2004;272:173–180. doi: 10.1007/s00438-004-1046-0. [DOI] [PubMed] [Google Scholar]
- 68.Tomaska L, Makhov AM, Griffith JD, Nosek J. t-loops in yeast mitochondria. Mitochondrion. 2002;1:455–459. doi: 10.1016/s1567-7249(02)00009-0. [DOI] [PubMed] [Google Scholar]