Abstract
Individuals with BRCA2 mutations are predisposed to breast cancers owing to genome instability. To determine the functions of BRCA 2, the human protein was purified. It was found to bind selectively to single-stranded DNA (ssDNA), and to ssDNA in tailed duplexes and replication fork structures. Monomeric and dimeric forms of BRCA 2 were observed by EM. BRCA 2 directed the binding of RA D51 recombinase to ssDNA, reduced the binding of RA D51 to duplex DNA and stimulated RA D51-mediated DNA strand exchange. These observations provide a molecular basis for the role of BRCA 2 in the maintenance of genome stability.
Breast cancer represents one of the most common causes of cancer-associated female mortality in the world today. Inheritable breast and ovarian cancers can be linked with mutations in the BRCA1, BRCA2 and PALB2 genes, all of which affect the efficiency of double-strand break repair mediated by homologous recombination1. The BRCA2 gene encodes a protein of 3,418 amino acids, containing (i) eight conserved BRC motifs that interact with the RAD51 recombinase2, (ii) a binding site for the meiosis-specific recombinase DMC1 (ref. 3) and (iii) a DNA-binding domain4. BRCA2’s interactions with RAD51 and DMC1 are required for the localization of both proteins to nuclear foci that mark sites of DNA breakage. Additionally, there is an unrelated RAD51-binding site at the C terminus of BRCA2 that may provide more complex levels of control during the cell cycle and in response to DNA damage5,6.
Efforts to understand the functions of BRCA2 have been hampered by the difficulty in purifying the full-length human protein. Consequently, most studies have focused on protein fragments or on small BRCA2-like proteins found in simple organisms. For example, the Brh2 protein from the fungus Ustilago maydis and the BRC-2 protein from the nematode Caenorhabditis elegans each contain a single BRC motif and can exert a stimulatory effect on DNA binding by RAD51 and/or RAD51’s ability to promote DNA-DNA interactions7,8. Similar effects have been seen with fragments of human BRCA2 (refs. 9–12).
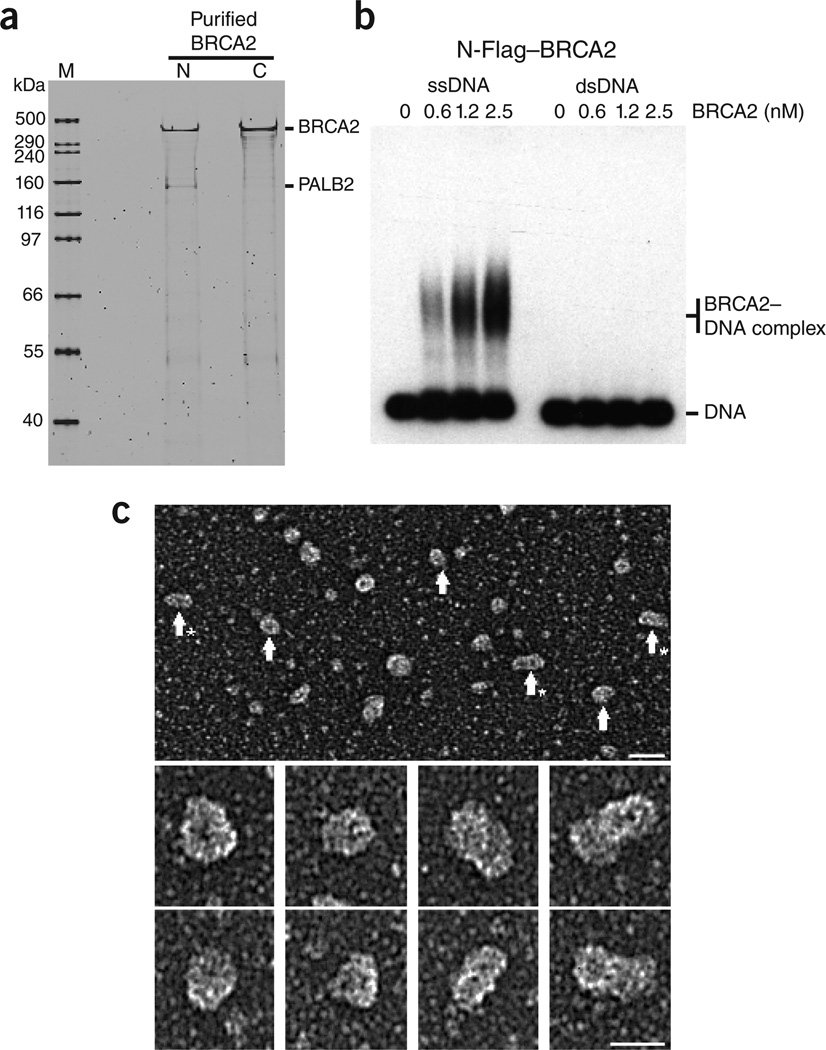
Here we have purified full-length human BRCA2 from HeLa cells carrying a human bacterial artificial chromosome (BAC), in which the BRCA2 gene, complete with upstream and downstream regulatory elements, was FLAP tagged at either the N- or C-terminus13. The FLAP tag is composed of green fluorescent protein (GFP), S- and Flag-affinity tags separated by PreScission- and TEV-protease sites (Supplementary Fig. 1). The N- and C-terminally tagged BRCA2 proteins were purified by Flag- and GFP-affinity purification, and the GFP and S tags were removed by TEV cleavage, leaving just a small, 1.3 kDa Flag tag (Supplementary Methods). The purified proteins were soluble and SDS-PAGE revealed that both protein preparations were of high quality (Fig. 1a). A small amount of PALB2, a high-affinity BRCA2 interaction partner14, co-purified with the N-terminally tagged BRCA2, whereas much less was associated with the C-terminally tagged protein. The reason for this difference is unknown, and both preparations behaved similarly in the experiments described here.
Figure 1.
Purified BRCA2 binds specifically to ssDNA. (a) N- or C-terminally FLAG-tagged BRCA2 was purified from human cells and analyzed by SDS-PAGE and SyPro Ruby staining. M, size marker. (b) Purified N-terminally Flag-tagged BRCA2 was incubated with 5′-32P end-labeled ssDNA or dsDNA, and complexes were analyzed by agarose gel electrophoresis and visualized by autoradiography. (c) Electron microscopic visualization of BRCA2. Top: large field of N-terminally Flag-tagged BRCA2 protein, rotary shadowcast with tungsten. Scale bar, 40 nm. Arrows, ball-shaped BRCA2 particles; arrows with asterisks, rod-shaped particles. Below: individual protein particles at higher magnification. Scale bar, 25 nm. Images are shown in reverse contrast.
To determine whether BRCA2 binds to DNA, N-terminally Flag-tagged BRCA2 was incubated with 5′-32P end-labeled ssDNA or double-stranded DNA (dsDNA). BRCA2 bound ssDNA in a concentration-dependent manner (Fig. 1b, left), whereas it did not form stable complexes with dsDNA (Fig. 1b, right). Similar results were obtained with C-terminally Flag-tagged BRCA2 (Supplementary Fig. 2). These results show that full-length BRCA2 is an ssDNA-binding protein, as suggested by previous studies of a fragment of BRCA2 containing the DNA-binding domain4.
When Flag-BRCA2 (385 kDa) was visualized by electron microscopy after rotary tungsten shadowcasting, we observed a mixture of ball-shaped (that is, roughly spherical) particles and elongated rod-shaped particles with the appearance of a dumbbell (Fig. 1c). The balls had a mean diameter of 15 ± 2 nm (including tungsten coating), whereas the rods were 16 ± 2 nm wide and 27 ± 3 nm long. In some instances longer BRCA2 rods were observed. The masses of the ball- and rod-shaped particles were determined by comparison with ferritin (440 kDa), revealing relative masses of 337 and 860 kDa, respectively (Supplementary Fig. 3), consistent with BRCA2 monomers and dimers. These observations were confirmed with unfixed BRCA2 protein using negative staining (data not shown).
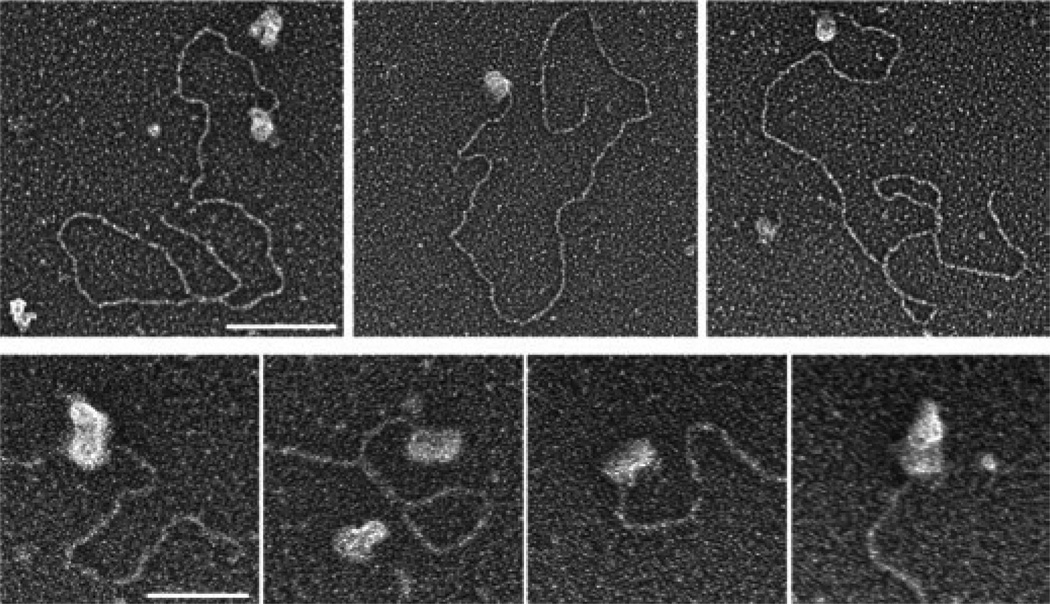
Because tailed duplex DNA provides a good model substrate for structures that initiate homologous recombination–mediated double-strand break (DSB) repair, we visualized the binding of BRCA2 to a 3.5-kb linear duplex containing a 54-nt 3′-extended ssDNA tail at one end and a blunt end at the other. Examination of 129 molecules with bound protein (out of a total of 310 DNA molecules) showed that 91% had BRCA2 bound exclusively to one end of the DNA substrate (Fig. 2). No example was found in which BRCA2 bound to both DNA termini. Only infrequently (9%) did we observe the apparent association of BRCA2 with internal duplex regions of DNA. Inspection of the end-bound BRCA2 proteins showed that these particles had dimensions of 19 ± 4 nm wide and 34 ± 6 nm long (n = 15), consistent with the rod-shaped BRCA2 dimers observed in the absence of DNA.
Figure 2.
Binding of BRCA2 to tailed duplex DNA. N-terminally Flag-tagged BRCA2 protein was incubated with linear duplex DNA (3.5 kb long) containing a 54-nt ssDNA overhang at one end. Samples were fixed, mounted on glow-discharged carbon grids and rotary shadowcast with tungsten. Top: BRCA2 localizes to ssDNA tails of the duplex DNA substrate. Scale bar, 100 nm. Bottom: higher magnification images of dumbbell-shaped BRCA2 dimers bound to ssDNA tails. Scale bar, 50 nm. Images are shown in reverse contrast.
We also analyzed the interaction of BRCA2 with replication fork structures containing a 25-nt-long region of ssDNA at the fork. Upon scoring 124 molecules, we found that 50 DNA molecules had bound BRCA2 protein and that in 75% of these cases BRCA2 was bound to the junction (Supplementary Fig. 4). Less frequently, BRCA2 appeared to be attached to internal duplex DNA (16%) or at the ends of the junction arm (10%). Given the preference of BRCA2 for binding ssDNA, these results are consistent with the interaction of BRCA2 to the ssDNA present at the replication fork. With the fork substrate, we observed the binding of both ball- and rod-shaped BRCA2 particles.
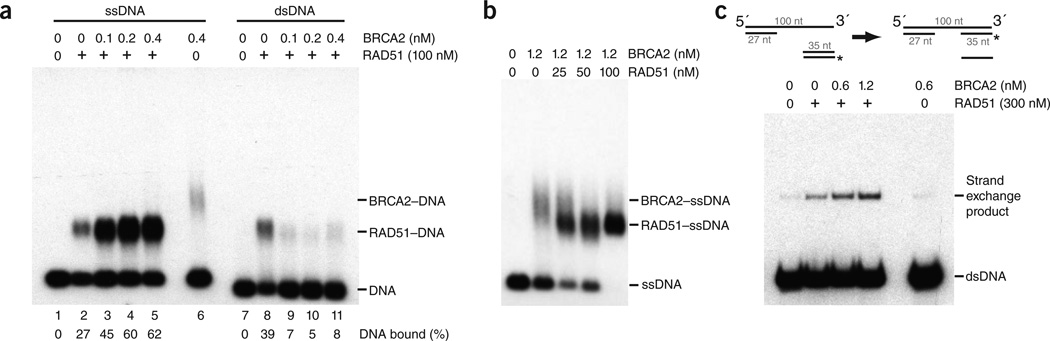
Whereas Escherichia coli RecA protein shows a high affinity for ssDNA compared with duplex DNA, RAD51 protein binds both ssDNA and dsDNA15. This lack of specificity is problematic for recombination because it is known that homologous recombination is initiated by the binding of RAD51 to resected ssDNA tails at sites of DNA breaks, which then interact and pair with RAD51-free duplex DNA. These observations led to suggestions that RAD51 requires a targeting factor that mediates its interaction with ssDNA. To determine whether BRCA2 modulates the DNA binding affinities of RAD51, we conducted DNA binding assays under conditions of low ionic strength, in which BRCA2 binds ssDNA (Supplementary Fig. 5), whereas RAD51 shows the same affinity for both ssDNA and dsDNA (Supplementary Fig. 6). The reactions included both Mg2+ and ATP to facilitate the formation of extended RAD51 nucleoprotein filaments, which are known to be the functionally active form of the protein. We found that the presence of low amounts of BRCA2 resulted in a significant increase in ssDNA binding by RAD51 (Fig. 3a, lanes 2–5). Conversely, BRCA2 reduced the ability of RAD51 to bind dsDNA, thus providing the differential targeting necessary for homologous recombination (Fig. 3a, lanes 8–11). Further analysis of the way that BRCA2 stimulates RAD51 nucleoprotein filament formation revealed no evidence of a ternary BRCA2–RAD51–DNA complex, suggesting that BRCA2 may act as a molecular chaperone for RAD51 loading (Fig. 3b).
Figure 3.
BRCA2 targets RAD51 to ssDNA and stimulates DNA strand exchange. (a) BRCA2 and RAD51 were mixed in binding buffer for 5 min at 24 °C, before the addition of 32P-labeled ssDNA or dsDNA. Incubation was then continued for 10 min, before complexes were fixed and analyzed by agarose gel electrophoresis and autoradiography. (b) BRCA2 was incubated for 5 min at 24 °C with 32P-labeled ssDNA, before RAD51 addition. Incubation was then continued for a further 5 min. Complexes were analyzed as in a. (c) Stimulation of RAD51-mediated strand exchange by BRCA2. Tailed duplex DNA was incubated with BRCA2 for 5 min before the addition of RAD51. After a further 5 min, 5′-32P end-labeled (*) duplex DNA (35 bp) was added and incubation continued for 10 min. The products were deproteinized and analyzed by 10% PAGE, followed by autoradiography.
Because BRCA2 targets the binding of RAD51 to ssDNA rather than dsDNA, a reaction likely to be important for RAD51 activity, we next analyzed whether BRCA2 could stimulate RAD51-mediated strand exchange. To do this, we first bound BRCA2 to tailed duplex DNA and then added RAD51 protein and a 32P-labeled duplex. The presence of BRCA2 resulted in the transfer of the 32P-labeled strand of the duplex to the tailed substrate (Fig. 3c). These results indicate that full-length BRCA2 protein stimulates RAD51-mediated DNA strand exchange.
The results presented here provide our first insights into the actions and structure of this important tumor suppressor. We have shown that BRCA2 associates with ssDNA, promotes the specific targeting of RAD51 to ssDNA tails on duplex DNA molecules resembling structures that initiate homologous recombination in vivo and stimulates RAD51-mediated DNA strand exchange. These reactions are likely to be important in terms of BRCA2’s biological functions in the maintenance of genome stability. Electron microscopic analyses revealed two distinct particle sizes, with masses suggesting monomeric and dimeric forms of BRCA2. Dimer formation would be consistent with the crystallographic dimer observed during X-ray analysis of the DNA-binding domain of BRCA2 (ref. 4) and the observed dimeric forms of U. maydis Brh2 (ref. 16). Monomeric and dimeric forms of BRCA2 were visualized binding to regions of ssDNA present within tailed or replication fork substrates.
The association of multiple BRCA2 units (either monomeric or dimeric) with regions of ssDNA suggests a rapid mechanism for the loading of RAD51 onto ssDNA. Given that we do not observe a ternary complex containing BRCA2–RAD51–ssDNA, our results indicate that BRCA2 acts as a molecular chaperone that facilitates the binding of RAD51 and then dissociates itself from the ssDNA. RAD51 monomers associate to form heptameric ring structures in solution17, and it is possible that the BRC repeats might collectively fold in such a way as to favor associations with RAD51 rings. As such, the actual amount of RAD51 bound by each BRCA2 unit could be considerably greater than the number of BRC repeats. How BRCA2 blocks binding of RAD51 to dsDNA is at present unknown but, given that there are eight BRC motifs per BRCA2 monomer, one scenario would be that BRCA2 binds RAD51 such that the critical concentration of free RAD51 in solution drops to the point where dsDNA binding does not take place.
The role of BRCA2 in the targeting of RAD51 is analogous to the way that Rad52 delivers Rad51 to ssDNA substrates for homologous recombination in yeast. In yeast, the recruitment of Rad51 to resected DSBs is dependent upon Rad52 in both mitotic and meiotic cells18, and in vitro studies show that Rad52 facilitates the formation of Rad51 filaments on RPA-bound ssDNA19,20. However, a defined mediator role for RAD52 in human cells has not been demonstrated, which may account for the lack of a clear DSB repair–defective phenotype in Rad52−/− mice21,22. It is possible that the eight BRC motifs in BRCA2 provide a rapid mechanism for the loading of RAD51 to ssDNA, such that the evolutionary development of BRCA2 has relegated RAD52 to a more minor, or alternative, role in DNA repair and genome maintenance.
ACKNOWLEDGMENTS
We thank T. Hyman (Max Planck Institute, Dresden) for providing the BAC modification cassettes. This work was supported by grants to S.C.W. (Cancer Research UK, the Breast Cancer Campaign, the Louis-Jeantet Foundation, Swiss Bridge and the European Research Council) and to J.D.G. and S.A.C. (US National Institutes of Health). T.T. was supported by the Alfred Benzon Foundation and the Carlsberg Foundation and S.L. by a European Molecular Biology Organization fellowship.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
T.T. and S.C.W. designed the study; S.L. and M.P. made the BRCA2 constructs; T.T. and M.J.M. made the RAD51 expression vectors, purified the proteins and carried out the biochemical analyses; and S.A.C. and J.D.G. visualized BRCA2 by electron microscopy. S.C.W. wrote the manuscript with contributions from T.T., S.A.C. and J.D.G.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Moynahan ME, Jasin M. Nat. Rev. Mol. Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bork P, Blomberg N, Nilges M. Nat. Genet. 1996;13:22–23. doi: 10.1038/ng0596-22. [DOI] [PubMed] [Google Scholar]
- 3.Thorslund T, Esashi F, West SC. EMBO J. 2007;26:2915–2922. doi: 10.1038/sj.emboj.7601739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang H, et al. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 5.Esashi F, et al. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 6.Ayoub N, et al. Curr. Biol. 2009;19:1075–1085. doi: 10.1016/j.cub.2009.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H, Li Q, Holloman WK, Pavletich NP. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 8.Petalcorin MIR, Sandall J, Wigley DB, Boulton SJ. J. Mol. Biol. 2006;361:231–242. doi: 10.1016/j.jmb.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 9.Shivji MKK, et al. Nucleic Acids Res. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shivji MKK, et al. Proc. Natl. Acad. Sci. USA. 2009;106:13254–13259. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carreira A, et al. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tal A, Arbel-Goren R, Stavans J. J. Mol. Biol. 2009;393:1007–1012. doi: 10.1016/j.jmb.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Lekomtsev S, Guizetti J, Pozniakovsky A, Gerlich DW, Petronczki M. J. Cell Sci. 2010;123:1395–1400. doi: 10.1242/jcs.068015. [DOI] [PubMed] [Google Scholar]
- 14.Sy SMH, Huen MSY, Chen JJ. Proc. Natl. Acad. Sci. USA. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson FE, Stasiak A, West SC. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Q, et al. Mol. Cell. Biol. 2007;27:2512–2526. doi: 10.1128/MCB.01907-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin DS, et al. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasior SL, et al. Proc. Natl. Acad. Sci. USA. 2001;98:8411–8418. doi: 10.1073/pnas.121046198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song BW, Sung P. J. Biol. Chem. 2000;275:15895–15904. doi: 10.1074/jbc.M910244199. [DOI] [PubMed] [Google Scholar]
- 20.Sugiyama T, Kowalczykowski SC. J. Biol. Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 21.de Vries FAT, et al. DNA Repair (Amst.) 2005;4:1121–1128. doi: 10.1016/j.dnarep.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Rijkers T, et al. Mol. Cell. Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]