Abstract
Hemoglobins are ubiquitous in Eukarya and Bacteria but, until now, have not been found in Archaea. A phylogenetic analysis of the recently revealed microbial family of globin-coupled heme-based sensors suggests that these sensors descended from an ancient globin-only progenitor, or a protoglobin (Pgb). Here, we report the discovery and characterization of two Pgbs from the Archaea: ApPgb from the obligately aerobic hyperthermophile Aeropyrum pernix, and MaPgb from the strictly anaerobic methanogen Methanosarcina acetivorans. Both ApPgb and MaPgb bind molecular oxygen, nitric oxide, and carbon monoxide by means of a heme moiety that is coordinated to the protein through the F8 histidine (histidine 120). We postulate that these archaeal globins are the ancestors of contemporary hemoglobins.
Keywords: globin-coupled sensor, oxygen sensor, NO sensor, myoglobin
Globins are heme-containing globular proteins that bind and transport molecular oxygen. It has been proposed that all globins evolved from an ≈17-kDa ancestral redox protein (1). The globin fold was initially characterized by eight α-helices, designated A through H (2, 3). An alignment of 700 vertebrate and nonvertebrate globins based on secondary structure assignments, following the approach of Lesk and Chothia (3) and Bashford et al. (4), showed that only two residues were invariant, the proximal histidine in the F helix (His F8) and a phenylalanine in the CD loop (Phe CD1) (5). Discovery of the truncated globins provided evidence of globins with the total number of helices less than eight (6-8). For a recently discovered group of globin-coupled sensors, Hou et al. (9) showed that the first 195 residues in the aerotaxis transducer HemAT-Hs (from Halobacterium salinarum) were sufficient to retain heme and its reversible O2-binding properties, whereas crystallization of HemAT-Bs from Bacillus subtilis confirmed the globin fold (10). A phylogenetic analysis of the microbial family of globin-coupled sensors (GCSs) suggests that the GCSs descend from an ancient globin-only progenitor, or protoglobin (Pgb), in the Archaea (11). Alternatively, it has been postulated that such Pgbs might have disappeared from Archaea, rendering their identification impossible (12). Here, we show that two proteins ApPgb and MaPgb, identified from the Archaea Aeropyrum pernix and Methanosarcina acetivorans, respectively, conform to the globin sequence motifs, contain heme, and demonstrably bind O2, CO, and NO.
Materials and Methods
Cloning and C-Terminal His-Tag Construction of the Pgb from A. pernix K1 (ApPgb). Genomic DNA from A. pernix served as the template for PCR amplification of the Pgb gene. NdeI and HindIII sites (underlined) were introduced in the primers for the PCR amplification (5′-CACATATGACCCCAAGTGATATCCC-3′; 5′-CTAAGCTTCCAGTCGTTTTCTTTTGTGTATGGATGGC-3′). The PCR product was cloned into the TOPO Blunt-PCR vector (Invitrogen) and then subcloned into the Escherichia coli expression vector pET27 as a NdeI-HindIII fragment.
Cloning and N-Terminal Maltose-Binding Protein (MBP) Fusion of the Pgb from M. acetivorans C2A (MaPgb). The M. acetivorans Pgb gene was amplified by PCR, with EcoRI and XbaI sites introduced during the PCR amplification. (5′-ATGAATTCAACTGGGGGGTAAGACCCGTGTCT-3′; 5′-GCTCTAGAGAAATCACCATATTTCACATATG-3). The PCR product was cloned into TOPO-Blunt-PCR vector, and then subcloned into Expression vector pMAL-c2 as an EcoRI-XbaI fragment. A pLysS E. coli host was transformed with the plasmid carrying the MBP fusion construction and induced according to the manufacturer (New England Biolabs).
ApPgb Proximal Histidine Mutant. The plasmid carrying the A. pernix globin gene was used as the template for the PCR based site-directed mutagenesis according to the protocol from Stratagene. The mutated gene in the pET27 expression vector was confirmed by DNA sequencing. The final construction containing the single mutation was transformed into E. coli pLysS cells for expression.
Expression and Purification of the Recombinant Globins in E. coli. Expression and one-step purification of the C-terminal His-tag Pgb were performed according to Piatibratov et al. (13). The MaPgb-MBP fusion protein was purified by affinity chromatography (amylose resin) according to the protocol from the manufacturer (New England Biolabs).
Preparation of Deoxy, Ferrous-Nitrosyl, and Ferric Globin. Both Ap- Pgb and MaPgb purify in the ferric form; however, the last step in ApPgb purification involves elution with 250 mM imidazole, resulting in the imidazole-bound form. Removal of the imidazole was accomplished by mixing the proteins with concentrated dithionite (≈250 mM), removing this reducing agent by gel filtration on a Sephadex G-25 (Pharmacia) column, and collecting the deoxy protein, all within an anaerobic glove box (Coy Laboratory Products, Ann Arbor, MI). Another form of preparation incorporates lower concentrations of dithionite (≈10 mM) with methyl viologen as an electron shuttle. Ferrous-nitrosyl Pgb was produced by adding a 15:1 NO to protein molar ratio. Ferric Pgb was prepared either by allowing the reduced samples to autoxidize in air or by adding a 1:1 ratio of potassium ferricyanide to protein.
Absorption Spectra, Ligand Binding, and Autoxidation. Absorption spectra were measured with a Cary 4000 UV-Vis spectrophotometer (Varian). Laser-flash photolysis and stopped-flow measurements were done with an LKS.60 laser kinetic spectrometer fitted with a PiStar stopped-flow drive unit (Applied Photophysics, Leatherhead, U.K.). For sample excitation, the LKS.60 spectrometer was coupled to a Quantel Brilliant B Nd:YAG laser with second-harmonic generation. Data acquisition was provided by an Agilent (Palo Alto, CA) 54830B digital oscilloscope for fast measurements or a 12-bit analog-to-digital converter (ADC) card within the workstation for slow measurements.
Ligand association and dissociation rates were measured for protein in 100 or 200 mM sodium phosphate (pH 7.0) at 25°C at the wavelength of maximum difference between the initial and final species. The carbon monoxide association rate was measured by laser-induced photodissociation of CO-saturated protein whereas the absorbance change was monitored at 421 nm. For the NO association rate, CO-saturated protein was photolyzed in the presence of dissolved NO, with absorbance changes monitored at 423 nm. At each ligand concentration, the observed association rate was measured at least three times, and the true rate constants were calculated from linear plots of the observed rates, kobs, vs. ligand concentration.
The CO off rate was determined by oxidation of CO-saturated protein with potassium ferricyanide (100 μM). The entire process was monitored at 422 nm over a time course of ≈6 min in the Cary 4000 spectrophotometer.
Oxy-protoglobin spectra were obtained in the Cary spectrophotometer and verified by performing flash-flow experiments on CO-bound protein injected with O2-saturated buffer. In addition, spectra were obtained with a fiber optic spectrometer from Ocean Optics (Orlando, FL) by using a detector with a 2048-element linear silicon charge-coupled-device (CCD) array that gives almost instantaneous spectra, although lower quality (data not shown). Autoxidation rates were measured after the addition of air-saturated buffer to deoxy ApPgb in air.
Sequence Alignment and Phylogenetic Analysis. Pgbs and representative globin protein sequences were aligned in multiple stages. Groups of similar globins were first aligned separately and then combined together in a profile-based alignment by using clustalx (14). The globin sequences were then manually adjusted in DNASTAR's MEGALIGN (DNASTAR, Madison, WI) to preserve the integrity of both the solved and JPRED predicted (15) secondary structures. Consensus sequences were calculated with consensus (www.bork.embl-heidelberg.de/Alignment/consensus.html). The HemAT globin domain sequence limits are based on the crystal structure of HemAT-Bs (Protein Data Bank ID code 1OR6). Bootstrapped (10,000 replicates) trees were created using clustalx, and exported from treeview (16) and modified in Adobe illustrator (Adobe Systems, Mountain View, CA) for presentation. Sequences outside the traditional globin fold (A-H helices) were not included in the construction of the trees.
3D Homology Model. genemine was used to create the ApPgb 3D homology model (truncated to 161 residues) based on the structure of the HemAT-Bs sensor domain (PDB ID code 1OR6) as guided by the alignment in Fig. 1A (10; www.bioinformatics.ucla.edu/~genemine/). Lack of a suitable structural template precluded us from including the N-terminal 44 residues that are not part of the traditional globin fold. hyperchem was used to correct manually the bonding in the heme group, including energy minimization by using the Polak-Ribiere algorithm, terminating with a 0.1 kcal/(Å mol) gradient (www.hyper.com). Illustrations were created by using molscript and raster3d.
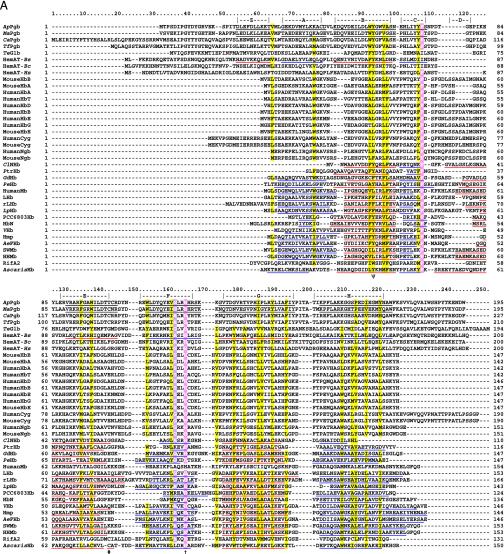
Fig. 1.
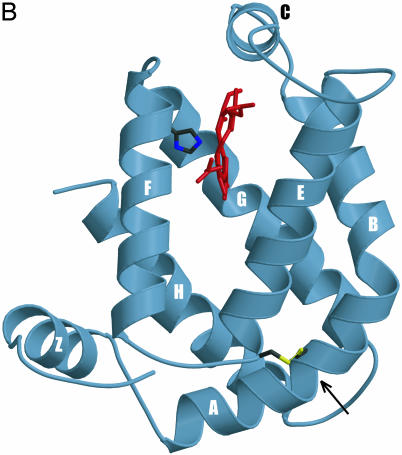
(A) Globin structural alignment with Pgbs. Secondary structure elements in globins with known 3D structures are underlined by color and labeled A through H whereas helices predicted by jpred are underlined without color. The pre-A helix is labeled Z, according to the HemAT-Bs structure. Positions at least 50% identical are indicated by bold characters, with the following notations: dagger (†), proximal F8 histidine; pound sign (#), E19 cysteine; psi (ψ), B10 Tyr. Color-coded amino acids are based on an 80% consensus sequence, and colored highlights are assigned to amino acid groups as follows: polar (p, KRHEDQNST) in red; turn-like (t, ACDEGHKNPQRST) in green; bulky hydrophobic (h, ACLIVMHYFW), and aliphatic (l, LIVM) in yellow; aromatic (a, FHWY) in white on pink background; small (s, ACDGNPSTV) in purple; and tiny (u, AGS) in white on purple background. The following sequences and structures were used (PDB IDs in parentheses): HumanMb (5MBN); HHMb (1WLA); SWMb (1MBO); AeFHb (1CQX); Hmp (1GVH); VHb (1VHB); rLHb (1D8U); LHb (2GDM); LpHb (1EBT); GdHb (1VRF); ClNHb (1KR7); PtrHb (1DLW); HbN (1IDR); PCC6803Hb (1MWB); PeHb (1H97); and AscarisHb (1ASH). (B) 3D homology model of ApPgb including the proximal histidine (dark green) and heme (red). The helices are labeled A through H with the disulfide bridge (yellow) indicated with an arrow. MouseHbA, -HbB, -HbE, -Cyg, and -Ngb stand for mouse Hb α, mouse Hb β, mouse Hb ε, mouse cytoglobin, and mouse neuroglobin. HumanHbA, -HbB, -HbG, -HbD, -HbE, -HbT, -Cyg, and -Ngb stand for human Hb α, human Hb β, human Hb γ, human Hb δ, human Hb ε, human Hb θ1, human cytoglobin, and human neuroglobin.
Molecular Dynamics Simulations (MDS) (Force Field and ab Initio MDS). Force field molecular dynamics used the CHARMM force field in the VMD/NAMD package (www.ks.uiuc.edu/Research/vmd/ and www.ks.uiuc.edu/Research/namd/) from the Theoretical and Computational Biophysics Group at the University of Illinois. Ab Initio molecular dynamics using the atom-centered density matrix propagation (ADMP) Method with 5,000-fs steps was used to characterize NO, CO, and H2S binding (available in gaussian 03, www.Gaussian.com). Density Functional Theory (Becke 3 Lee Yang Parr or B3LYP) with 6-31G basis set was applied to the Fe and ligand, and Universal Force Field was applied to the rest of the heme group and a few of the surrounding protein residues, including the proximal Histidine. The protein model with His-120 covalently attached to the heme Fe was solvated in a water bath cube of ≈9,700 water molecules with cyclic boundary conditions throughout the simulation with cysteines 45 and 102 joined by a disulfide link. NO, CO, and H2S all formed stable bonds with Fe at B3LYP/6-31G (see Figs. 6 and 7, which are published as supporting information on the PNAS web site).
Results and Discussion
Using the BLAST algorithm, all available complete and incomplete microbial genomes were queried with a consensus sequence corresponding to the heme-binding domain of the globin-coupled sensors (11). Initially, an ≈23-kDa (194-residue) protein was identified in the thermophilic cyanobacterium Thermosynechococcus elongatus. This sequence was further used to query the National Center for Biotechnology Information's (nr) protein database with PSI-BLAST. After two iterations, the search results identified the globin-coupled sensors, the flavohemoglobins, and additional proteins with molecular masses ranging from ≈23 kDa (194 residues) to ≈27 kDa (227 residues). The additional proteins belonged to the archaea A. pernix and M. acetivorans, an Actinobacterium Thermobifida fusca, and the Green non-sulfur bacterium Chloroflexus aurantiacus (Fig. 1 A). The archaeal Pgbs, ApPgb and MaPgb, were exactly 195 aa long: the minimum length found to be required for heme and reversible O2 binding by the globin-coupled aerotaxis transducer HemAT-Hs (9).
To determine experimentally whether ApPgb and MaPgb bind heme, both genes were expressed in E. coli from recombinant vectors, and the corresponding proteins were purified by His-tag and MBP affinity chromatography, respectively, according to Piatibratov et al. (13) (Fig. 2). Electronic absorption spectra of the liganded and unliganded ferrous (Fe2+) (Fig. 3 A and E) and ferric (Fe3+) (Fig. 3 B-D and F) forms of ApPgb absorb in the near ultra-violet and visible regions, a characteristic of heme proteins. Difference spectra (reduced minus oxidized) resulted in a peak at 564 nm, indicative of a b-type heme (data not shown). Similar results were found for MaPgb (data not shown).
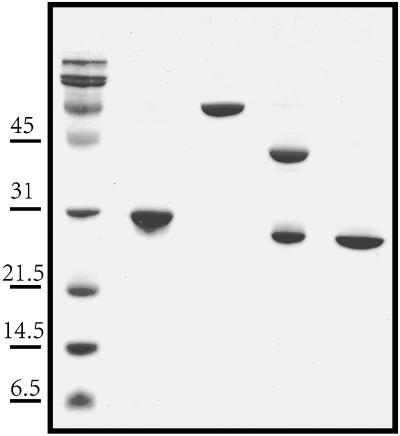
Fig. 2.
Electrophoresis of purified ApPgb and MaPgb. Lane 1, protein molecular weight markers; lane 2, ApPgb6x-His from A. pernix; lane 3, MaPgb-MBP fusion from M. acetivorans; lane 4, MaPgb-MBP digested with Factor Xa; lane 5, purified MaPgb. The apparently larger molecular weight of ApPgb6x-His is attributed to the His tag construction.
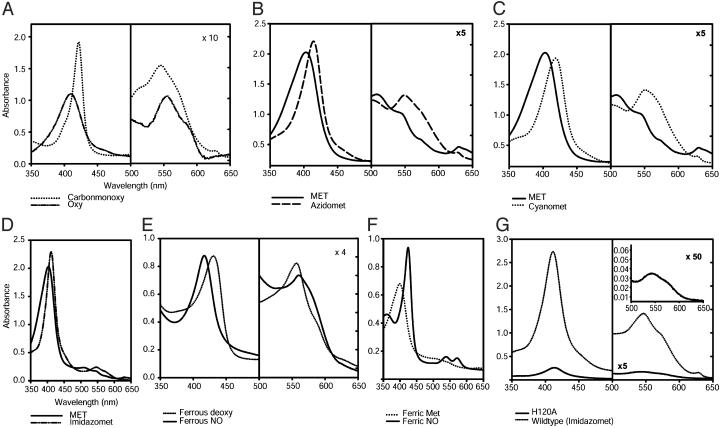
Fig. 3.
Absorption spectra of ApPgb in different redox states and bound to various ligands. (A) Ferrous (Fe2+) ApPgb in the CO-bound (dotted line) and the O2-bound (dashed and dotted line) forms. (B) Ferric (Fe3+) met (solid line) and azido (N3-, dashed line) ApPgb. (C) Cyano (CN-, dotted line) ApPgb. (D) Imidazole-bound (dashed and dotted line) ApPgb. (E) Ferrous deoxy (dotted line) ApPgb saturated with NO to give the Fe2+-NO (solid line) species. (F) Ferric met (dotted line) and the Fe3+-NO (solid line) species. When NO is added to ferrous deoxy ApPgb in approximately a 20:1 ratio, a spectrum identical to the Fe3+-NO spectrum results. All ligands were added to saturation. MaPgb absorption spectra are virtually identical except for an ≈2-nm blue shift at each λmax. (G) Comparison of wild-type ApPgb bound with imidazole (solid line) to the H120A mutant in the presence of imidazole (dashed and dotted line). The H121A mutant possesses a wild-type spectrum (data not shown). All measurements were done at 25°C and pH 7.0 in either 100 or 200 mM sodium phosphate buffer.
Ferrous deoxy ApPgb bound molecular oxygen (O2), carbon monoxide (CO), and nitric oxide (NO) to form the corresponding oxy-, carbonmonoxy- (Fig. 3A) and nitrosyl-protoglobin (Fig. 3E) species. Addition of O2 to the deoxy species caused the Soret band to shift from 433 nm to 410 nm (Fig. 3A), autoxidizing rapidly with half-lives of 4.3 (ApPgb) and 3.6 (MaPgb) minutes (Fig. 4C). In contrast, typical globins are known to autoxidize with half-lives on the order of hours to days (17, 18), and both the full-length and truncated versions of the HemAT-Hs protein have been shown to form stable O2-bound complexes (9). The relatively rapid oxidation of ApPgb in air suggests that this protein may have a role in transferring electrons to O2 under oxic conditions, possibly as a strategy for O2 detoxification.
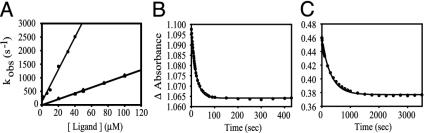
Fig. 4.
Parameters for binding of ligands to ferrous ApPgb at 25°C and pH 7.0. (A) Association rates for binding of CO (triangles) and NO (circles), as determined by laser-flash photolysis. (B) Dissociation rate of CO, as determined by oxidation of CO-saturated protein with potassium ferricyanide. (C) Autoxidation of ApPgb on exposure of the deoxy form to air. Details are in Materials and Methods.
Carbon monoxide binding to ApPgb produced a typically intense Soret absorption band at 422 nm (Fig. 3A). The rate constants for CO association and dissociation at 25°C and pH 7 were 11 μM-1·s-1 (Fig. 4A) and 47 ms-1 (Fig. 4B and Table 1), respectively. Similar values for those parameters have been reported for Hbs from plants and other species. For example, the CO association rate constant for ApPgb falls within the range reported for rice Hb (7.2 μM-1·s-1) and Parasponia Hb (14 μM-1·s-1) (19, 20). The CO dissociation rate constant for ApPgb is similar to values reported for the heme-based sensor kinase BjFixL from Bradyrhizobium japonicum (45 ms-1) and the phosphodiesterase AxPDEA1 from Acetobacter xylinum (60 ms-1) (21, 22). The equilibrium association constant for binding of CO to ApPgb (230 μM-1) falls between the values measured for elephant Hb (140 μM-1) and barley Hb (520 μM-1) (17, 23).
Table 1. Ligand binding properties of ApPgb compared with sperm whale myoglobin and the Ascaris hemoglobin.
The ferrous forms of the Pgbs were also found to bind NO, which is now known to be a physiological ligand of bacterial as well as eukaryotic Hbs (24-26). Binding of NO to deoxy ApPgb caused a blue-shift of the Soret absorption band from 433 nm to 419 nm (Fig. 3E). The association rate constant for NO binding to ferrous ApPgb was 59 μM-1·s-1 (Fig. 4A and Table 1), approximately three times higher than for sperm-whale Mb (SWMb) but three times lower than for leghemoglobin (27, 28). Ferric ApPgb was also found to bind NO, yielding an absorption spectrum with a Soret band at 425 nm and clear α and β bands at 574 and 542 nm, respectively (Fig. 3F). Similar results in ligand binding were obtained with MaPgb (data not shown).
The Pgbs feature three residues that are thought to have an ancient origin: a proximal histidine (His F8), a cysteine at the E19 position (Cys E19), and a distal tyrosine (B10 Tyr). In an alignment of SWMb with the truncated Hbs, the proximal histidine (His F8) was the only residue found to be absolutely conserved (29). In the ApPgb and MaPgb proteins, there are two histidine residues near the site of heme binding, His 120 and His 121. To determine which of those histidines coordinates to the heme iron, each of those two histidine residues in ApPgb was independently changed to alanine by site-directed mutagenesis. The H121A ApPgb retained a wild-type absorption spectrum whereas the H120A ApPgb almost entirely lost its visible absorption (Fig. 3G). A 3D homology model, created for ApPgb based on the ferric unliganded HemAT-Bs sensor domain (PDB ID code 1OR6) (Fig. 1B) and refined by force-field molecular-dynamics simulations, confirmed the identity of the proximal histidine as His 120.
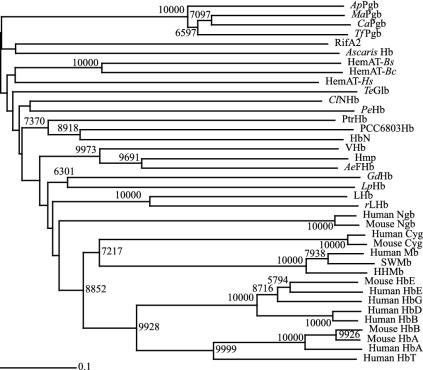
Molecular modeling of ApPgb also identified Cys-102 as being analogous to the E19 cysteine near the E-helix terminus of both the Ascaris suum Hb (AscarisHB) and the H2S-binding annelid Hb from Riftia pachyptila. This modeling further indicated that Cys-102 and Cys-45 at the A-B helical junction are precisely positioned to form a disulfide bridge (Fig. 1B). Such a disulfide linkage should contribute to the stability of those thermophilic Pgbs. Interestingly, a Neighbor-Joining tree (Fig. 5) shows that the Pgbs and the Hbs from A. suum and R. pachyptila group within the same branch. In the A. suum Hb, the positioning of the Cys E19 near the distal pocket has been shown to play a role in its NADPH-dependent NO-activated deoxygenase function (24). In the annelid Hb from R. pachyptila, however, Cys E19 is thought to be important for H2S binding (30). The possibility of cysteine thiols binding atypical ligands and bringing about diseased states has been suggested as a possible driving force for the evolutionary loss of H2S-binding in Hbs from organisms living in sulfide-free habitats (31). This hypothesis is consistent with ancient globins working to detoxify sulfide and nitric oxide, and the Cys E19 being absent from modern globins that have adapted to oxic environments.
Fig. 5.
Globin amino acid neighbor-joining phylogenetic tree based on the alignment in Fig. 1. Bootstraps ≥5,000 are indicated at each node. The horizontal scale bar represents 0.1 substitution per site. For abbreviations, see Fig. 1 legend.
In the Mycobacterium tuberculosis Hb, HbN, a distal tyrosine (Tyr B10) is thought to be an ancient adaptation for the scavenging and detoxification of NO, and its gradual loss during evolution has been proposed as a possible requirement for efficient O2 transport by Hbs (32). In the A. suum Hb, the globin-coupled sensors, and several other known globins, Tyr B10 is thought to play a central role in O2 binding due to stabilization of the bound O2 by hydrogen bonding (6, 7, 11, 18, 33-37). Replacement of the Tyr B10 with phenylalanine in the A. suum Hb resulted in rapid autoxidation of this protein (33). For both the ApPgb and MaPgb Pgbs, molecular modeling indicates that, although a Tyr B10 is present, this residue is unlikely to stabilize O2 binding because the lowest-energy orientation of the side chain is parallel to the heme plane rather than directed at the site of ligand coordination. This result is consistent with the high autoxidation rate observed for the Pgbs when exposed to O2 (Fig. 4C).
In summary, we have identified globins from Archaea that bind O2, CO, and NO. Mutagenesis experiments coupled with our 3D homology model of ApPgb indicate that heme is covalently bound to the apo-globin by means of His-120. In addition, the model predicts an intramolecular disulfide bridge aiding in ApPgb thermostability. Moreover, the B10 distal tyrosine is oriented parallel to the heme plane rather than directed toward the bound ligand, decreasing the Pgb's capacity to bind O2. In summary, we conclude that ApPgb and MaPgb are prototypes for contemporary Hbs. They may serve several biological functions, including protection from nitrosative and oxidative stress.
Supplementary Material
Acknowledgments
This work is dedicated to Dr. Oskar Zaborsky. We thank Dr. Yutaka Kawarabayasi (Center for Glycoscience, Advanced Industrial Science and Technology, Japan) for providing the A. pernix genomic DNA, and Dr. William W. Metcalf (Department of Microbiology, University of Illinois at Urbana-Champaign) for providing the M. acetivorans genomic DNA. This investigation was supported by National Science Foundation Grant MCB0080125 and by a University of Hawaii intramural grant (to M.A.) and by U.S. Public Health Service Grant HL-64038 (to M.-A.G.-G.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Mb, myoglobin; FHb, flavohemoglobin; NHb, nerve Hb; trHb, truncated Hb; HemAT, heme-based aerotaxis transducer; Pgb, protoglobin; HHMb, horse-heart Mb; SWMb, sperm-whale Mb; AeFHb, Alcaligenes eutrophus FHb; Hmp, E. coli FHb; VHb, Vitreoscilla stercoraria Hb; LHb, Lupinus luteus leghemoglobin; rLHb, nonsymbiotic rice leghemoglobin; LpHb, Lucina pectinata Hb I; GdHb, Glycera dibranchiata Hb component IV; ClNHb, Cerebratulus lacteus NHb; PtrHb, Paramecium caudatum trHb; HbN, Mycobacterium tuberculosis trHb-N; PCC6803Hb, Synechocystis Sp-PCC6803 trHb; PeHb, Paramphistomum epiclitum Hb; RifA2, Riftia pachyptila extracellular Hb A2; AscarisHb, Ascaris suum Hb; ApPgb, Aeropyrum pernix Pgb; MaPgb, Methanosarcina acetivorans Pgb; CaPgb, Chloroflexus aurantiacus Pgb; TfPgb, Thermobifida fusca Pgb; HemAT-Bc, Bacillus cereus HemAT; HemAT-Bs, Bacillus subtilis HemAT; HemAT-Hs, Halobacterium salinarum HemAT; TeGlb, Thermosynechococcus elongatus globin.
References
- 1.Moens, L., Vanfleteren, J., Van de Peer, Y., Peeters, K., Kapp, O., Czeluzniak, J., Goodman, M., Blaxter, M. & Vinogradov, S. (1996) Mol. Biol. Evol. 13, 324-333. [DOI] [PubMed] [Google Scholar]
- 2.Perutz, M. F (1970) Nature 228, 726-739. [DOI] [PubMed] [Google Scholar]
- 3.Lesk, A. M. & Chothia, C. (1980) J. Mol. Biol. 136, 225-270. [DOI] [PubMed] [Google Scholar]
- 4.Bashford, D., Chothia, C. & Lesk, A. M. (1987) J. Mol. Biol. 196, 199-216. [DOI] [PubMed] [Google Scholar]
- 5.Kapp, O. H., Moens, L., Vanfleteren, J., Trotman, C. N., Suzuki, T. & Vinogradov, S. N. (1995) Protein Sci. 4, 2179-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milani, M., Pesce, A., Ouellet, Y., Ascenzi, P., Guertin, M. & Bolognesi, M. (2001) EMBO J. 20, 3902-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pesce, A., Couture, M., Dewilde, S., Guertin, M., Yamauchi, K., Ascenzi, P., Moens, L. & Bolognesi, M. (2000) EMBO J. 19, 2424-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarricone, C., Galizzi, A., Coda, A., Ascenzi, P. & Bolognesi, M. (1997) Structure 5, 497-507. [DOI] [PubMed] [Google Scholar]
- 9.Hou, S., Freitas, T., Larsen, R. W., Piatibratov, M., Sivozhelezov, V., Yamamoto, A., Meleshkevitch, E. A., Zimmer, M., Ordal, G. W. & Alam, M. (2001) Proc. Natl. Acad. Sci. USA 98, 9353-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, W. & Phillips, G. N., Jr. (2003) Structure 11, 1097-1110. [DOI] [PubMed] [Google Scholar]
- 11.Freitas, T., Hou, S. & Alam, M. (2003) FEBS Lett. 552, 99-104. [DOI] [PubMed] [Google Scholar]
- 12.Hardison, R. (1999) Am. Sci. 87, 126-137. [Google Scholar]
- 13.Piatibratov, M., Hou, S., Brooun, A., Yang, J., Chen, H. & Alam, M. (2000) Biochim. Biophys. Acta 1524, 149-154. [DOI] [PubMed] [Google Scholar]
- 14.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuff, J. A., Clamp, M. E., Siddiqui, A. S., Finlay, M. & Barton, G. J. (1998) Bioinformatics 14, 892-893. [DOI] [PubMed] [Google Scholar]
- 16.Page, R. D. (1996) Comput. Appl. Biosci. 12, 357-358. [DOI] [PubMed] [Google Scholar]
- 17.Zhao, X., Vyas, K., Nguyen, B. D., Rajarathnam, K., La Mar, G. N., Li, T., Phillips, G. N., Jr., Eich, R. F., Olson, J. S., Ling, J. & Bocian, D. F. (1995) J. Biol. Chem. 270, 20763-20774. [DOI] [PubMed] [Google Scholar]
- 18.Couture, M., Yeh, S. R., Wittenberg, B. A., Wittenberg, J. B., Ouellet, Y., Rousseau, D. L. & Guertin, M. Proc. Natl. Acad. Sci. USA. 96, 11223-11228. [DOI] [PMC free article] [PubMed]
- 19.Arrendondo-Peter, R., Hargrove, M. S., Sarath, G., Moran, J. F., Lohrman, J., Olson, J. S. & Klucas, R. V. (1997) Plant Physiol. 115, 1259-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wittenberg, J. B., Wittenberg, B. A., Gibson, Q. H., Trinick, M. J. & Appleby, C. A. (1986) J. Biol. Chem. 261, 13624-13631. [PubMed] [Google Scholar]
- 21.Gilles-Gonzalez, M. A., Gonzalez, G., Perutz, M. F., Kiger, L., Marden, M. C. & Poyart, C. (1994) Biochemistry 33, 8067-8073. [DOI] [PubMed] [Google Scholar]
- 22.Chang, A. L., Tuckerman, J. R., Gonzalez, G., Mayer, R., Weinhouse, H., Volman, G., Amikam, D., Benziman, M. & Gilles-Gonzalez, M. A. (2001) Biochemistry 40, 3420-3426. [DOI] [PubMed] [Google Scholar]
- 23.Duff, S. M., Wittenberg, J. B. & Hill, R. D. (1997) J. Biol. Chem. 272, 16746-16752. [DOI] [PubMed] [Google Scholar]
- 24.Minning, D. M., Gow, A. J., Bonaventura, J., Braun, R., Dewhirst, M., Goldberg, D. E. & Stamler, J. S. (1999) Nature 401, 497-502. [DOI] [PubMed] [Google Scholar]
- 25.Flogel, U., Merx, M. W., Godecke, A., Decking, U. K. & Schrader, J. (2001) Proc. Natl. Acad. Sci. USA 98, 735-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poole, R. K. & Hughes, M. N. (2000) Mol. Microbiol. 36, 775-783. [DOI] [PubMed] [Google Scholar]
- 27.Eich, R. F., Li, T., Lemon, D. D., Doherty, D. H., Curry, S. R., Aitken, J. F., Mathews, A. J., Johnson, K. A., Smith, R. D., Phillips, G. N., Jr., & Olson, J. S. (1996) Biochemistry 35, 6976-6983. [DOI] [PubMed] [Google Scholar]
- 28.Hargrove, M. S., Barry, J. K., Brucker, E. A., Berry, M. B., Phillips, G. N., Jr., Olson, J. S., Arredondo-Peter, R., Dean, J. M., Klucas, R. V. & Sarath, G. (1997) J. Mol. Biol. 266, 1032-1042. [DOI] [PubMed] [Google Scholar]
- 29.Wittenberg, J. B., Bolognesi, M., Wittenberg, B. A. & Guertin, M. (2002) J. Biol. Chem. 277, 871-874. [DOI] [PubMed] [Google Scholar]
- 30.Bailly, X., Jollivet, D., Vanin, S., Deutsch, J., Zal, F., Lallier, F. & Toulmond, A. (2002) Mol. Biol. Evol. 19, 1421-1433. [DOI] [PubMed] [Google Scholar]
- 31.Bailly, X., Leroy, R., Carney, S., Collin, O., Zal, F., Toulmond, A. & Jollivet, D. (2003) Proc. Natl. Acad. Sci. USA 100, 5885-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouellet, H., Ouellet, Y., Richard, C., Labarre, M., Wittenberg, B., Wittenberg, J. & Guertin, M. (2002) Proc. Natl. Acad. Sci. USA 99, 5902-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Baere, I., Perutz, M. F., Kiger, L., Marden, M. C. & Poyart, C. (1994) Proc. Natl. Acad. Sci. USA 91, 1594-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardner, A. M., Martin, L. A., Gardner, P. R., Dou, Y. & Olson, J. S. (2000) J. Biol. Chem. 275, 12581-12589. [DOI] [PubMed] [Google Scholar]
- 35.Das, T. K., Weber, R. E., Dewilde, S., Wittenberg, J. B., Wittenberg, B. A., Yamauchi, K., Van Hauwaert, M. L., Moens, L. & Rousseau, D. L. (2000) Biochemistry 39, 14330-14340. [DOI] [PubMed] [Google Scholar]
- 36.Yeh, S. R., Couture, M., Ouellet, Y., Guertin, M. & Rousseau, D. L. (2000) J. Biol. Chem. 275, 1679-1684. [DOI] [PubMed] [Google Scholar]
- 37.Ouellet, H., Juszczak, L., Dantsker, D., Samuni, U., Ouellet, Y. H., Savard, P. Y., Wittenberg, J. B., Wittenberg, B. A., Friedman, J. M. & Guertin, M. (2003) Biochemistry 42, 5764-5774. [DOI] [PubMed] [Google Scholar]
- 38.Gibson, Q. H., Regan, R., Olson, J. S., Carver, T. E., Dixon, B., Pohajdak, B., Sharma, P. K. & Vinogradov, S. N. (1993) J. Biol. Chem. 268, 16993-16998. [PubMed] [Google Scholar]
- 39.Gibson, Q. H. & Smith, M. H. (1965) Proc. R. Soc. London B Biol. Sci. 163, 206-214. [DOI] [PubMed] [Google Scholar]
- 40.Davenport, H. E. (1949) Proc. R. Soc. London B Biol. Sci. 136, 255-270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.