Abstract
Mdm4 is a critical inhibitor of the p53 tumor suppressor. Mdm4 null mice die early during embryogenesis due to increased p53 activity. In this study, we explore the role that Mdm4 plays in the intestinal epithelium by crossing mice carrying the Mdm4 floxed allele to mice with the Villin Cre transgene. Our data show that loss of Mdm4 (Mdm4intΔ) in this tissue resulted in viable animals with no obvious morphological abnormalities. However, these mutants displayed increased p53 levels and apoptosis exclusively in the proliferative compartment of the intestinal epithelium. This phenotype was completely rescued in a p53 null background. Notably, the observed compartmentalized apoptosis in proliferative intestinal epithelial cells was not due to restricted Mdm4 expression in this region. Thus, in this specific cellular context, p53 is negatively regulated by Mdm4 exclusively in highly proliferative cells.
Keywords: p53 stability, cell cycle arrest, apoptosis, Mdmx
Introduction
The p53 tumor suppressor plays an essential role in inhibiting tumorigenesis by transcriptionally activating genes involved in cell cycle arrest, apoptosis and senescence in abnormal cells (Vousden 2000). However, p53 activity must be kept under control in healthy cells to avoid detrimental effects to the organism such as unnecessary cellular death. Two major players involved in the regulation of p53 activity are Mdm2 and Mdm4.
The Mdm2 oncogene inhibits p53 activity principally by acting as an E3 ubiquitin ligase to target p53 for 26S proteasome-dependent protein degradation (Honda et al 1997, Kubbutat et al 1997). The importance of Mdm2 in regulating p53 function in vivo was demonstrated with the generation of Mdm2 null mice. These mutants die before implantation due to increased apoptosis, while concomitant loss of Mdm2 and p53 completely rescued this phenotype (Chavez-Reyes et al 2003, Jones et al 1995, Montes de Oca Luna et al 1995). Interestingly, Mdm2 is also a transcriptional target of p53 itself and, thus, it participates in a negative feedback loop with p53 (Barak et al 1993, Wu et al 1993). On the other hand, Mdm4 inhibits p53 mainly by physically binding to p53 and interfering with its transcriptional activity (Shvarts et al 1996). Although Mdm4 possesses a RING finger domain in its structure, it cannot ubiquitinate and target p53 for protein degradation (Jackson & Berberich 2000, Sharp et al 1999). Notably, loss of Mdm4 leads to embryonic lethality that can be completely rescued in a p53 null background (Finch et al 2002, Migliorini et al 2002, Parant et al 2001). Together, these data indicate that both Mdm2 and Mdm4 are critical inhibitor of p53 in vivo.
The importance of these genes in specific cell types in vivo has been further demonstrated with the use of Mdm2 and Mdm4 conditional mouse alleles (Grier et al 2006, Grier et al 2002). For example, deletion of Mdm2 or Mdm4 in cells of the central nervous system leads to p53-dependent embryonic lethal phenotypes (Francoz et al 2006, Xiong et al 2006). Simultaneous deletion of both genes in proliferative and quiescent cells of the central nervous system yields a more dramatic phenotype, indicating that these two proteins synergize to inhibit p53 activity in both dividing and non-dividing cells (Francoz et al 2006, Xiong et al 2006). Furthermore, Mdm2 deletion in embryonic cardyomyocytes or in quiescent adult smooth muscle cells also results in lethal phenotypes due to increased p53-dependent apoptosis (Boesten et al 2006, Grier et al 2006), whereas loss of Mdm4 in these two cell types leads to a much weaker or complete absence of cellular defects, respectively (Boesten et al 2006, Grier et al 2006, Xiong et al 2007). Finally, in contrast to the aforementioned studies, deletion of Mdm2 in the intestinal epithelium results in multiple abnormalities in this tissue early in life, yet mice survive due to the ability of the tissue to compensate for cellular death mediated by p53 activation (Valentin-Vega et al 2008). Thus far, these studies together suggest that Mdm2 inhibits p53-activity in both proliferative and fully differentiated cells in all tissues analyzed. However, it appears that Mdm4 is required to restrain p53 activity specifically depending on the cellular context.
To obtain more insights about the role of Mdm4 toward p53 regulation in cells with different differentiation potentials, we sought to delete this oncogene in the intestinal epithelium. The architecture of this organ provides an excellent model to study the function of genes in cells at various stages of differentiation (Sancho et al 2004). The intestinal epithelial-specific Villin Cre mouse model was used in this study as this transgene targets both highly proliferative and differentiated intestinal epithelial cells (Madison et al 2002). Our data show that Mdm4 regulates p53 levels and activity exclusively in the highly proliferative compartment of this tissue. Interestingly, this compartmentalized regulation is not due to a restricted expression of Mdm4 in proliferative intestinal epithelial cells.
Materials and Methods
Mice
All animal protocols were approved by the Institutional Animal Care and Use Committee. All mice were >90% C57BL/6. VilCre mice were kindly provided by Dr. Gumucio (Madison et al 2002). VilCre mice were crossed with Mdm4FX conditional mice (Grier et al 2006). The following primers were used to distinguish between the Mdm4FX and Mdm4 null (Mdm4intΔ) alleles: A, 5'-TAGAATCTGGAATTACAGACAG-3'; B, 5'-TGTCTTTAGCATTTACTAAGAGCT-3'; and C, 5'-TATCCAGTGTCCTCTTCTGGCTT-3' (Grier et al 2006). Intestines of postnatal day 3 (P3) pups were dissected and washed in Phosphate Buffered Saline (PBS), rolled, incubated in 10% formalin for up to 48 hours, and embedded in paraffin. Intestines of 8-week old mice were cut into sections (duodenum, jejunum, ileum, and colon), and luminal contents flushed with cold PBS prior to rolling. Tissues were cut in 4μ-thick sections, stained with hematoxylin and eosin (H&E) and analyzed by pathology.
β-galactosidase activity assays
Postnatal intestines were dissected and quickly washed in cold PBS followed by fixation in 4% paraformaldehyde for 2 hours at 4°C. Then, samples were cryopreserved in sucrose and embedded in Optimum Cutting Temperature compound (Sakura). β-galactosidase activity assays were performed as previously described (Valentin-Vega et al 2008).
Immunohistochemistry, immunofluorescence, TUNEL, and BrdU incorporation assays
Immunohistochemistry and immunofluorescence assays were performed as described, respectively (Evans et al 2001, Valentin-Vega et al 2008). TUNEL assay was performed by using the TdtT Fragel DNA fragmentation kit (Oncogene) as indicated by the company except that the Diaminobenzidine (DAB) substrate supplied in the kit was substituted for the DAB distributed by the Vector Laboratories Company. The following antibodies were used for immunohistochemistry and immunofluorescence studies: rabbit anti-p53 (1:200, CM5, Vector Laboratories), rabbit anti-Caspase-3 (1:100, Cell Signaling,), rabbit anti-Ki-67 (1:50, Abcam), and mouse anti-pan cytokeratin (1:200, Sigma). BrdU incorporation analyses were performed as previously described (Parant et al 2001).
Immunoprecipitation and western blot analyses
Protein extracts were isolated from 3-day old whole intestines using NP-40 lysis buffer with proteinase inhibitors (50 mM Tris-HCL (pH 7.5), 150 mM NaCl, 0.5% NP-40.50 mM NaF, 1 mM Na3VO4, 1 mM DTT, 100 mM PMSF). For immunoprecipitation, 500 µg of total protein were pre-cleared with Protein A/G-conjugated beads (Santa Cruz Biotechnology), and then immunoprecipitated with 4 µg of anti-Mdm4 antibody (mouse monoclonal anti-Mdm4, Sigma) overnight at 4°C. Protein A/G-conjugated beads (Santa Cruz Biotechnology) were added next and samples were incubated for 1 hr at 4°C. Samples were then washed with lysis buffer and beads were collected. Proteins were subjected to 8% SDS-polyacrylamide gel electrophoresis. Mdm4 was detected by western blot using mouse anti-Mdm4 from Sigma (overnight, 1:1000 dilution). 50 µg of total protein was separated by SDS-PAGE to analyze equal loading in gel. Vinculin was used as loading control (mouse anti-Vinculin, 1:2000, Sigma). Protein in film was quantified using the ImageQuant program (Version 5.2, Molecular Dynamics). Mdm4 protein levels were normalized to the value of Vinculin.
In situ hybridization
3-day old pups were sacrificed and intestines removed. Samples were washed in cold PBS, rolled and embedded in OCT. In situ hybridization (ISH) was performed at the Baylor College of Medicine ISH core facility using digoxigenin-labelled Mdm4 probe on 25 µm sections (Yaylaoglu et al 2005).
Quantification of apoptotic and proliferative cells
Quantification of apoptotic bodies in the intervillus compartment of P3 mice was performed by counting the number of apoptotic cells in more than 150 intervillus units per mouse (n = 3 per group). Apoptotic bodies were identified by H&E staining. Similarly, cellular proliferation in P3 mice was quantified by counting the number of Ki-67 positive cells per intervillus unit. At least 200 units were counted per mouse, n ≥ 3 per group).
Reverse transcriptase RT-PCR analysis
Reverse transcriptase real time-PCR analysis of the p53 target gene, p21 was performed as described (Valentin-Vega et al 2008).
Results
Mice lacking Mdm4 in the intestinal epithelium display normal intestinal morphology
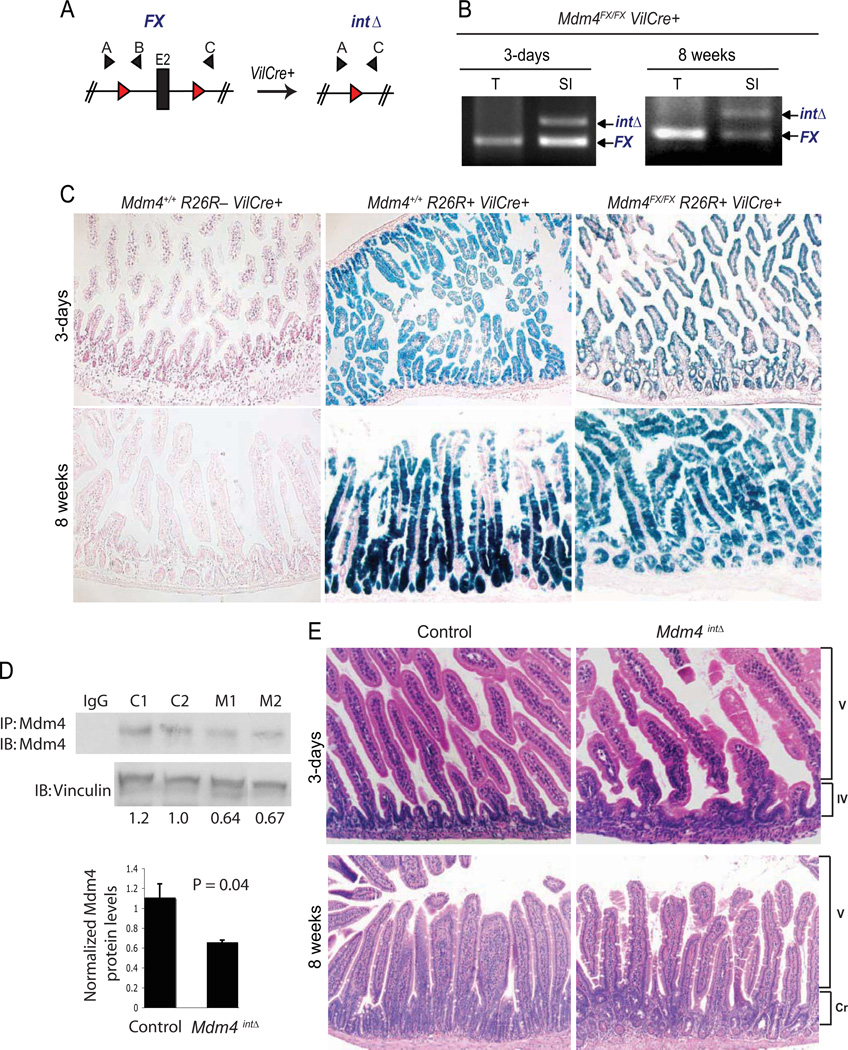
Mice lacking Mdm4 in the whole body die during early embryogenesis, a phenotype that is completely rescued by loss of p53 (Finch et al 2002, Migliorini et al 2002, Parant et al 2001). In order to understand the function of Mdm4 in non-embryonic tissues, an Mdm4 conditional allele (or FX allele) has been generated (Fig. 1A) (Grier et al 2006). Disruption of this conditional Mdm4FX allele in the whole mouse phenocopies the phenotypes observed in the original Mdm4-null mouse (Grier et al 2006). To delete Mdm4 in the intestinal epithelium, the intestinal epithelial-specific Villin Cre (VilCre) transgenic mouse was used in this study (Madison et al 2002). A series of genetic crosses were performed to generate Mdm4FX/Δ2 and Mdm4FX/FX mice with or without VilCre. Since Mdm4 mutants of both genotypes showed equivalent phenotypes, we labeled Mdm4FX/FX VilCre+ or Mdm4FX/Δ2 VilCre+ mice as Mdm4intΔ to simplify the nomenclature. Analysis of progeny at weaning indicates that Mdm4intΔ mice are born at the expected Mendelian ratio (Table 1). To investigate if homologous recombination occurred in the intestine of Mdm4intΔ mice, we first performed a PCR-based analysis using a set of primers that recognize the non-recombined (FX) or the recombined (intΔ) alleles (Fig. 1A). As expected, recombination was observed exclusively in the intestine of both young and adult Mdm4intΔ mice (Fig. 1B). The observed non-recombined alleles may originate from cells carrying the non-recombined allele or from contaminated non-epithelial cells present in the sample. Recombination efficiency was also analyzed by breeding Mdm4intΔ mice to the ROSA26R reporter mouse (Soriano 1999) followed by-galactosidase activity assay. High frequency of recombination was observed in both, highly proliferative and differentiated compartments of the small intestine of 3-day and 8-week old Mdm4FX/FX R26R VilCre+ mice (Fig. 1C). Finally, to investigate whether Mdm4 protein levels were lost in the intestine of Mdm4intΔ mice, we performed IP-western blot analysis against Mdm4 protein. Intestines of Mdm4intΔ mice showed a significant decrease in the protein levels of Mdm4 as compared to control intestines (Fig. 1D). The remaining Mdm4 protein observed in Mdm4intΔ mice could be due to contamination with non-epithelial cells in the sample as whole intestine was used in this experiment. Together, these data indicate that Mdm4 is lost in both proliferative and differentiated intestinal epithelial cells of Mdm4intΔ mice and, thus, loss of Mdm4 in this tissue yields viable animals.
Figure 1. Mice survive with loss of Mdm4 in the intestinal epithelium and display normal intestinal morphology.
(A) Schematic diagram of the non-recombined (FX) and recombined (intΔ) Mdm4 alleles. The Mdm4 conditional allele targets exon 2 (E2), which contains the translation start site. Red triangles represent loxP sites flanking exon 2; Black triangles represent primer sets used to identify the FX and intΔ alleles (primers A and B amplify the non-recombined (FX) allele while primers A and C amplify the recombined (intΔ) allele). (B) PCR analysis in the intestine of 3-day old Mdm4FX/FX VilCre+ mice using primers A, B, and C. SI, small intestine; T, tail. (C) β-galactosidase activity assays in the small intestine of 3-day and 8-week old mice. Blue denotes cells with β-galactosidase activity (original magnification X100). Mdm4+/+ R26R− VilCre+, negative control; Mdm4+/+ R26R+ VilCre+, positive control; Mdm4FX/FX R26R+ VilCre+, Mdm4intΔ mutant. (D) Mdm4 protein expression in 3-day old whole intestines. Mdm4 protein was immunoprecipitated and then detected by western blot analysis. Mdm4 protein levels were quantified and normalized to the value of Vinculin (loading control). Normalized Mdm4 protein levels were graphed and the p-value was calculated using the Student’s t-test. IgG, Immunoglobulin G (negative control); C1, control #1; C2, control #2; M1, Mdm4intΔ mutant #1; M2, Mdm4intΔ mutant #2. (E) H&E staining of the small intestine of 3-day old pups or intestinal jejunum region of 8-week old mice (original magnification X200 or X100, respectively). V, villus region, IV, intervillus region, Cr, crypt compartment. Intervillus and crypt compartments are regions of undifferentiated cells in young and adult intestines, respectively. The villus compartment contains differentiated intestinal epithelial cells.
Table 1.
Analysis of the progeny from Mdm4FX/FX × Mdm4+/Δ2 VilCre+ cross at weaning
| Genotype | Number of mice n=186 |
|---|---|
| Mdm4FX/+ VilCre− | 47 |
| Mdm4FX/+ VilCre+ | 46 |
| Mdm4FX/Δ2 VilCre− | 49 |
| Mdm4FX/Δ2 VilCre+ | 44 |
Abreviations: Mdm4Δ2, Mdm4-null allele; Mdm4FX, Mdm4 conditional allele; n, total number of mice analyzed, Chi square, p = 0.969
As Mdm4intΔ mice survive, we next examined the intestinal morphology of these mutants. We did not detect obvious morphological defects in the intestine of 3-day old Mdm4intΔ pups (Fig. 1E). Similarly, analysis of the intestinal epithelium of adult (8-week old) mice also showed no significant morphological changes in the intestine of Mdm4intΔ mutants (Fig. 1E). Furthermore, we aged Mdm4intΔ mice and observed that these mutants display a normal lifespan as compared to control littermates (data not shown). Collectively, depletion of Mdm4 in the intestinal epithelium does not significantly alter the architecture of this tissue.
Absence of Mdm4 results in increased p53-dependent apoptosis exclusively in highly proliferative cells of the small intestine
Since Mdm2 is an E3 ubiquitin ligase that targets p53 for 26S proteosome-dependent protein degradation (Honda et al 1997, Kubbutat et al 1997), loss of Mdm2 in cells always results in high p53 protein levels (Boesten et al 2006, Francoz et al 2006, Grier et al 2006, Xiong et al 2006). On the other hand, Mdm4 does not directly affect p53 degradation but interacts with Mdm2 to regulate p53 function (Sharp et al 1999, Stad et al 2000). To elucidate the role of Mdm4 in regulating p53 activity in the intestinal epithelium, we first investigated if absence of Mdm4 affects p53 protein levels in this tissue. p53 immunohistochemistry revealed that Mdm4intΔ mice showed increased p53 levels exclusively in the proliferative compartment (Fig. 2A). High levels of p53 in the absence of Mdm4 have been previously reported only in cells of the central nervous system (Xiong et al 2006). The apoptotic response was then examined in the intestines of Mdm4intΔ mice. 3-day old Mdm4intΔ mice displayed a significant increased in apoptotic response in the intestinal epithelium as compared to control animals (control, 0.053 ± 0.029; Mdm4intΔ, 0.573 ± 0.22; t-Test, P = 0.01; 200 intervillus units scored per mouse; n = 3 per group) (Fig. 2B–D). Like 3-day old mutants, adult Mdm4intΔ mice also showed a comparable increase in apoptosis in all regions of the intestinal epithelium (duodenum, jejunum, etc.) (data not shown). Interestingly, in contrast to 3-day old Mdm2intΔ mice that exhibit apoptosis in both prolifeartive and differentiated cells of the intestinal epithelium (Valentin-Vega et al 2008), 3-day old Mdm4intΔ mice showed a restricted apoptotic response in the proliferative compartment of this tissue (Fig. 2B–C). Collectively, loss of Mdm4 in the intestinal epithelium results in increased apoptotic response exclusively in highly proliferative cells of the intestinal epithelium.
Figure 2. Mdm4 regulates p53 specifically in highly proliferative cells of the small intestine.
(A) Immunohistochemistry against p53 in the small intestine of 3-day old mice (original magnification X200). Box shows higher magnification of the intervillus (proliferative) region. (B) Caspase-3 immunofluorescence in 3-day old small intestines. Green staining shows caspase-3 positive cells (original magnification X400). (C) TUNEL assay performed in 3-day old small intestines. Black staining illustrates TUNEL positive cells (original magnification X200). (D) Quantification of apoptotic cells in the intervillus pockets of 3-day old small intestines scored by H&E (n = 3 per group). Mdm4intΔ p53−/− genotype represents mice with loss of Mdm4 in the intestine that also lack p53 in the whole body. V, villus (differentiated) region, IV, intervillus (proliferative) region.
To investigate if the increased apoptotic response observed in Mdm4intΔ mice is p53-dependent, we bred these mutants into a p53-null background. As expected, a complete rescue of the apoptotic response was observed in neonatal Mdm4intΔ mice that simultaneously lack p53 (Mdm4intΔ p53−/−) (Fig. 2D). These data indicate that the increased apoptotic response observed in the absence of Mdm4 in the intestinal epithelium is dependent on p53 activity.
Previous studies have shown that unlike Mdm2, loss of Mdm4 in the whole mouse or in the central nervous system results in p53-mediated cell cycle arrest (Francoz et al 2006, Migliorini et al 2002, Parant et al 2001, Xiong et al 2006). To investigate if p53 also triggers cell cycle arrest in the intestinal epithelium in the absence of Mdm4, we first performed immunofluorescence against the proliferative marker, Ki-67. Our data showed no difference in Ki-67 positive cells between Mdm4intΔ and control mice (control, 10.4 ± 0.189; Mdm4intΔ, 9.72 ± 0.858; t-Test, P = 0.231; 200 intervillus units scored per mouse; n = 3 per group) (Fig. 3A). Additionally, we performed BrdU incorporation analyses in 8-week old mouse intestines and again observed no significant difference in the number of BrdU labeled cells between both groups (control, 13.21 ± 0.799; Mdm4intΔ, 15.5 ± 0.14; t-Test, P = 0.06; 100 intervillus units scored per mouse; n = 3 per group) (Fig. 3B).
Figure 3. Cellular proliferation is intact in the small intestine of Mdm4intΔ mice.
(A) Ki-67 immunofluorescence in 3-day old small intestines. Orange fluorescence, cytokeratin (epithelial marker); turquoise fluorescence, Ki-67 (proliferation marker); blue fluorescence, TOPRO-3 (nuclear counterstain); original magnification X400. Graph represents the quantification of the Ki-67 positive staining. (B) Analysis of BrdU incorporation in 8-week old mouse intestines. Mice were injected with 100 µg BrdU/g body weight and sacrificed 2 hr later. BrdU labeled cells (brown) were quantified and values were graphed. The p-value was obtained using Student’s t-test (n = 3 per group); original magnification X200. (C) Reverse transcription RT-PCR analysis of the p53 target, p21. Five mice per group were analyzed and the fold change in p21 mRNA levels was graphed. The p-value was obtained using Student’s t-test.
We also analyzed the mRNA levels of the p53 target gene, p21, which is involved in cell cycle arrest. Interestingly, even though the proliferation capacity of the intestinal epithelium in Mdm4intΔ mice was intact, we detected a significant increase in p21 mRNA levels in the mutant intestines (control, 1.05 ± 0.086; Mdm4intΔ, 2.38 ± 0.19; t-Test, P = 0.03; n = 5 per group (Fig. 3C). In summary, in contrast to embryonic tissues and cells of the central nervous system, loss of Mdm4 in the intestinal epithelium did not lead to defects in the proliferation capacity of these cells despite the increase in p21 mRNA levels. Thus, Mdm4 does not regulate p53-dependent cell cycle arrest in this cellular context.
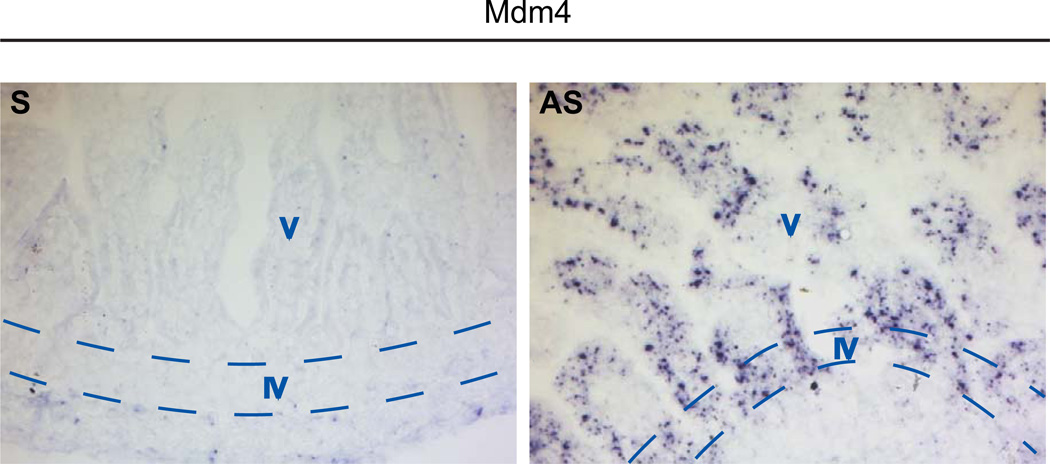
Increase in p53-dependent apoptosis exclusively in the proliferative compartment of 3-day old Mdm4intΔ mice correlated with the expression pattern of p53 protein in these mutants (compare Fig. 2A with Fig. 2B–C). A possible explanation for this phenomenon is that in the small intestine, Mdm4 is expressed only in highly proliferative cells. To investigate this possibility, we performed in situ hybridization against Mdm4 mRNA in 3-day old wild-type intestine given that Mdm4 is not detectable at the protein level in vivo. Unexpectedly, Mdm4 mRNA was detected in both proliferative and differentiated compartments of the intestinal epithelium (Fig. 4). This finding indicates that the expression pattern of Mdm4 is not responsible for the regulation of p53 exclusively in the intervillus compartment.
Figure 4. Mdm4 RNA is expressed in the entire intestinal epithelium of 3-day old mice.
In situ hybridization against Mdm4 in 3-day old small intestines of wild-type mice. Black dots represent deposition of Mdm4 RNA in the tissue. S, sense probe; AS, anti-sense probe (original magnification X200). V, villus (differentiated) region, IV, intervillus (proliferative) region.
Discussion
The data presented in this study indicate that like Mdm2, Mdm4 is also an important modulator of the p53 tumor suppressor in the intestinal epithelium. Importantly, while Mdm2 regulates p53 in both highly proliferative and differentiated intestinal epithelial cells (Valentin-Vega et al 2008), Mdm4 regulates p53 exclusively in the proliferative compartment of this tissue. Interestingly, our results demonstrate that this compartmentalized regulation is not due to a restricted expression of Mdm4 in the intestinal epithelium as Mdm4 RNA was clearly detected in both highly proliferative and differentiated cells of the tissue. Perhaps, posttranslational modifications of Mdm4 are necessary to achieve its full activation and inhibition of p53 in specific cell populations of the intestine.
An interesting observation from this study is that, in contrast to mice lacking Mdm2 in the intestinal epithelium (Valentin-Vega et al 2008), Mdm4intΔ mice did not display morphological defects in this organ despite the increase in p53-dependent apoptosis. The intestinal epithelium is one of the most dynamic and highly proliferative organs in the body (Sancho et al 2004). Therefore, it is likely that newly formed cells are replacing the dying ones. Consequently, the morphology of the tissue is preserved in Mdm4intΔ mice. However, mice lacking Mdm2 in the intestine display a drastic morphological defect probably because the resultant apoptotic index is much higher than in the absence of Mdm4. In these mutants, the intestinal epithelium needs to increase its proliferation potential to compensate for cellular loss and guarantee organismal survival (Valentin-Vega et al 2008).
Noticeably our data, together with previous reports, indicate that Mdm2 is not only a stronger p53 inhibitor than Mdm4 in all cell types analyzed (Boesten et al 2006, Francoz et al 2006, Grier et al 2006, Xiong et al 2007, Xiong et al 2006), but it also regulates p53 activity in both proliferative and differentiated cells whereas Mdm4-mediated p53 regulation appears to be tissue specific. For example, while Mdm4 is required to modulate p53 activity in both proliferative and quiescent cells of the central nervous system (Francoz et al 2006, Xiong et al 2006), it is dispensable for regulating p53 in quiescent smooth muscle cells and adult red blood cells (Boesten et al 2006, Maetens et al 2007). In contrast, mice with loss of Mdm4 in cardyomyocytes do not show developmental defects, but acquire heart defects during adulthood indicating that Mdm4 is important in fully differentiated cardyomyocytes (Xiong et al 2007). In summary, these data suggest that Mdm2 is the major inhibitor of p53 function in vivo, while Mdm4 may only be required to regulate p53 in certain cell types at specific time points.
An important observation from our study is that loss of Mdm4 in the intestinal epithelium leads to increased p53 levels in the proliferative compartment exclusively, yet Mdm4 is not an E3 ubiquitin ligase (Jackson & Berberich 2000, Sharp et al 1999). It has been demonstrated in vitro that Mdm4 and Mdm2 interact through their RING finger domains, which results in Mdm2 stabilization due to the inability of Mdm2 to self-ubiquitinate (Sharp et al 1999, Stad et al 2000). In this system, we could not detect Mdm2 protein levels (data not shown) and could not therefore address this possibility. However, Mdm2 haplo-insufficiency (a 2-fold difference) affects p53-dependent apoptosis and transformation (Terzian et al 2007). Thus, it is possible that loss of Mdm4 causes self-ubiquitination and degradation of Mdm2 and subsequently results in p53 stabilization in this cell type.
Elucidating the tissue-specific functions of the p53/Mdm2/Mdm4 pathway has important therapeutic implications. Tumor cells that retain wild type p53 might be treated by interfering with the Mdm2/p53 or Mdm4/p53 complexes to activate p53 in such cells. Recently, molecules such as Nutlin-3 and Rita have been identified and shown to disrupt the association between Mdm2 and p53 resulting in death of cancer cell lines and xenografts (Issaeva et al 2004, Sarek et al 2007, Tovar et al 2006, Vassilev et al 2004). However, given that Mdm2 affects a wide variety of cell types, the administration of such therapeutic molecules in cancer patients might be detrimental for the organism since it might target normal cells as well. Administration of Mdm4 inhibitors instead might be more beneficial for the individual since Mdm4 is a less potent p53 inhibitor in normal cells in vivo. A recent study shows that although retinoblastoma tumors generally lack p53 mutations, the activity of wild type p53 is frequently abrogated by the ability of the tumor to select for cells with high Mdm4 expression (Laurie et al 2006). This study suggests that indeed Mdm4 might serve as a good chemotherapeutic target for retinoblastoma patients. As Mdm4 inhibitors have not yet been identified, it would be worth to put more efforts on the development of these novel anti-cancer drugs.
Acknowledgements
This study was supported by NIH grant CA47296 to GL. YAVV was partially supported by the Schissler, and Sowell-Huggins Foundations. We thank Drs. Shunbin Xiong and Susan Henning for guidance. We also thank the Baylor College of Medicine ISH core facility for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. Embo J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesten LS, Zadelaar SM, De Clercq S, Francoz S, van Nieuwkoop A, et al. Mdm2, but not Mdm4, protects terminally differentiated smooth muscle cells from p53-mediated caspase-3-independent cell death. Cell Death Differ. 2006;13:2089–2098. doi: 10.1038/sj.cdd.4401973. [DOI] [PubMed] [Google Scholar]
- Chavez-Reyes A, Parant JM, Amelse LL, de Oca Luna RM, Korsmeyer SJ, Lozano G. Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer Res. 2003;63:8664–8669. [PubMed] [Google Scholar]
- Evans SC, Viswanathan M, Grier JD, Narayana M, El-Naggar AK, Lozano G. An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene. 2001;20:4041–4049. doi: 10.1038/sj.onc.1204533. [DOI] [PubMed] [Google Scholar]
- Finch RA, Donoviel DB, Potter D, Shi M, Fan A, et al. mdmx is a negative regulator of p53 activity in vivo. Cancer Res. 2002;62:3221–3225. [PubMed] [Google Scholar]
- Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A. 2006;103:3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol. 2006;26:192–198. doi: 10.1128/MCB.26.1.192-198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier JD, Yan W, Lozano G. Conditional allele of mdm2 which encodes a p53 inhibitor. Genesis. 2002;32:145–147. doi: 10.1002/gene.10066. [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. Mol Cell Biol. 2000;20:1001–1007. doi: 10.1128/mcb.20.3.1001-1007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- Maetens M, Doumont G, Clercq SD, Francoz S, Froment P, et al. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood. 2007;109:2630–2633. doi: 10.1182/blood-2006-03-013656. [DOI] [PubMed] [Google Scholar]
- Migliorini D, Denchi EL, Danovi D, Jochemsen A, Capillo M, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22:5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- Sancho E, Batlle E, Clevers H. Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- Sarek G, Kurki S, Enback J, Iotzova G, Haas J, et al. Reactivation of the p53 pathway as a treatment modality for KSHV-induced lymphomas. J Clin Invest. 2007;117:1019–1028. doi: 10.1172/JCI30945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DA, Kratowicz SA, Sank MJ, George DL. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem. 1999;274:38189–38196. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. Embo J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stad R, Ramos YF, Little N, Grivell S, Attema J, et al. Hdmx stabilizes Mdm2 and p53. J Biol Chem. 2000;275:28039–28044. doi: 10.1074/jbc.M003496200. [DOI] [PubMed] [Google Scholar]
- Terzian T, Wang Y, Van Pelt CS, Box NF, Travis EL, Lozano G. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol. 2007;27:5479–5485. doi: 10.1128/MCB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega YA, Okano H, Lozano G. The intestinal epithelium compensates for p53-mediated cell death and guarantees organismal survival. Cell Death Differ. 2008;15:1772–1781. doi: 10.1038/cdd.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vousden KH. p53: death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Xiong S, Van Pelt CS, Elizondo-Fraire AC, Fernandez-Garcia B, Lozano G. Loss of Mdm4 results in p53-dependent dilated cardiomyopathy. Circulation. 2007;115:2925–2930. doi: 10.1161/CIRCULATIONAHA.107.689901. [DOI] [PubMed] [Google Scholar]
- Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci U S A. 2006;103:3226–3231. doi: 10.1073/pnas.0508500103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaylaoglu MB, Titmus A, Visel A, Alvarez-Bolado G, Thaller C, Eichele G. Comprehensive expression atlas of fibroblast growth factors and their receptors generated by a novel robotic in situ hybridization platform. Dev Dyn. 2005;234:371–386. doi: 10.1002/dvdy.20441. [DOI] [PubMed] [Google Scholar]