Abstract
Background
Developments in biotechnology and genomics have increased the focus of biostatisticians on prediction problems. This has led to many exciting developments for predictive modeling where the number of variables is larger than the number of cases. Heterogeneity of human diseases and new technology for characterizing them presents new opportunities and challenges for the design and analysis of clinical trials.
Purpose
In oncology, treatment of broad populations with regimens that do not benefit most patients is less economically sustainable with expensive molecularly targeted therapeutics. The established molecular heterogeneity of human diseases requires the development of new paradigms for the design and analysis of randomized clinical trials as a reliable basis for predictive medicine[1, 2].
Results
We have reviewed prospective designs for the development of new therapeutics with candidate predictive biomarkers. We have also outlined a prediction based approach to the analysis of randomized clinical trials that both preserves the type I error and provides a reliable internally validated basis for predicting which patients are most likely or unlikely to benefit from the new regimen.
Conclusions
Developing new treatments with predictive biomarkers for identifying the patients who are most likely or least likely to benefit makes drug development more complex. But for many new oncology drugs it is the only science based approach and should increase the chance of success. It may also lead to more consistency in results among trials and has obvious benefits for reducing the number of patients who ultimately receive expensive drugs which expose them risks of adverse events but no benefit. This approach also has great potential value for controlling societal expenditures on health care. Development of treatments with predictive biomarkers requires major changes in the standard paradigms for the design and analysis of clinical trials. Some of the key assumptions upon which current methods are based are no longer valid. In addition to reviewing a variety of new clinical trial designs for co-development of treatments and predictive biomarkers, we have outlined a prediction based approach to the analysis of randomized clinical trials. This is a very structured approach whose use requires careful prospective planning. It requires further development but may serve as a basis for a new generation of predictive clinical trials which provide the kinds of reliable individualized information which physicians and patients have long sought, but which have not been available from the past use of post-hoc subset analysis.
INTRODUCTION
Today is an exciting time for cancer research. Developments in biotechnology and genomics have provided tools for understanding cancer biology and identification of important molecular targets. It has become clear that cancers of the same primary site and stage are diverse in terms of their oncogenesis, pathogenesis and responsiveness to therapy. It is also an exciting time for biostatistics and statistical bioinformatics. Large clinical trials to identify small average treatment effects in heterogeneous groups of patients have resulted in practice standards in which many patients are treated with toxic drugs to which they do not benefit. Biostatisticians now have the opportunity to develop new approaches to clinical trial design and analysis that enables a new era of predictive medicine in which appropriate treatments can be matched to appropriate patients in a reliable manner.
The challenge of developing new statistical methods for prediction problems where the number of candidate variables (p) is much greater than the number of cases (n) has been much discussed. Many statistical methods address problems of inference rather than prediction and even prediction methods such as regression analysis are often used for purposes of inference. The objective of developing a prediction model should be, however, to predict accurately with independent data, not to make inferences about which variables are statistically significant. Statistical significance and measures of association such a hazard ratios or odds ratios are not appropriate measures of prediction accuracy[3]. Many important prediction methods that have been developed are not directly applicable to p>n problems[4]. It is not just that technical problems arise, such as that the sample covariance matrix becomes singular; but there are also more fundamental problems of how to avoid over-fitting and how to evaluate model performance. Standard approaches to model development based on maximal likelihood estimates of regression coefficients, selecting variables using stepwise regressions or using the sample correlation matrix often lead to over-fitting and poor prediction accuracy in p>n problems. For p>n problems one can no longer use the concept of goodness of fit to judge model adequacy. For example, in p>n binary classification problems it is almost always possible to find a hyper-plane that separates the classes perfectly in the data used for model development. Consequently it becomes essential to separate the data used for model development from the data used for evaluating model prediction accuracy using either simple sample splitting or a re-sampling method based on cross-validation or the bootstrap[5].
The new challenges with regard to clinical trial design have received less attention. The standard paradigm for the design of phase III oncology clinical trials prescribes use of broad eligibility criteria and basing conclusions on the test of the overall null hypothesis that the average treatment effect is zero. The emphasis on broad eligibility criteria has been based on concern that drugs found effective in clinical trials might subsequently be used in broader patient populations[1, 2]. Some clinical trials even abandoned formal eligibility criteria in favor of the “uncertainty principle” which stated that if the individual physician was uncertain about which treatment might be better for a patient, then that patient was eligible [6]. The focus on the overall null hypothesis was based on concern about the multiple testing involved in the commonly practiced exploratory post-hoc subset analysis and the assumption that qualitative interactions are unlikely [6, 7]. The advice was perform subset analyses, but don’t believe them and the famous subset analysis of the ISIS-2 trial based on patient astrological sign is still prominent in the minds of many statisticians [8].
Developments in cancer biology have raised important questions about some aspects of the traditional approach to conducting clinical trials in oncology. Tumors of the same stage and primary site often differ in key ways and so emphasis on representativeness and assumptions that qualitative interactions are unlikely become less compelling. Today we are challenged to develop a paradigm of clinical trial design and analysis that enables development of a predictive medicine that is science based and reliable. Most of the oncology drugs being developed have intended molecular targets and the traditional diagnostic classification schemes include patients whose tumors are and are not driven by deregulation of those targets. For some drugs, the targets are well understood and there is a compelling biological basis for focusing development on a restricted subset of patients whose tumors are characterized by deregulation of the drug target. For other drugs there is more uncertainty about the target, and how to measure whether the target is driving tumor invasion/progression in an individual patient[9]. It is clear that the primary analysis of the new generation of oncology clinical trials must in many cases consist of more than just testing the null hypothesis of no average effect. But it is also clear that the tradition of post-hoc data dredging subset analysis is not an adequate basis for predictive oncology. We need prospective analysis plans that provide for both preservation of the type I experiment-wise error rate and for focused predictive analyses that can be used to reliably select patients in clinical practice for use of the new regimen. These two objectives are not inconsistent, and clinical trials should be sized for both purposes.
The following sections summarize some of the designs my colleagues and I have been developing for the new generation of cancer clinical trials. Developing new treatments with companion diagnostics or predictive biomarkers for identifying the patients who benefit does not make drug development simpler, quicker or cheaper as is sometimes claimed. Actually it makes drug development more complex and probably more expensive. But for many new oncology drugs it is the only science based approach and should increase the chance of success. It may also lead to more consistency in results among trials and has obvious benefits for reducing the number of patients who ultimately receive expensive drugs which expose them risks of adverse events but no benefit. This approach also has great potential value for controlling societal expenditures on health care.
Prognostic and Predictive Biomarkers
Because of the complexity of cancer biology and because of the gap between where basic research leaves off and clinical development starts, development of a new drug with predictive biomarkers for identifying the patients most or least likely to benefit from the drug is often complex. A recent example of both the developmental complexity and the economic impact of predictive biomarkers was the recent finding that the anti-EGFR antibodies that were approved for the treatment of advanced colorectal cancer are not effective for patients with K-ras mutations[10]. Although the result was biologically compelling, the hypothesis was not developed until the phase III trials of the antibodies were either underway or completed and this complicated their assessment. Nevertheless, the results based on re-analysis of multiple previously conducted randomized clinical trials were compelling and a press release from the American Society of Clinical Oncology (ASCO) indicated that using K-ras mutation testing to inform the use of one of the anti-EGFR antibody, cetuximab, was expected to save over 600 million dollars per year in the US alone.
Both predictive biomarkers and prognostic biomarkers can be useful for informing treatment decisions. A “prognostic biomarker,” is a biological measurement made before treatment to indicate long-term outcome for patients either untreated or receiving standard treatment. Prognostic biomarkers can be useful for identifying patients who have a good enough prognosis with standard therapy (e.g. surgery) that they don’t require cytotoxic chemotherapy. For example, the Oncotype DX and MammaPrint gene expression signatures are used to identify patients with stage I, estrogen receptor positive breast cancer who have a very low recurrence risk without chemotherapy[11, 12]. A “predictive biomarker,” is a biological measurement made before treatment to identify which patient is likely or unlikely to benefit from a particular treatment. Prognostic or predictive classifiers can be based on a single gene or protein measurements, such as HER-2 amplification or K-ras mutation, or based on a score that summarize the expression level of multiple genes. In the following sections we will discuss some of the issues in designing therapeutic clinical trials with candidate predictive biomarker classifiers.
Design Issues
The standard clinical trial approaches are based on the assumption that qualitative treatment by subset interactions are unlikely. This means that if the new treatment is better than control for one type of eligible patient, then the new treatment will be better than control for the other subsets of eligible patients although the degree of improvement in outcome may differ. Most cancer drugs being developed today are targeted at the protein products of specific deregulated genes. Although the biology of drug disease interactions is often incompletely understood, the heterogeneity of cancers of the same primary site results in qualitative treatment by subset interactions being likely. Consequently, the basic underpinning of doing broad-based clinical trials and then doing post-hoc subset analyses, but not believing them, no longer really is what we should be doing. How then can we develop new drugs in a manner more consistent with modern tumor biology and obtain reliable information about what regimens work for what kind of tumors and have a greater chance of success of showing that the drugs really do work for the right patient?
The ideal approach is prospective drug development with a companion diagnostic[13]. This approach, which is being used extensively today in oncology involves (i) Development of a promising completely specified genomic classifier using pre-clinical and early phase clinical studies. The classifier may be based on mutation or amplification of a single gene, over-expression of a single protein, or a composite index incorporating the levels of expression of multiple RNA transcripts. (ii) Development of an analytically validated test for measurement of that classifier. Analytically validated means that the test accurately measures what it is supposed to measure, or if there is no gold-standard measurement, that the test is reproducible and robust. (iii) Use of that completely specified classifier and analytically validated test to design and analyze a new clinical trial to evaluate the effectiveness of that drug and how the effectiveness relates to the classifier. The guiding principle is that the data used to develop the classifier should be distinct from the Phase III data used to test hypotheses about treatment effects and subsets determined by the classifier as distinct from post-hoc assessment. This is in contrast to doing a trial with everything measured and then analyzing it to death and deriving conclusions that are not credible to the investigators of the scientific community.
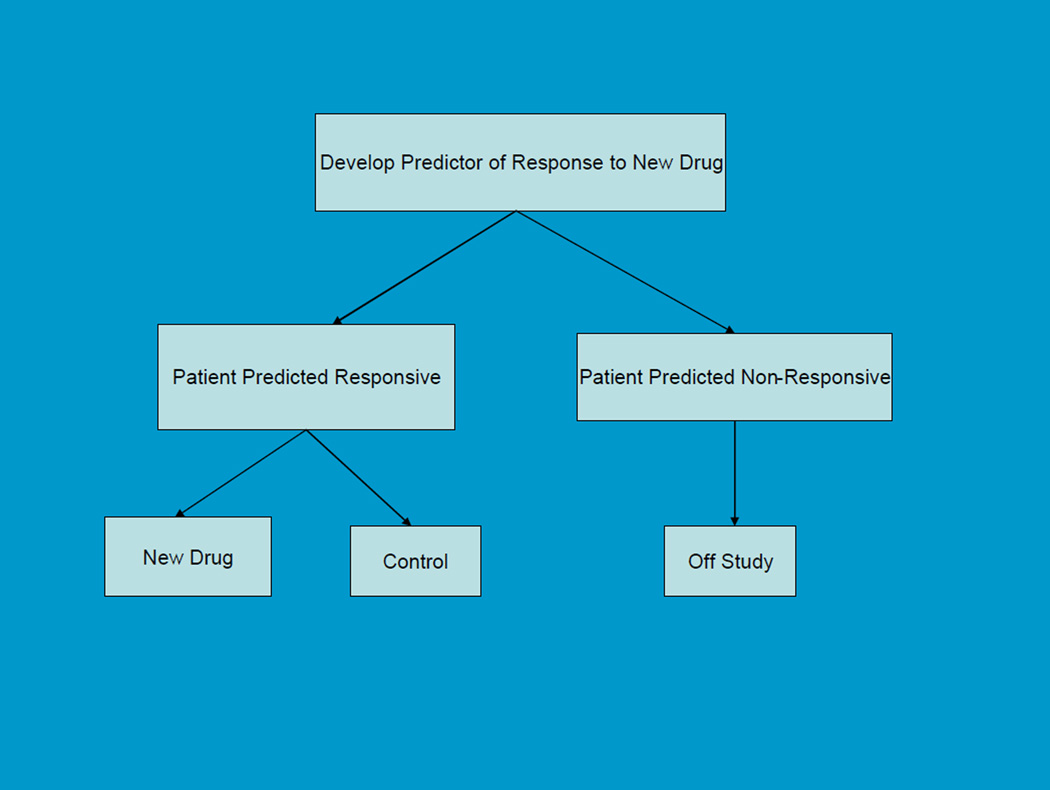
In cases where there is compelling biological or phase II data that the biomarker identifies patients who are very unlikely to benefit from the new treatment, the phase III randomized clinical trial may exclude such patients (Figure 1). Maitournam and Simon [14–16] have shown that this approach can dramatically reduce the number of randomized patients needed compared to the traditional approach with broad eligibility and focus on the overall test of average treatment effect for all randomized patients even when the test is quite imperfect.. This was the approach successfully used for the development of Herceptin for breast cancer where it reduced the required sample size by an order of magnitude. With a strong biological basis for the biomarker, it may be unacceptable to expose test-negative patients to the new drug. With this “targeted” or “enrichment” design, analytical validation of test accuracy or reproducibility, biological rationale, and perhaps phase 2 data provide the basis for regulatory approval of the test if that is required. We have developed web-based sample size planning tools, that enable users to evaluate the number of patients needed to randomize and to screen in using the targeted enrichment design compared to the traditional unselected design as a function of the performance of the test and the selectivity of the treatment (http://brb.nci.nih.gov).
Figure 1.
Targeted Design
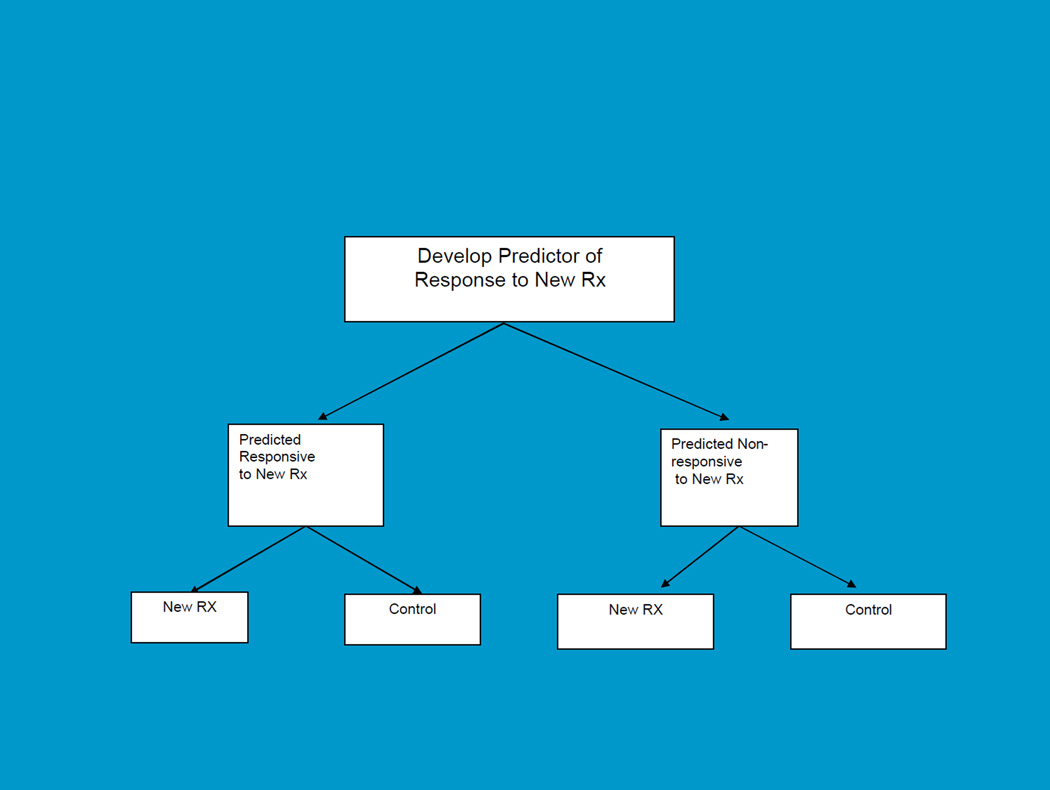
For settings where there is a candidate biomarker but not sufficient basis for using it to restrict eligibility, the biomarker stratified design is more appropriate. This is shown in Figure 2. In this case the RCT comparing the new treatment to control includes both test-positive and test-negative patients, but a prospective primary analysis plan stipulating how the test will be used in the analysis of treatment effect is defined in the protocol. It’s not just a matter of using stratification to balance the randomization. Stratifying the randomization is useful primarily because it ensures that you actually have the test done on all the patients in order for them to get into the trial. What really is crucial, however, is having a prospective analysis plan that defines how the test result will be used in the primary analysis, how the experiment-wise type 1 error will be controlled, and how the trial will be sized for these primary objectives. The purpose of the trial is to have the biomarker used as part of the primary analysis, not relegated to exploratory hypothesis generation. The purpose of the trial is also not to modify, optimize or re-develop the classifier if it is based on a gene expression signature.
Figure 2.
Biomarker Stratified Design
These stratified designs can have several kinds of analysis plans. These have been discussed in more detail elsewhere[17, 18]. These plans can be investigated for specific clinical trials using the web-based software on our website described above. One of the analysis plans is a “fall-back” analysis for situations where one does not have strong confidence in the biomarker. This analysis plan involves a two-step analysis. The first step is an overall test of average treatment effect for all randomized patients, but the threshold of significance used (α1) is less than the traditional 0.05 two sided level; e.g. 0.03. If the overall test is not significant at that reduced level (α1), then one pre-defined subset analysis in the biomarker-positive patients at the reduced significance level 0.05 - α1 (e.g. 0.02) is permitted so that the overall type 1error is preserved, at the 0.05 level. Wang et al. [19] have pointed out that this proposal is conservative and the levels used can be sharpened by taking into consideration the correlation between the two tests. In taking the correlation into account, however, one should pay attention to the structure of the testing procedure. The subset test is done only when the overall test is not significant. Doing the subset test should not be predicated on having the overall test significant at a pre-specified level as that would not be appropriate for the context in which lack of benefit of the new treatment for test negative patients should have no bearing on the potential for substantial benefit for a minority of test positive patients.
The web based programs provide sample size planning for the various analysis plans provided. For example, suppose that the trial is planned for having 90% power for detecting a uniform 33% reduction in overall hazard using a two-sided significance level of .03. Then 297 events are required instead of the 256 events needed for a similar trial based on a two-sided significance threshold of .05. If, however, the overall test of treatment effect is not significant at the .03 level, the test positive subset will include approximately 75 events if only 25% of the patients are test positive and the event rates in the two groups are equal. In that case, the test of treatment effect performed at the two-sided .02 level for the test positive patients has power .75 for detecting a 50% reduction in hazard. By delaying the treatment evaluation in the test positive patients, 80% power can be achieved when there are 84 events and 90% power when there are 109 events in the test positive subset..
Adaptive Designs
The prospective drug and companion diagnostic test approach is being used today in the development of many new cancer drugs where the biology of the drug target is well understood. Because of the complexity of cancer biology, however, there are many cases in which the biology of the target is not well understood at the time that the phase III trials are initiated. We have been developing adaptive designs for these settings. The designs are adaptive, not with regard to sample size or randomization ratio, but rather with regard to the subset in which the new treatment is evaluated relative to the control.
For example with the adaptive threshold design we assumed that a predictive biomarker score was prospectively defined in a randomized clinical trial comparing a new treatment T to a control C. The score is not used for restricting eligibility and no cut-point for the score is prospectively indicated. A fallback analysis begins as described above by comparing T to C for all randomized patients using a significance threshold α1, say .03, less than the traditional .05. If the treatment effect is not significant at that level, then one finds the cut-point s* for the biomarker score which leads to the largest treatment effect in comparing T to C restricted to patients with score greater than s*. Jiang et al. [20] employed a log-likelihood measure of treatment effect and let L* denote the log-likelihood of treatment effect when restricted to patients with biomarker level above s*. The null distribution of L* was determined by repeating the analysis after permuting the treatment and control labels a thousand or more times. If the permutation statistical significance of L* is less than .05-α1 (e.g. .02), then treatment T is considered superior to C for the subset of the patients with biomarker level above s*. Jiang et al. provided bootstrap confidence intervals for s*. They provided an approach to sample size planning for a trial based on this fallback strategy and also upon a more powerful strategy that does not utilize a portion of the total type I error for a test of the overall null hypothesis of average treatment effect.
The analysis plan used in the adaptive threshold design is based on computing a global test based on a maximum test statistic. For the adaptive threshold design, the maximum is taken over the set of cut-points of a biomarker score. The idea of using a global maximum test statistic is much more broadly applicable, however. For example, suppose multiple candidate binary tests, B1, …, BK are available at the start of the trial. These tests may or may not be correlated with each other. Let Lk denote the log-likelihood of treatment effect for comparing T to C when restricted to patients positive for biomarker k. Let L* denote the largest of these values and let k* denote the test for which the maximum is achieved. As for the adaptive threshold design, the null distribution of L* can be determined by repeating the analysis after permuting the treatment and control labels a thousand or more times. If the permutation statistical significance of L* is less than .05-α1 (e.g. .02), then treatment T is considered superior to C for the subset of the patients positive for biomarker test k*. The stability of the indicated set of patients who benefit from T (i.e. k*) can be evaluated by repeating the computation of k* for bootstrap samples of patients.
Freidlin and Simon[21] also published an adaptive signature design based on the fallback design. If the overall treatment effect is not significant at a reduced level α1, the patients in the clinical trial are partitioned into a training set and a validation set. A classifier is developed in the training set. The classifier identifies the patients who appear to benefit from the new treatment T compared to the control C. Freidlin and Simon provided methods for developing this classifier based on whole genome transcript expression data, but the analysis approach can be used much more broadly. For example, the training set can be used just to select among a set of candidate single gene/protein classifiers or to optimize a pre-defined classifier with regard to a new platform for measurement. In any case, the classifier defined on the training set is used to classify the patients in the validation set as either sensitive, i.e. predicted likely to benefit from the new treatment T relative to C or not sensitive. One then compares outcomes for the sensitive patients in the validation set who received T versus the sensitive patients in the validation set who received C. Let L denote the log-rank statistic (if outcomes are time-to-event) for this comparison of T vs C of sensitive patients in the validation set. The null distribution of L is determined by repeating the entire analysis after permuting the treatment and control labels a thousand or more times. If the permutation statistical significance of L is less than .05-α1 (e.g. .02), then treatment T is considered superior to C for the subset of the patients predicted to be sensitive using the classifier developed in the training set. Freidlin et al.[22] recently demonstrated that the power of this approach can be substantially increased by embedding the classifier development and validation process in a 10 fold cross-validation.
Predictive analysis of clinical trials (PACT)
The idea of embedding the classifier development and validation in a ten-fold cross-validation is very powerful and much more broadly applicable than in the context described by Freidlin et al.[22] The concept is to prospectively define an algorithm for classifying patients as likely or not likely to have better outcome on the new treatment T compared to the control C. The algorithm A is applied to a set of data D={(y,z,x)} consisting of an outcome y, a treatment indicator z, and a covariate vector × for a training set of cases. With survival data, the outcome y would be a pair, survival time and censoring indicator for the case. The algorithm can be based on whatever baseline variables are of interest and can be of any type e.g. Bayesian[23] or frequentist, linear model or decision tree based, etc. When the algorithm is applied to a set of data it must completely define a binary classifier C(x|A,D) which takes value 1 if a patient with covariate vector × is predicted to have better outcome on T than on C and takes value 0 otherwise. In the notation C(x|A,D) the A denotes the algorithm and the D denotes the dataset used to train the algorithm A to produce the classifier C.
Let P denote the set of patients in the clinical trial. Let P be partitioned into K disjoint subsets P1, …, PK. Let the i’th training set consist of the full set of patients except for the i’th subset; i.e. the complement of Pi in P, Di=P-Pi. Let C(x|A,Di) denote the binary classifier developed by applying the algorithm A to training set Di. Use this classifier to classify the patients in Pi as either sensitive to T or insensitive to T. Let vj= C(xj|A,Di) denote the predictive classification for patients j in Pi. vj =1 if the patient is predicted to be sensitive to the new treatment T relative to control C, and zero otherwise. Since the patients in Pi were not included in the training set Di used to train C(x|A,Di), this classification is of a predictive type, not just evaluating goodness of fit to the same data used to develop the classifier. Since each patient appears in exactly one Pi, each patient is classified exactly once and that classification is done with a classifier developed using a training set not containing that patient.
Let Scv denote the set of patients j for whom vj=1, i.e.who are predicted to be sensitive to the new treatment. We can evaluate the predictive value of our algorithm by comparing outcomes of the patients in Scv who received treatment T to the outcomes for the patients in Scv who received the control C. Let L(Scv) denote a measure of difference in outcomes for that comparison; e.g. a log-rank statistic if outcomes are time-to-event. We can generate the null distribution of L by repeating the entire analysis for thousands of random permutations of the treatment labels. This test can be used as the primary significance test of the clinical trial to test the strong null hypothesis that the new treatment and control are equivalent for all patients on the primary endpoint of the trial. Alternatively, it can be used as a fall-back test as described in the previous sections.
Having rejected the strong null hypothesis described above, the application of the algorithm A to the full dataset P provides a decision tool C(x|A,P) that can be used by physicians in deciding which of their patients should receive the new regimen T. If this classifier is used for patients whose distribution of covariate vectors is the same as the patients P in the clinical trial, then the expected t year survival can be estimated as
| (1) |
Where n denotes the total number of patients in the trial, nsens denotes the number of patients in Scv, KM(t, Scv ∩ T) is the Kaplan Meier estimate of survival beyond time t for patients in Scv who received treatment T, and KM(t, S̅cv ∩ C) denotes the Kaplan Meier estimate of survival beyond time t for patients in the complement of Scv in P, i.e. the patients predicted to by non-sensitive based on the cross-validation, who received treatment C.
The predicted outcome (1) using the algorithm can be compared to the expected outcome based on the standard analysis which indicates that all patients should receive the new treatment T if the average treatment effect is significantly better compared to the control C, and if not, then no patients should receive T. If the usual overall log-rank test is significant at the 0.05 level, then the expected outcome for this standard analysis would be KM(t, P ∩ T). Otherwise, the expected outcome would be KM(t, P ∩ C). Depending on the outcome of the clinical trial, the expected outcome can be compared to that given by (1) for evaluating whether the algorithm provides improved utility as a decision tool for applying the results of the trial to individual patients. Improved utility may result from identifying a subset of patients who benefit from the new treatment in cases where the average treatment effect is not significant or from identifying a subset of patients who do not benefit from the new treatment in cases where the average treatment effect is significant.
Confidence intervals for the performance of classifier based treatment assignment (1) can be obtained by repeating the analysis with bootstrap samples of the patients P. For a given non-parametric bootstrap sample, the entire analysis is repeated. The algorithm A is the same, but the resulting classifier and the sensitive Scv subset subset changes. For each bootstrap sample, expression (1) is re-computed and the empirical distribution of the values of (1) provide a bootstrap confidence interval. Confidence intervals for the performance of the standard analysis method, i.e. either KM(t, P ∩ T) or KM(t, P ∩ C) can be computed by standard frequentist methods. By employing the same bootstrap samples to as for re-development of the classifier, however, one can also obtain a confidence interval for the difference between the two approaches to therapeutic decision making with regard to survival of a subsequent cohort of patients.
Applying algorithm A to bootstrap samples of the patients P in the trial also provides information about the stability of the subset who benefit from the new treatment T. Although the precision of the identification of this sensitive subset will be limited by the size of the clinical trial, information about specificity of treatment benefit may be substantially greater than with standard methods in which all future patients are presumed to benefit or not benefit from one treatment or the other. The effectiveness of the decision tool based on C(x| A,P) will, however, depend on the algorithm A. Algorithms that over-fit the data will provide classifiers that make poor predictions and the expected outcomes may be substantially worse than using the traditional approach. Algorithms based on Bayesian models with many parameters and non-informative priors may be as prone to over-fitting as frequentist models with many parameters. The effectiveness of an algorithm will also depend on the dataset, i.e. the unknown truth about how treatment effect varies among patient subsets. A strong advantage of the proposed approach, however, is that an almost unbiased estimate of the performance of a defined algorithm can be obtained from the dataset of a clinical trial. This is clearly preferable to performing exploratory analysis on the full dataset without any cross-validation, reporting the very misleading goodness of fit of the model to the same data used to develop the model, and cautioning that the results need testing in future clinical trials. Clearly, many aspects of the prediction paradigm introduced here for the design and analysis of clinical trials warrant and require further development.
Conclusion
Developments in biotechnology and genomics have increased the focus of biostatisticians on prediction problems. This has led to many exciting methodologic developments for predictive modeling where the number of variables is larger than the number of cases. Heterogeneity of human diseases and new technology for characterizing them presents new opportunities and challenges for the design and analysis of clinical trials. In oncology, treatment of broad populations with regimens that do not benefit most patients is less economically sustainable with expensive molecularly targeted therapeutics. The established molecular heterogeneity of human diseases requires the development of new paradigms for the use of randomized clinical trials as a reliable basis predictive medicine[1, 2]. We have presented here prospective designs for the development of new therapeutics with candidate predictive biomarkers. We have also outlined a prediction based approach to the design and analysis of randomized clinical trials which appears greatly superior to post-hoc subset analysis and may serve as a starting point for further research.
Acknowledgments
The Conference was funded by NIH/NCI (R13-CA132565) and co-sponsored by The American Statistical Association and the Society for Clinical Trials.
Questions from the audience
[QUESTION]:
Leon Chiu from Bristol-Myers-Squibb. You indicated in your talk the importance of having a certain level of belief in your classifier in using optimal designs. That belief would be based perhaps on some Phase II or a significant biological argument. For the K-ras mutation there is a substantial amount of literature so there is plenty of evidence, but in new drug development that may not be the case. How much evidence is necessary to have a reasonable level of belief in your classifier?
[ANSWER]:
For oncology I believe that predictive biomarkers for new drug development will generally not be black-box classifiers from gene expression profiling, but will be biologically based on genomic changes like K-ras mutations. I agree with you though that the complexity of cancer biology and uncertainty about drug mechanism often makes it very difficult to know the correct biomarker with great confidence prior to initiating the phase III trial. Compelling biological or empirical evidence would be necessary for using the biomarker to restrict eligibility, but that level of confidence is not necessary for most of the other designs I described. Many of the designs are motivated by the lack of that level of confidence, utilize a fallback analysis plan that preserves type I error and provides the opportunity to benefit from the use of classifiers for which you have less than complete confidence. There is a cost in the design of adequate phase III trials using predictive biomarkers, and so some reasonable degree of confidence should be present for their inclusion in the primary analysis plan.
[QUESTION]:
Nancy Geller from the National Heart, Lung and Blood Institute (NHLBI.
I’d like to pursue your last point about embedding subset selections in cross-validation. Suppose I just do an ordinary clinical trial, not anything particularly sexy and I embed subset selection in cross-validation. Are you now giving me freedom to dredge without enumerating even what variables I would do the subset analysis on?
[ANSWER]:
In order to apply the method I’ve proposed, the “data dredging” algorithm must be completely defined in advance. Otherwise, it cannot be applied in a K-fold validation manner. The variables need to be specified in advance and the way those variables will be combined or modeled for the subset analysis needs to be specified in advance. The entire way the subset analysis will be conducted must be extensively defined to result in a binary classifier, C(x|A,D) using the notation I presented, which given a covariate vector × provides an indication of whether the patient is predicted to benefit from the new treatment T relative to C or not. The algorithm must be specified in this way so that it can be applied K times in a K-fold cross-validation. There has to be a completely defined algorithm which can be applied from scratch on each of the K training sets defined in the cross-validation. The × vector could just consist of clinical and histopathologic covariates; it doesn’t have to involve genomics at all.
[QUESTION]
Roger Day from University of Pittsburgh.
In a way, maybe every drug will end up being an orphan drug, and the question is, how can you do drug development when for each drug the potential patient population becomes much smaller? Is there now a much bigger role for, for example, nonprofit foundations or AHRQ treatment evaluations that take over because the companies really can’t muster the incentives to do this kind of research, and it becomes more difficult for them to have profitable products?
[ANSWER]:
I think it’s too early to tell whether every drug will be an orphan drug. My own guess is it won’t be that way. When people talk about personalized medicine they’re thinking that treatment or prevention decisions should be personalized based on the individual genome. For therapeutics in oncology, I don’t think that’s the way it’s going to work out. More likely cancers will be classified based on the oncogenic mutations and de-regulated genetic pathways that drive their growth and invasion. Although tumors are quite individualized in terms of all the mutations they contain, many of these mutations are late events that reflect genomic instability and do not have important phenotypes. For most primary sites, I believe that the number of relevant categories will be quite limited.
Footnotes
Presented at the Annual University of Pennsylvania Conference on Statistical Issues on Clinical Trials: Statistical Issues in Developing Targeted Therapies, April 29, 2009.
References
- 1.Simon R. An agenda for clinical trials: Clinical trials in the genomic era. Clinical Trials. 2004;1:468–470. doi: 10.1191/1740774504cn046xx. [DOI] [PubMed] [Google Scholar]
- 2.Simon R. New challenges for 21st century clinical trials. Clinical Trials. 2007;4:167–169. doi: 10.1177/1740774507076800. [DOI] [PubMed] [Google Scholar]
- 3.Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in guaging the performance of a diagnostic, prognostic or screening marker. American Journal of Epidemiology. 2004;159:882–890. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- 4.Harrell FE. Regression Modeling Strategies. New York: Springer-Verlag; 2001. [Google Scholar]
- 5.Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: A comparison of resampling methods. Bioinformatics. 2005;21(15):3301–3307. doi: 10.1093/bioinformatics/bti499. [DOI] [PubMed] [Google Scholar]
- 6.Peto R, Pike MC, Armitage P. Design and analysis of randomized clinical trials requiring prolonged observation of each patient: I. Introduction and design. British Journal of Cancer. 1976;34:585. doi: 10.1038/bjc.1976.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peto R, Pike MC, Armitage P. Design and analysis of randomized clinical trials requiring prolonged observation of each patient: II Analysis and examples. British Journal of Cancer. 1977;35:1. doi: 10.1038/bjc.1977.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Group I-C. Randomised trial of IV streptokinase, oral aspirin, both or neither among 17187 cases of suspected acute myocardial infarction. Lancet. 1988:349–360. [PubMed] [Google Scholar]
- 9.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 10.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008 Oct 23;359(17):1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 11.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. New England Journal of Medicine. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 12.van-de-Vijver MJ, He YD, Veer LJvt, Dai H, Hart AAM, Voskuil DW, et al. A gene expression signature as a predictor of survival in breast cancer. New England Journal of Medicine. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 13.Simon R. A roadmap for developing and validating therapeutically relevant genomic classifiers. Journal of Clinical Oncology. 2005;23:7332–7341. doi: 10.1200/JCO.2005.02.8712. [DOI] [PubMed] [Google Scholar]
- 14.Maitournam A, Simon R. On the efficiency of targeted clinical trials. Statistics in Medicine. 2005;24:329–339. doi: 10.1002/sim.1975. [DOI] [PubMed] [Google Scholar]
- 15.Simon R, Maitournam A. Evaluating the efficiency of targeted designs for randomized clinical trials. Clinical Cancer Research. 2005;10:6759–6763. doi: 10.1158/1078-0432.CCR-04-0496. [DOI] [PubMed] [Google Scholar]
- 16.Simon R, Maitournam A. Evaluating the efficiency of targeted designs for randomized clinical trials: Supplement and Correction. Clinical Cancer Research. 2006;12:3229. doi: 10.1158/1078-0432.CCR-04-0496. [DOI] [PubMed] [Google Scholar]
- 17.Simon R. Using genomics in clinical trial design. Clinical Cancer Research. 2008;14:5984–5993. doi: 10.1158/1078-0432.CCR-07-4531. [DOI] [PubMed] [Google Scholar]
- 18.Simon R. Designs and adaptive analysis plans for pivotal clinical trials of therapeutics and companion diagnostics. Expert Review of Molecular Diagnostics. 2008;2(6):721–729. doi: 10.1517/17530059.2.6.721. [DOI] [PubMed] [Google Scholar]
- 19.Wang SJ, O'Neill RT, Hung HMJ. Approaches to evaluation of treatment effect in randomized clinical trials with genomic subset. Pharmaceutical Statistics. 2007;6:227–244. doi: 10.1002/pst.300. [DOI] [PubMed] [Google Scholar]
- 20.Jiang W, Freidlin B, Simon R. Biomarker adaptive threshold design: A procedure for evaluating treatment with possible biomarker-defined subset effect. Journal of the National Cancer Institute. 2007;99:1036–1043. doi: 10.1093/jnci/djm022. [DOI] [PubMed] [Google Scholar]
- 21.Freidlin B, Simon R. Adaptive signature design: An adaptive clinical trial design for generating and prospectively testing a gene expression signature for sensitive patients. Clinical Cancer Research. 2005;11:7872–7878. doi: 10.1158/1078-0432.CCR-05-0605. [DOI] [PubMed] [Google Scholar]
- 22.Freidlin B, Jiang W, Simon R. The cross-validated adaptive signature design for predictive analysis of clinical trials. Clinical Cancer Research. 2010 doi: 10.1158/1078-0432.CCR-09-1357. (In Press). [DOI] [PubMed] [Google Scholar]
- 23.Simon R. Bayesian subset analysis: application to studying treatment-by-gender interactions. Statistics in Medicine. 2002;21:2909–2916. doi: 10.1002/sim.1295. [DOI] [PubMed] [Google Scholar]