Abstract
Chronic obstructive pulmonary disease (COPD) is linked to cardiovascular disease; however, there are few studies on the associations of cardiovascular genes with COPD.
We assessed the association of lung function with 2,100 genes selected for cardiovascular diseases among 20,077 European-Americans and 6,900 African-Americans. We performed replication of significant loci in the other racial group and an independent consortium of Europeans, tested the associations of significant loci with percent emphysema, and examined gene expression in an independent sample. We then tested the association of a related lipid biomarker with FEV1/FVC and percent emphysema.
We identified one new polymorphism for FEV1/FVC (rs805301) in European-Americans (p=1.3×10−6) and a second (rs707974) in the combined European-American and African-American analysis (p=1.38×10−7). Both SNPs flank the gene for apolipoprotein M (apoM), a component of HDL. Both replicated in an independent cohort. SNPs in a second gene related to apoM and HDL, PCSK9, were associated with FEV1/FVC among African-Americans. rs707974 was associated with percent emphysema among European-Americans and African-Americans, and APOM expression was related to FEV1/FVC and percent emphysema. Higher HDL levels were associated with lower FEV1/FVC and greater percent emphysema.
These findings suggest a novel role for the APOM/HDL pathway in the pathogenesis of COPD and emphysema.
Keywords: Apolipoproteins; Cholesterol; Percent Emphysema; Polymorphism, Single Nucleotide; Pulmonary Disease, Chronic Obstructive
INTRODUCTION
Chronic obstructive pulmonary disease (COPD) is a leading cause of death globally[1] and is characterized by persistent airflow obstruction.[2,3] Emphysema is defined anatomically by permanent enlargement of airspaces distal to terminal bronchioles with destruction of alveolar walls.[4]
Familial studies suggest a genetic influence on COPD.[5-7] Recent genome-wide association studies (GWAS) have identified loci associated with the ratio of forced expiratory volume in one second to forced vital capacity (FEV1/FVC) among participants of European ancestry.[8-13] Many of these genes have been shown to influence susceptibility to COPD;[14-16] however, they explained little more than 3% of the variance in lung function.
Emphysema also has a familial predisposition.[17] However, understanding of the genetic basis for emphysema, beyond alpha1-antirypsin deficiency, is more limited. A GWAS identified one genetic locus for radiologist-defined emphysema on computed tomography (CT) but none for quantitatively assessed emphysema.[18] Candidate gene association studies have identified additional genes for emphysema.[19-24]
Complimentary genotyping strategies to better delineate the genetic basis of COPD and emphysema are therefore warranted. One such strategy is a “gene-centric” genotyping chip, which includes a large panel of candidate genes and often better gene coverage than GWAS chips. No such chips have been designed specifically for lung disease; however, the ITMAT/Broad/CARe (IBC) chip[25] includes 2,100 candidate genes primarily selected for cardiovascular disease.
Respiratory and cardiac function are tightly linked at cellular,[26,27] physiologic,[28] structural,[29] and anatomic levels. For example, endothelial dysfunction is implicated in the pathogenesis of atherosclerosis[30] and emphysema in animal models[31-34] and humans,[35] the later via ceramide-mediated endothelial cell apoptosis.[36-38] High-density lipoprotein (HDL) may also be relevant to COPD and emphysema, as HDL increases in vitro ceramide levels.[39] HDL levels and function are affected by apolipoprotein M (apoM).[40-42]
We examined associations of FEV1/FVC on the IBC chip in European-American and African-American participants in the Candidate-gene Association Resource (CARe) consortium.[43] Findings were replicated in the SpiroMeta consortium.[11] We performed additional analyses of identified genes with the percentage of emphysema-like lung (percent emphysema), of gene expression, and of HDL with lung function and percent emphysema in the Multi-Ethnic Study of Atherosclerosis (MESA) SNP Health Association Resource (SHARe) and MESA COPD Study.
METHODS
Study Samples
Analyses of Lung Function
The association of genes and lung function were assessed in the seven CARe cohorts that measured spirometry: Atherosclerosis Risk in Communities (ARIC), Coronary Artery Risk Development in young Adults (CARDIA), Cleveland Family Study (CFS), Cardiovascular Health Study (CHS), Framingham Heart Study (FHS), Jackson Heart Study (JHS) and the subset of MESA with spirometry. These cohorts have been previously described[44-53] and are summarized in the supplement. Exclusion criteria were lack of valid spirometric or genetic data, age less than 23 years and a restrictive pattern of spirometry, defined as FVC less than the lower limit of normal[54] and FEV1/FVC of greater than 0.70.
Replication of Lung Function SNPs
Replication for FEV1/FVC was performed in the SpiroMeta consortium, a large independent sample of 14 GWAS studies.[11] Replication in airflow obstruction was performed using publically available data from the SpiroMeta and CHARGE consortia,[9] which partly overlaps with European-American participants in the CARe consortium. Details are provided in the supplement.
Analyses of Percent Emphysema
Percent emphysema was examined among all participants in MESA SHARe, which comprises all participants who consented to genetic analyses in MESA,[44] MESA Family[55] and MESA Air Pollution[56] studies. Spirometry was not required.
Gene Expression Analyses
mRNA expression was examined in peripheral blood mononuclear cells in MESA COPD Study, an independent sample described in the supplement.
Appropriate Institutional Review Boards approved study protocols and written informed consent was obtained from all participants.
Phenotypic Measures
Spirometry
Pre-bronchodilator spirometry was performed by trained and certified spirometry technicians in accordance with the American Thoracic Society guidelines. Spirometry methods and equipment were highly standardized and in some cases identical across cohorts, as described in the supplement.
Percent emphysema
Percent emphysema was assessed in MESA SHARe on lung fields of cardiac CT scans, which image approximately 70% of lung volume from the carina to the lung bases, at a single center by trained readers, as previously described and validated compared to full-lung scans.[57] Percent emphysema was defined as percentage of total voxels in the lung less than −950 Hounsfield Units (HU). The MESA COPD Study used the same approach on full-lung scans using Apollo (Vida Diagnostics) software.
HDL
HDL was measured in EDTA plasma using the cholesterol oxidase method (Roche Diagnostics Corporation, Indianapolis, IN) after precipitation of non-HDL with magnesium/dextran.[58]
Genotyping
All CARe participants were genotyped with the IBC Illumina iSELECT array, a 50,000 gene-centric SNP array.[25] All genotyping was performed at a single center. Quality control methods are described in the supplement.
MESA SHARe participants were genotyped using the Affymetrix Genome-Wide Human SNP Array 6.0 platform at a single center.
Statistical Analyses
Analyses of candidate genes with FEV1/FVC employed linear regression, stratified by race and adjusted for age, age2, height, height2, sex, smoking status, pack-years, pack-years2, site (if applicable), and the first 10 principal components (PCs) for ancestry. Association testing of rank-normalized residuals was performed under an additive genetic model.[59-61]
Cohort-specific association results were meta-analyzed, again stratified by race, using inverse variance weighting in METAL[62] with cohort-specific and overall genomic control. A priori, we planned to replicate our top loci identified among European-Americans in the African-American cohorts and vice-versa as distinct cohorts. Race-specific results were then meta-analyzed in METAL[62] for the combined European-American and African-American analyses. The Bonferroni-adjusted thresholds for statistical significance in CARe were 1.31×10−6 in European-Americans and 1.13×10−6 in African-Americans and combined analyses, which are exceedingly conservative for the IBC chip.
Analyses for log-transformed percent emphysema used the same analytical approach supplemented with a linear mixed effects model for family-based-data,[60] and adjustment for age, sex, site, scanner, height, weight, tube current, cigarettes per day, pack-years, asthma and PCs.
Analysis details for gene expression and HDL association studies with lung function and percent emphysema are provided in the supplement.
To address multiple comparisons, we considered analysis of percent emphysema to be analogous to a modified Holm’s procedure[63] on the pathway of apoM. We hypothesized that SNPs rs805301 and rs707974 are in linkage disequilibrium (LD) with causative APOM variant that affects percent emphysema, which affects FEV1/FVC, thus the Holm-Bonferonni corrected threshold for statistical significance for subsequent analyses was set at 0.025.
RESULTS
The mean age of the 26,977 CARe participants with spirometry was 54+/−13 years, 52% ever-smoked, with median pack-years of 20. Additional characteristics of the 20,077 European-American and 6,900 African-American participants are shown by cohort and race in table 1.
Table 1.
Characteristics of European-American and African-American CARe participants with spirometry in the Candidate-gene Association Resource (CARe) cohorts
| European-American Participants | African-American Participants | Combined | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARIC | FHS | CHS | CARDIA | MESA | CFS | All | ARIC | JHS | CHS | CARDIA | MESA | CFS | All | ||
| Number | 8,825 | 5,316 | 3,479 | 1,228 | 1,049 | 180 | 20,077 | 2,740 | 1,597 | 503 | 1,052 | 799 | 209 | 6,900 | 26,977 |
| Age (years) mean±SD |
54±6 | 48±13 | 73±6 | 36±3 | 65±10 | 51±15 | 55±13 | 53±6 | 50±12 | 73±6 | 34±4 | 65±10 | 48±14 | 52±13 |
54±13 |
| Male Sex(%) | 47 | 46 | 44 | 47 | 50 | 47 | 46 | 37 | 38 | 35 | 40 | 47 | 41 | 39 | 44 |
| Height(cm) mean±SD |
169±9 | 170±9 | 165±9 | 172±9 | 169±10 | 170±10 | 169±10 | 168±9 | 169±9 | 164±9 | 170±10 | 168±10 | 169±10 | 168±9 | 169±10 |
| BMI# mean±SD |
27±5 | 27±5 | 26±4 | 26±5 | 28±5 | 32±8 | 27±5 | 30±6 | 32±7 | 28±5 | 29±7 | 30±6 | 34±8 | 30±7 | 28±6 |
| Cigarette Smoking(%) Never Former Current |
41 36 23 |
52 35 13 |
47 43 10 |
61 20 19 |
40 51 9 |
50 21 29 |
46 36 18 |
49 23 28 |
69 17 14 |
51 35 |
62 11 27 |
40 45 15 |
40 35 25 |
55 23 22 |
48 33 19 |
| Pack years¶ median(IQR) |
26 (12-40) |
16 (7-30) |
30 (13-50) |
10 (4-19) |
19 (8-35) |
14 (5-32) |
23 (10-38) |
17 (8-30) |
14 (7-26) |
21 (10-38) |
6 (3-11) |
15 (6-28) |
11 (5-24) |
14 (6-27) |
20 (9-36) |
| ppFEV1 mean±SD |
94±16 | 98±14 | 89±22 | 99±11 | 92±16 | 96±18 | 94±17 | 97±17 | 95±15 | 92±24 | 100±14 | 94±19 | 92±19 | 96±17 |
95±17 |
| FEV1/FVC mean±SD |
0.74 ±0.08 |
0.76 ±0.07 |
0.69 ±0.10 |
0.79 ±0.06 |
0.73 ±0.09 |
0.77 ±0.07 |
0.74 ±0.09 |
0.76 ±0.08 |
0.80 ±0.09 |
0.70 ±0.11 |
0.81 ±0.06 |
0.75 ±0.10 |
0.78 ±0.08 |
0.77 ±0.09 |
0.75±0.09 |
Abbreviations: ARIC = Atherosclerosis Risk in Communities, CARDIA = Coronary Artery Risk Development in Young Adults, CFS = Cleveland Family Study, CHS = Cardiovascular Health Study, FHS = Framingham Heart Study, MESA = Multi-Ethnic Study of Atherosclerosis, JSH = Jackson Heart Study, SD = Standard Deviation, cm= centimeters, BMI = Body Mass Index, IQR = Inter Quartile Range, ppFEV1 = percent predicted forced expiratory volume in one second, FEV1/FVC = ratio of forced expiratory volume in one second over forced vital capacity.
BMI is the weight in kilograms divided by the square of the height in meters
In ever smokers
Association Study of 2,100 Candidate Genes with Lung Function in CARe
Among European-Americans, we identified one new SNP (rs805301) for FEV1/FVC (figure 1a). Among African-Americans, no SNPs were significantly associated with FEV1/FVC using the Bonferroni cutoff; however, three SNPs were significant with the less conservative cutoff (p<x10−5; figure 1b). In the combined European-American and African-American analysis, we identified a second new SNP (rs707974) for FEV1/FVC (figure 1c; table 2).
Figure 1.
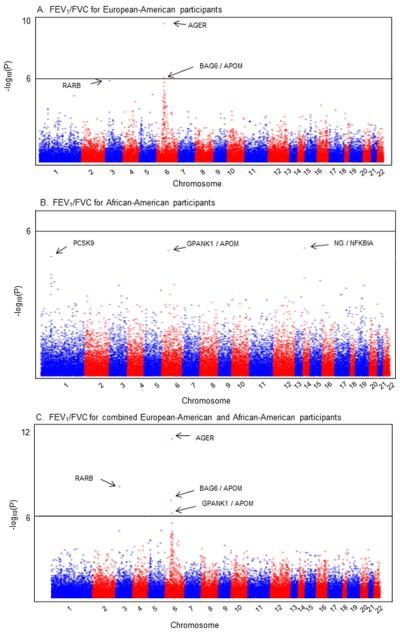
Manhattan Plots of association results for FEV1/FVC stratified by race and combined.
Manhattan Plots ordered by chromosome position of association results for FEV1/FVC. Top 3 loci are labeled with arrows. (A) Meta-analysis of 38,294 SNPs among 20,077 European-American participants. The solid black line represents 1 × 10−6. (B) Meta-analysis of 44,416 SNPs among 6,900 African-American participants. The solid black line represents 1 × 10−6. (C) Meta-analysis of SNPs among combined European-American & African-American participants. The solid black line represents 1 × 10−6.
Abbreviations: FEV1/FVC = ratio of forced expiratory volume in one second over forced vital capacity.
Table 2.
Top five SNPS associated with the FEV1/FVC ratio among European-American and African-American participants in the Candidate-gene Association Resource (CARe) cohorts
| European-American Participants (n=20,077) | ||||||
|---|---|---|---|---|---|---|
| SNP ID (function#) |
Chr. | Gene¶ | Coded allele |
Allele freq.+ |
β§ (SE) | p-value |
| rs2070600 (ns) |
6 | AGER/RNF5 | T | 0.05 | 0.162 (0.026) | 2.19 × 10−10 |
| rs805301 (intron) |
6 | BAG6/APOM | C | 0.37 | − 0.054 (0.011) | 1.32 × 10−6 |
| rs1286664 (intron) |
3 | RARB | T | 0.17 | 0.067 (0.014) | 2.07 × 10−6 |
| rs6941112 (intron) |
6 | STK19/C4B | A | 0.33 | 0.053 (0.011) | 2.90 × 10−6 |
| rs3117582 (upstream) |
6 | BAG6 | T | 0.89 | 0.081 (0.018) | 5.23 × 10−6 |
| African-American Participants (n=6,900) | ||||||
|---|---|---|---|---|---|---|
| SNP ID (function#) |
Chr. | Gene¶ | Coded allele |
Allele freq.+ |
β§ (SE) | p-value |
| rs1951269 (unknown) |
14 | NG/ NFKBIA | A | 0.78 | −0.097 (0.021) | 4.08 × 10−6 |
| rs707974 (3′UTR) |
6 | GPANK1/APOM | G | 0.02 | 0.291 (0.064) | 5.17 × 10−6 |
| rs565436 (intron) |
1 | PCSK9 | A | 0.60 | 0.08 (0.018) | 9.32 × 10−6 |
| rs533375 (intron) |
1 | PCSK9 | A | 0.25 | −0.083 (0.021) | 5.29 × 10−5 |
| rs7156874 (intron) |
14 | PSMA6 | A | 0.02 | 0.271 (0.068) | 7.33 × 10−5 |
| Combined European-American and African-American Participants (n=26,977) | ||||||
|---|---|---|---|---|---|---|
| SNP ID (function#) |
Chr. | Gene¶ | Coded allele |
Allele freq.+ |
β§ (SE) | p-value |
| rs2070600 (ns) |
6 | AGER/RNF5 | T | 0.04 | 0.168 (0.025) | 8.37 × 10−12 |
| rs1286664 (intron) |
3 | RARB | T | 0.18 | 0.067 (0.014) | 1.60 × 10−8 |
| rs805301 (intron) |
6 | BAG6/ APOM | C | 0.43 | −0.050 (0.009) | 1.26 × 10−7 |
| rs707974 (3′UTR) |
6 | GPANK1/ APOM | G | 0.10 | 0.088 (0.016) | 1.38 × 10−7 |
| rs6941112 (intron) |
6 | STK19/C4B | A | 0.30 | 0.052 (0.011) | 1.22 × 10−6 |
The European-Americans SNPs represent five loci (r2 range; 0.005-0.20).
Abbreviations: SNP= single nucleotide polymorphism, FEV1/FVC= ratio of forced expiratory volume in one second over forced vital capacity, Chr = chromosome, Ref = reference, β = effect estimate, SE = standard error, ns=non-synonymous coding SNP, UTR = untranslated.
Gene abbreviations: AGER = advanced glycosylation end product-specific receptor (also known as RAGE), RNF5 = ring finger protein 5, BAG6= BCL2-associated athanogene 6 (also known as BAT3), APOM = apolipoprotein M, RARB = retinoic acid receptor, beta, STK19 = serine/threonine kinase 19, C4B = complement component 4B (Chido blood group), NG = near gene, NFKBIA = nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha, GPANK1 = G patch domain and ankyrin repeats 1, PCSK9 = proprotein convertase subtilisin/kexin type 9, PSMA6 = proteasome (prosome, macropain) subunit, alpha type, 6.
Function reported is reported dbSNP Genome Build 37.3 (http://www.ncbi.nlm.nih.gov/projects/SNP/)
If two genes are listed for the SNP, the first gene listed was annotated using dbSNP (Genome Build 37.3) and the second gene was annotated using the IBC chip (annotation from Genome Build 36). If there is only one gene listed, there was no discrepancy between Genome Builds 37.3 and Build 36.
Frequency of allele labeled in table as “coded allele”
rank-normalized residuals of FEV1/FVC adjusted for age, age2, height, height2, sex, smoking status, pack-years, pack-years2 the first 10 principal components (PCs) for ancestry, genomic control at the cohort and meta-analysis level. A cohort-specific site covariate was included in the regression model for cohorts with multiple sites (ARIC, CARDIA, CHS, and MESA).
The new SNP (rs805301) identified in European-Americans was selected for the IBC chip as a variant in APOM based upon Genome Build 36 and is annotated in BAG6 on Genome Build 37.3, which is the upstream flanking gene of APOM. It replicated among African-Americans (p=0.036) and remained significant in the combined meta-analysis (table 2). The risk allele (C) was associated with a decrease in FEV1/FVC in both racial groups.
The new SNP identified in the combined European-American and African-American analysis, rs707974, was the second most significant SNP in African-Americans (table 2) and would have been significant with less stringent Bonferroni cutoff. It was also selected as an APOM variant and is now annotated in GPANK1, the downstream flanking gene of APOM separated by an open-reading frame, C6orf47. It was not significant for FEV1/FVC in European-Americans (p=2.84×10−5).
SNPs rs805301 and rs707974 were not in high LD in European-Americans or African-Americans (r2=0.07 and r2=0.03, respectively), suggesting that they are separate loci (Figure 2). They were also not in high LD with the previously described AGER SNP rs2070600 in European-Americans[11,12] (r2=0.035 and r2=0.37, respectively). In addition, rs805301 remained associated with FEV1/FVC after adjustment for rs2070600 (p=6.82×10−4) and rs2070600 was only nominally associated with FEV1/FVC among African-Americans (p=0.009). These findings suggest that associations of rs805301and rs707974 with lung function are unrelated to AGER.
Figure 2.
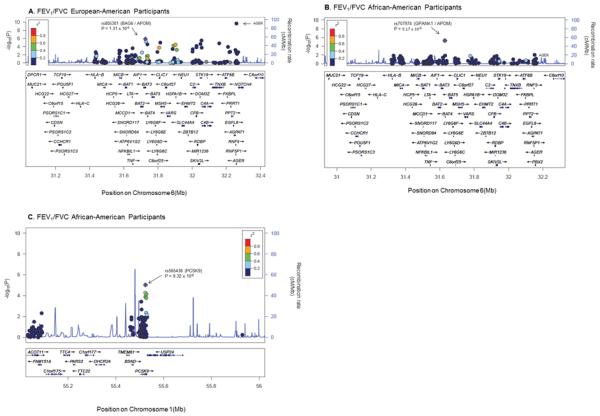
Regional Association Plots of top SNPs flanking APOM (rs805301, rs707974) and PCSK9 SNPs for FEV1/FVC
The selected SNPs with the lowest p value are illustrated by the purple diamond. The correlations (r2) of surrounding SNPs in the region are indicated by the colors shown on the graph. For the SNPs flanking APOM(rs805301 and rs707974), a 600kb flanking size was selected to include the AGER SNP on the plot whereas 500kb flanking size was selected for the PCSK9 SNP. Plots were generated using LocusZoom.[97] The Genome builds/LD populations implemented were hg 18/HapMap Phase II CEU and hg/19 1000 Genomes Nov 2010 AFR for European-American and for African-American participants, respectively.
Abbreviations: SNPs = single nucleotide polymorphisms, APOM = apolipoprotein M, PCSK9 = proprotein convertase subtilisin/kexin type 9, FEV1/FVC = ratio of forced expiratory volume in one second over forced vital capacity.
Sensitivity analyses restricted to participants free of clinical cardiovascular disease, age 55 years or less, and free of asthma yielded similar results, as did analyses additionally adjusted for diabetes, hypertension and asthma (supplement). Analyses stratified by smoking status yielded similar results (supplement).
The other top loci in African-Americans were in NFKBIA and PCSK9 (table 2). PCSK9 is related to apoM[64] and six of the top 30 SNPs for FEV1/FVC in African-Americans were in PCSK9 (figure 2). SNPs in neither gene replicated in European-Americans. Regional association plots for additional loci are displayed in supplementary figure 3.
Results for the FEV1 are displayed in supplementary figures 4, 5 and 6. Top SNPs associated with FEV1/FVC and FEV1 are presented in supplementary tables 1 and 2.
Replication of SNPs Flanking APOM in SpiroMeta
Both rs805301 and rs707974 replicated for FEV1/FVC in 20,288 European participants in the SpiroMeta consortium in a consistent direction (β=−0.03, p=0.02 and β=0.05, p=0.02, respectively).
We reviewed publically available results from the SpiroMeta-CHARGE GWAS meta-analysis of airflow obstruction.[9] SNP rs805301 was associated with airflow obstruction (p=0.004) and rs707974 was nominally associated with airflow obstruction in individuals without asthma (p=0.026; supplement).
Association of SNPs Flanking APOM with Percent Emphysema in MESA
SNP rs707974 was significantly associated with percent emphysema among 2,551 European-Americans and 2,457 African-Americans (p=4.74×10−4 and p=0.009, respectively) and in combined analyses (p=1.67×10−5; table 3) in MESA. The characteristics of these participants are shown in supplementary table 3. The direction of the association of rs707974 with percent emphysema and lung function was consistent: risk allele (A) was associated with greater percent emphysema and a lower FEV1/FVC.
Table 3.
Association of SNPs flanking APOM with percent emphysema among European-American and African-American participants in MESA.
| European-American Participants (n = 2,552) | ||||
|---|---|---|---|---|
| SNP | Ref. allele | Allele freq.# | β¶ (SE) | p-value |
| rs805301 | C | 0.37 | +0.019 (0.018) | 0.29 |
| rs707974 | G | 0.10 | −0.098 (0.028) | 4.74 × 10−4 |
| African-American Participants (n = 2,483) | ||||
|---|---|---|---|---|
| SNP | Ref. allele | Allele freq.# | β¶ (SE) | p-value |
| rs805301 | C | 0.58 | + 0.013 (0.018) | 0.46 |
| rs707974 | G | 0.02 | −0.160 (0.061) | 0.009 |
| Combined European-American and African-American Participants (n = 5,035) | ||||
|---|---|---|---|---|
| SNP | Ref. allele | Allele freq.+ | β¶ (SE) | p-value |
| rs805301 | C | +0.015 (0.001) | 0.14 | |
| rs707974 | G | −0.094 (0.022) | 1.67 × 10−5 | |
Abbreviations: SNP= single nucleotide polymorphism, No = number of participants, β = effect estimate, SE = standard error, HDL = high density lipoprotein cholesterol, PCs = principal components for ancestry
Reference allele frequency
Log transformed percent emphysema-950 adjusted for adjusted for age, sex, site, CT scanner, height, weight, weight greater than 220 pounds, cigarettes per day, pack-years, asthma, and PCs.
Not reported, combined across races
We performed association testing under an additive genetic model stratified by race and subsequently in the combined population controlling for race/ethnicity for each quantitative phenotype. Genome-wide significant set at 10×−6 using Bonferonni correction. The SNPs are not in tight LD (r2 = 0.059)
The association with percent emphysema persisted in an independent sample of 1,138 European-American and 1,563 African-American MESA participants who did not have spirometry measures and who were therefore excluded from the lung function analysis (p=0.02 and p=0.003; respectively). Additional adjustment for socioeconomic status yielded similar results whereas restriction to 418 European-Americans and 209 African-Americans with FEV1/FVC < 0.70 yielded non-significant results; however, the effect size was greater in African-Americans and similar in European-Americans in these groups compared to the overall MESA sample (supplement). SNP rs805301 was not significantly associated with percent emphysema.
PCSK9 was nominally associated with percent emphysema in European-Americans (p=0.04) but not African-Americans. AGER SNP rs2070600 was significantly associated with percent emphysema among European-Americans and African-Americans (p=2.54×10−4 and p=0.001, respectively).
Gene Expression of SNPs Flanking APOM in MESA COPD
APOM expression was significantly, inversely associated with FEV1/FVC (table 4) in an independent sample of 101 participants in the MESA COPD Study, the characteristics of which is described in the supplement.. We secondarily examined expression of GPANK1, BAG6 and PCSK9. GPANK1 expression was associated with FEV1/FVC (β=−0.096; 95%CI −0.175,−0.017; p=0.02) whereas BAG6 and PCSK9 expression were not associated with FEV1/FVC.
Table 4. Association of APOM gene expression with the FEV1/FVC and CT percent: emphysema in 101 participants in the MESA COPD Study.
| Mean difference in FEV1/FVC per increase in APOM gene expression# (95% CI) |
p-value | Mean difference in Percent Emphysema¶ per increase in APOM gene expression# (95% CI) |
p-value | |
|---|---|---|---|---|
| Model 1 | −0.091 (−0.178, −0.003) | 0.04 | 1.05 (0.10, 2.0) | 0.03 |
| Model 2 | −0.092 (−0.178, −0.003) | 0.04 | 1.04 (0.13, 1.95) | 0.03 |
| Model 3 | 0.76 (−0.14, 1.67) | 0.10 | ||
| Model 4 | 1.14 (0.20, 2.09) | 0.02 |
Abbreviations: FEV1/FVC= ratio of forced expiratory volume in one second over forced vital capacity, APOM = apolipoprotein M
Model 1: Adjusted for age, gender, cohort and race/ethnicity
Model 2: Additionally adjusted for smoking status, and pack-years
Model 3: Additionally adjusted for height, and weight
Model 4: Additionally adjusted BAG6 (probe-set 210208)
APOM probe-set 214910_s_at
Log-transformed CT percent emphysema below −950 HU
We tested the association of gene expression with FEV1/FVC and with percent emphysema in the MESA COPD Study implementing linear regression models weighted to account for the sampling schema as described in the supplement.
APOM expression was positively associated with percent emphysema in minimally adjusted models and after adjustment for BAG6 (table 4). BAG6 expression was not associated with percent emphysema except after adjustment for APOM (p=0.01). PCSK9 was significantly associated with percent emphysema (β=1.150; 95%CI 1.0, 1.32; p=0.016).
Association of HDL with Lung Function and Percent Emphysema in MESA
Among 3,044 participants with spirometry, higher HDL levels were independently associated with a lower FEV1/FVC (−0.24% per 10 mg/dl HDL; 95%CI:−0.45, −0.03; p=0.027).
Among 8,367 participants with percent emphysema, higher HDL levels were independently associated with greater percent emphysema (0.53% increase in percent emphysema per 10 mg/dl HDL; 95% CI: 0.34, 0.73; p<0.001). Figure 3 shows the multivariate relationship of HDL to percent emphysema, which was non-linear (p<0.001) with a plateau at HDL levels greater than 60 mg/dL.
Figure 3.
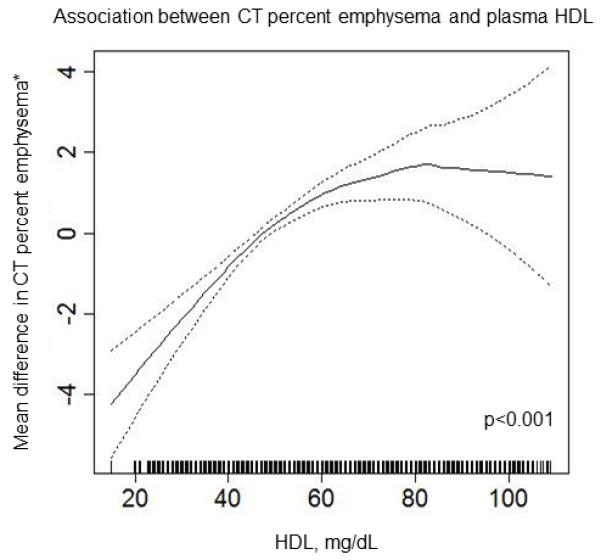
Multivariate association between HDL and percent emphysema
Results of multivariate analyses of the relationship between percent emphysema and plasma HDL among 8,367 MESA SHARe participants are shown. The solid line indicates smoothed regression line adjusted for age, sex, race/ethnicity, height, weight, educational attainment, scanner, tube current, total cholesterol, exercise, pack-years, cigarettes per day, alcohol use, inhaled steroids and use of statins. Figure and p value was produced using a loess smoothing function in a generalized additive model in R/GAM (R version 2.13.0).Dashed lines indicated 95% confidence intervals.
To assess for potential survival bias among older participants, we repeated the HDL-emphysema analysis among 5,241 participants 45-65 years old and found consistent results. Findings were also consistent within strata of gender, race and smoking history (supplement).
DISCUSSION
This large, biracial study identified two new SNPs for FEV1/FVC, one in European-Americans (rs805301) and one in the combined European-American and African-American analysis (rs707974). Both SNPs were originally selected as APOM polymorphisms and are now annotated in genes flanking APOM. Both replicated in an independent sample. In addition, rs707974 was significantly associated with percent emphysema in both European-Americans and African-Americans, APOM gene expression was associated with FEV1/FVC and percent emphysema and HDL was associated with FEV1/FVC and percent emphysema.
The identified SNPs flanking APOM are unlikely to be causative variants but might be linked with a functional APOM variant. Consistent with this thinking, the APOM promoter SNP rs805297 alters APOM expression[65] and is in weak linkage disequilibrium with rs707974 among African-Americans and European-Americans (r2=0.36 and r2=0.32, respectively) and rs805301 among European-Americans (r2=0.23; supplementary figure 7).
APOM encodes apoM, a lipoprotein-associated plasma protein.[66] The majority of apoM is found in HDL.[67] In murine models, modifying APOM gene expression changes apoM plasma concentration, which affects HDL levels, pre-β-HDL formation, reverse cholesterol transport and remodels plasma HDL.[40,67] Hence APOM gene expression alters the function and quality of HDL.
ApoM and HDL are relevant to the pathogenesis of COPD, particularly emphysema, via three related pathways. First, HDL inhibits tumor necrosis factor-stimulated sphingosine kinase activity in human endothelial cells thereby increasing ceramide and decreasing sphingosine-1-phosphate (S1P) cellular levels.[39,68] Ceramide, a second messenger molecule, modulates endothelial cell apoptosis and is implicated in emphysema pathogenesis.[37,38]
Second, HDL-associated-apoM is the plasma carrier for S1P and this HDL-apoM subclass presents S1P to the S1P1 endothelial cell receptor which is endothelium-protective.[42, 69] S1P has an essential role in maintaining endothelial barrier integrity in the lung and is implicated in emphysema pathogenesis.[42,70]
Third, HDL binds and incorporates alpha-1-antitrypsin. HDL-bound alpha-1-antitrypsin inhibits extracellular matrix degradation and apoptosis in vascular smooth muscle.[71,72]
The relevance of APOM and HDL to COPD pathogenesis is further reinforced by our findings that PCSK9 polymorphisms were associated with FEV1/FVC and, nominally, percent emphysema and that PCSK9 gene expression was associated with percent emphysema. PCSK9 augments the degradation of low density lipoprotein receptors[73] and gain-of-function mutations in PCSK9 cause familial hypercholesterolemia[74]. HDL levels in patients with PCSK9 mutations are generally increased[75-77] and most placebo-controlled trials of PCSK9 inhibitors have shown modest increases in HDL levels.[78-82] Furthermore, plasma levels of PCSK9 are associated with plasma apoM levels.[64]
The association of apoM and HDL with FEV1/FVC and emphysema are probably distinct from their relationships to cardiovascular disease and we speculate that the roles of HDL and apoM in the lungs are different from their roles in atherosclerosis. Although HDL has long been thought to be atheroprotective, the definitive epidemiologic study on HDL and cardiovascular disease suggested no benefit[83] and large-scale randomized clinical trials of cholesterol ester transfer proteins, which raise HDL levels, have yet to show a benefit on clinical cardiovascular events[84,85]. The literature on apoM in cardiovascular disease is relatively small and mixed, with animal studies suggesting atheroprotective effects [40,41]; however, in humans, plasma apoM levels were not associated with atherosclerotic disease[86].
Despite the strong mechanistic support implicating APOM in COPD, the latest genome build annotates the new SNPs in genes neighboring APOM, which raises the possibility that they are unrelated to APOM. GPANK1 may be involved in immunity[87] and BAG6 is implicated in apoptosis;[87] both are associated with lung cancer[88,89] and neither have been associated with cardiovascular disease. Given that genome builds change over time, the identification of the two new SNPs in the same region in separate racial groups, the gene expression findings, and the HDL associations all suggest that APOM rather than the other genes are implicated in COPD pathogenesis.
Prior studies of genetic risk for emphysema include one GWAS[18] and candidate gene association studies.[19-24] The GWAS identified BICD1 as associated with severe emphysema on radiologist interpretation but no loci for percent emphysema.[18] The candidate gene association studies did not include rs707974 or rs805301.
Two small studies found increased levels of HDL in severe COPD defined by spirometry.[90,91] Conversely, lower HDL was associated with lower FEV1 in a population-based study (which did not report the association for FEV1/FVC[92] and advanced COPD and emphysema patients from the Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points Study.[93]
The study has several potential limitations. The genomic inflation factor for the meta-analysis in European-Americans was 1.080, suggesting possible population stratification. However, we adjusted our analysis with 10 PCs and with cohort-specific and overall genomic control to address population stratification. Furthermore, we replicated the new APOM SNP identified in European-Americans (rs805301) in African-Americans and both SNPs replicated in an independent cohort and with gene expression, all of which makes population stratification less of a concern.
Although we analyzed two phenotypes of COPD in general population samples, these traits do not capture the entire phenotypic complexity of clinical COPD. Results for the two phenotypes, however, were consistent with each other, similar among patients with airflow limitation, and supported by gene expression in a study of clinical disease. Furthermore, multiple prior genes identified for lung function in population-based samples have been replicated in studies of clinical COPD.[14-16] Hence it is likely that the current findings apply to clinical COPD.
The association between HDL and percent emphysema may be subject to confounding and reverse causation, a small study suggested that HDL levels decrease in COPD patients undergoing lung transplantation.[94] However, we adjusted for multiple potential confounders in this well-phenotyped cohort and the genetic studies are unlikely to be subject to reverse causation.
Similar to other population-based GWAS, we used pre-bronchodilator spirometry for lung function measurement. Percent emphysema was measured on partial-lung CT scans however; we previously validated percent emphysema on partial-lung scans compared to full-lung scans in this cohort and have confirmed multiple prior hypotheses using them.[28,57] Percent emphysema, like lung function, is related to gender, body size, ancestry and socioeconomic status,[95] in addition to current smoking.[96] We adjusted, however, for all of these variables in the analyses.
In conclusion, we identified one new SNP related to FEV1/FVC among European-Americans and a second new SNP in the combined European-American and African-American analysis that was also associated with percent emphysema. Both new SNPs flank APOM, and APOM expression was associated with FEV1/FVC and percent emphysema. APOM encodes apoM, which is primarily bound to HDL, and higher levels of HDL were associated with lower FEV1/FVC and greater percent emphysema. Together, these findings suggest a novel effect of the APOM/HDL-cholesterol pathway in the pathogenesis of COPD and emphysema. Further examination of this pathway is warranted to determine if it is targetable to treat or prevent COPD, and ongoing clinical trials of PCSK9-inhibitors[78-82] and other medications that raise HDL levels[85] may consider monitoring for pulmonary effects.
Supplementary Material
Supplementary Figure 1 Cohort and race specific quantile-quantile plots of association results for FEV1/FVC.
Cohort and race specific quantile-quantile results for all SNPs associated with FEV1/FVC. (A) Cohort specific genomic inflation factors calculated after controlling for 10 principal components analyses for ancestry for European-American participants. (B) Cohort specific genomic inflation factors calculated after controlling for 10 principal components analyses for ancestry for African-American participants.
Supplementary Figure 7 Linkage Disequilibrium Plots based on population samples.
D’ pairwise linkage disequilibrium (LD) plots of population data from CEU-Utah residents with Northern and Western ancestry from CEPH collection (A) and ASW-African ancestry in Southwest USA (B). LD plots created with Haploview, HapMap 3 release 2. Red: D’ = 1 (LOD ≥ 2.0); blue: D’ = 1 (LOD < 2.0); pink: D’ < 1 (LOD ≥ 2.0); white: D’ < 1 (LOD < 2.0).
APOM SNP (rs805297), BAG6 SNP (rs805301) and GPANK1 SNP (rs707974) are circled in green. In the CEU population (a) Block 2 is 11kb in length and includes rs805301 and rs805297. Although rs707974 is in Block 3 (1kb), the D’=1 (LOD ≥ 2.0) for rs707974 and rs805297. In the ASW population (b) Block 3 is 11 kb in length and includes rs805297 and rs707974. SNP rs805301 is in Block 2 (2 kb) and the D’=0.78 (LOD<2.0) for rs805301 and rs805297.
APOM SNP (rs805297) is in LD with rs707974 among African-Americans and European-Americans (r2=0.36 and r2=0.32, respectively) and it is in LD with rs805301 among European-Americans (r2=0.23) but not African-Americans (r2=0.06) using HapMap3 release2.
LOD = logarithm of odds.
Supplementary Figure 2 Quantile-quantile plots of meta-analysis association results for FEV1/FVC stratified by race and across all races.
Quantile-quantile results are for meta-analysis of cohorts for SNPs associated with FEV1/FVC stratified by race and combined across all races. Genomic inflation factors calculated after controlling for 10 principal components for ancestry.
Supplementary Figure 3 Regional association plots of top SNPs associated with FEV1/FVC stratified by race and combined across all races.
Regional association plots of top SNPs for FEV1/FVC in European-American participants (A-D), African-American participants (E-H) and combined analysis of European-American and African-American participants (I-M). The selected SNPs with the lowest p value are illustrated by the purple diamond. The correlations (r2) of surrounding SNPs in the region are indicated by the colors shown on the graph. A 500Kb flanking size was selected for all plots. Plots were generated using LocusZoom. The Genome builds/LD populations implemented were hg 18/HapMap Phase II CEU for European-American and combined European-American and African-American analyses. The hg/19 1000 Genomes Nov 2010 AFR for African-American participants.
Abbreviations: SNPs = single nucleotide polymorphisms, FEV1/FVC = ratio of forced expiratory volume in one second over forced vital capacity.
Supplementary Figure 4 Manhattan Plots ordered by chromosome position of association results for FEV1.
(A) Meta-analysis of 38,294 SNPs among 20,077 European-American participants. (B) Meta-analysis of 44,416 SNPs among 6,900 African-American participants. (C) Meta-analysis of SNPs among combined European-American & African-American participants.
Abbreviations: FEV1 = forced expiratory volume in one second.
Supplementary Figure 5 Quantile-quantile plots of meta-analysis association results for FEV1 stratified by race and across all races.
Quantile-quantile results are for meta-analysis of cohorts for SNPs associated with FEV1 stratified by race and combined across all races. Genomic inflation factors calculated after controlling for 10 principal components.
Supplementary Figure 6 Regional association plots of top SNPs associated with FEV1 stratified by race and combined across all races.
Regional association plots of top SNPs for FEV1 in European-American participants (A-E), African-American participants (F-I) and combined analysis of European-American and African-American participants (J-N). The selected SNPs with the lowest p value are illustrated by the purple diamond. The correlations (r2) of surrounding SNPs in the region are indicated by the colors shown on the graph. A 500Kb flanking size was selected for all plots. Plots were generated using LocusZoom. The Genome builds/LD populations implemented were hg 18/HapMap Phase II CEU for European-American and combined European-American and African-American analyses. The hg/19 1000 Genomes Nov 2010 AFR for African-American participants.
Abbreviations: SNPs = single nucleotide polymorphisms, FEV1/FVC = ratio of forced expiratory volume in one second over forced vital capacity.
ACKNOWLEDGEMENTS
The authors wish to acknowledge the support of the National Heart, Lung, and Blood Institute and the contributions of the research institutions, study investigators, field staff and study participants in creating this resource for biomedical research. The following parent studies have contributed parent study data, ancillary study data, and DNA samples through the Broad Institute (N01-HC-65226) to create this genotype/phenotype data base for wide dissemination to the biomedical research community: ARIC, CARDIA, CFS, CHS, FHS, JHS, and MESA, MESA Family, MESA Air Pollution and MESA Lung studies. A full list of participating MESA Investigators and institutions can be found at http://www.mesa-nhlbi.org. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. This manuscript has been reviewed by the MESA Investigators for scientific content and consistency of data interpretation with previous MESA publications and significant comments have been incorporated prior to submission for publication. Dr. London is supported by the Division of Intramural Research, NIEHS, NIH, DHHS. The ARIC Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. CHS research was supported by NHLBI contracts, with additional contribution from NNDS and the NIA. The authors thank the research institutions, study investigators, field staff and study participants for the following studies contributed to SpiroMeta consortium ALSPAC, B58C-T1DGC, B58C-WTCCC, EPIC obese cases, EPIC population based, FTC, KORA S3, the Korcula study, NFBC1966, NSPHS, ORCADES, SHIP, the Twins UK study, the Vis study.
Funding: NIH/NHLBI grants RC1-HL100543, R01-HL077612, and R01-HL093081, in addition to: ARIC: HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; NIH/NHGRI contract U01HG004402 and National Institutes of Health contract HHSN268200625226C; N01-HC-55015, N01-HC-55016, N01-HC-55021, N01-HC-55019, N01-HC-55020, N01-HC-55017, N01-HC-55018; Broad Institute:N01-HC-65226; CARDIA: N01-HC-48047, N01-HC-48048, N01-HC-48049, N01-HC-48050, N01-HC-95095, N01-HC-45204, N01-HC-45205, N01-HC-05187, N01-HC-45134, N01-HC-95100; CFS: RO1 HL46380-01-16; CHS: N01-HC-85239, N01-HC-85079, N01-HC-85080, N01-HC-85081, N01-HC-85082, N01-HC-85083, N01-HC-85084, N01-HC-85085, N01-HC-85086, N01-HC-35129, N01 HC-15103, N01-HC-75150, N01-HC-45133, N01 HC-55222, U01 HL080295; AG-023629, AG-15928, AG-20098, and AG-027058 FHS: N01-HC-25195; JHS: N01-HC-95170, N01-HC-95171, N01-HC-95172; MESA: N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, R01-HL093081 and RR-024156. R01-HL-071051, R01-HL-071205, R01-HL-071250, R01-HL-071251, R01-HL-071252, R01-HL-071258, R01-HL-071259, EPA grant RD831697; SpiroMeta: see supplement.
Footnotes
AUTHOR CONTRIBUTION
Study design: KMB, SSR, RGB
Data collection: MSA, GLB, PLE, NH, DH, SRH, EAH, JDK, GTO, MP, CAP, LL, SJL, SR, JIR, LJS, MDT, MT, KW, WW, TRY, SSR, RGB Data analysis: KMB, AM, JBW, JK, TDP, YM, FSA, EO Obtaining funding: SSR, JIR, JDK, RGB Drafting manuscript: KMB Critical revision of manuscript: KMB, MSA, AM, JBW, FSA, GLB, PE, NH, DH, SRH, EAH, JDK, LL, SJL, YM, GTO, EO, CAP, MP, SR, JIR, LJS, MDT, MYT, KW, WW, TRY, SSR, RGB
References
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FGR, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo J-P, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KMV, Nasseri K, Norman P, O’Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA, 3rd, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh P-H, Yip P, Zabetian A, Zheng Z-J, Lopez AD, Murray CJL, AlMazroa MA, Memish ZA. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. Erratum appears in Lancet. 2013 Feb 23;381(9867):628 Note: AlMazroa, Mohammad A [added]; Memish, Ziad A [added] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petty TL, Weinmann GG. Building a national strategy for the prevention and management of and research in chronic obstructive pulmonary disease. National Heart, Lung, and Blood Institute Workshop Summary. JAMA. 1997;277:246–253. doi: 10.1001/jama.277.3.246. [DOI] [PubMed] [Google Scholar]
- 3.Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med. 2001;163:1256–1276. doi: 10.1164/ajrccm.163.5.2101039. [DOI] [PubMed] [Google Scholar]
- 4.McLean KH. The pathogenesis of pulmonary emphysema. The American Journal of Medicine. 1958;25(1):62–74. doi: 10.1016/0002-9343(58)90199-2. [DOI] [PubMed] [Google Scholar]
- 5.Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O’Donnell WJ, Reilly JJ, Ginns L, Mentzer S, Wain J, Speizer FE. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. American Journal of Respiratory & Critical Care Medicine. 1998;157(6 Pt 1):1770–1778. doi: 10.1164/ajrccm.157.6.9706014. [DOI] [PubMed] [Google Scholar]
- 6.Silverman EK, Palmer LJ, Mosley JD, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, Province MA, Rao DC, Reilly JJ, Ginns LC, Speizer FE, Weiss ST. Genomewide linkage analysis of quantitative spirometric phenotypes in severe early-onset chronic obstructive pulmonary disease. American Journal of Human Genetics. 2002;70(5):1229–1239. doi: 10.1086/340316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCloskey SC, Patel BD, Hinchliffe SJ, Reid ED, Wareham NJ, Lomas DA. Siblings of Patients With Severe Chronic Obstructive Pulmonary Disease Have a Significant Risk of Airflow Obstruction. Am J Respir Crit Care Med. 2001;164(8):1419–1424. doi: 10.1164/ajrccm.164.8.2105002. [DOI] [PubMed] [Google Scholar]
- 8.Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat Me, Tang W, Zhai G, Zhao JH, Smith AV, Huffman JE, Albrecht E, Jackson CM, Evans DM, Cadby G, Fornage M, Manichaikul A, Lopez LM, Johnson T, Aldrich MC, Aspelund T, Barroso I, Campbell H, Cassano PA, Couper DJ, Eiriksdottir G, Franceschini N, Garcia M, Gieger C, Gislason GK, Grkovic I, Hammond CJ, Hancock DB, Harris TB, Ramasamy A, Heckbert SR, Heliovaara M, Homuth G, Hysi PG, James AL, Jankovic S, Joubert BR, Karrasch S, Klopp N, Koch B, Kritchevsky SB, Launer LJ, Liu Y, Loehr LR, Lohman K, Loos RJF, Lumley T, Al Balushi KA, Ang WQ, Barr RG, Beilby J, Blakey JD, Boban M, Boraska V, Brisman J, Britton JR, Brusselle GG, Cooper C, Curjuric I, Dahgam S, Deary IJ, Ebrahim S, Eijgelsheim M, Francks C, Gaysina D, Granell R, Gu X, Hankinson JL, Hardy R, Harris SE, Henderson J, Henry A, Hingorani AD, Hofman A, Holt PG, Hui J, Hunter ML, Imboden M, Jameson KA, Kerr SM, Kolcic I, Kronenberg F, Liu JZ, Marchini J, McKeever T, Morris AD, Olin A-C, Porteous DJ, Postma DS, Rich SS, Ring SM, Rivadeneira F, Rochat T, Sayer AA, Sayers I, Sly PD, Smith GD, Sood A, Starr JM, Uitterlinden AG, Vonk JM, Wannamethee SG, Whincup PH, Wijmenga C, Williams OD, Wong A, Mangino M, Marciante KD, McArdle WL, Meibohm B, Morrison AC, North KE, Omenaas E, Palmer LJ, Pietilainen KH, Pin I, Polasek O, Pouta A, Psaty BM, Hartikainen A-L, Rantanen T, Ripatti S, Rotter JI, Rudan I, Rudnicka AR, Schulz H, Shin S-Y, Spector TD, Surakka I, Vitart V, Volzke H, Wareham NJ, Warrington NM, Wichmann HE, Wild SH, Wilk JB, Wjst M, Wright AF, Zgaga L, Zemunik T, Pennell CE, Nyberg F, Kuh D, Holloway JW, Boezen HM, Lawlor DA, Morris RW, Probst-Hensch N, Kaprio J, Wilson JF, Hayward C, Kahonen M, Heinrich J, Musk AW, Jarvis DL, Glaser S, Jarvelin M-R, Ch Stricker BH, Elliott P, O’Connor GT, Strachan DP, London SJ, Hall IP, Gudnason V, Tobin MD. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43(11):1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilk JB, Shrine NRG, Loehr LR, Zhao JH, Manichaikul A, Lopez LM, Smith AV, Heckbert SR, Smolonska J, Tang W, Loth DW, Curjuric I, Hui J, Cho MH, Latourelle JC, Henry AP, Aldrich M, Bakke P, Beaty TH, Bentley AR, Borecki IB, Brusselle GG, Burkart KM, Chen T-h, Couper D, Crapo JD, Davies G, Dupuis J, Franceschini N, Gulsvik A, Hancock DB, Harris TB, Hofman A, Imboden M, James AL, Khaw K-T, Lahousse L, Launer LJ, Litonjua A, Liu Y, Lohman KK, Lomas DA, Lumley T, Marciante KD, McArdle WL, Meibohm B, Morrison AC, Musk AW, Myers RH, North KE, Postma DS, Psaty BM, Rich SS, Rivadeneira F, Rochat T, Rotter JI, Soler Artigas M, Starr JM, Uitterlinden AG, Wareham NJ, Wijmenga C, Zanen P, Province MA, Silverman EK, Deary IJ, Palmer LJ, Cassano PA, Gudnason V, Barr RG, Loos RJF, Strachan DP, London SJ, Boezen HM, Probst-Hensch N, Gharib SA, Hall IP, O’Connor GT, Tobin MD, Stricker BH. Genome Wide Association Studies Identify CHRNA5/3 and HTR4 in the Development of Airflow Obstruction. American Journal of Respiratory and Critical Care Medicine. 2012 doi: 10.1164/rccm.201202-0366OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock DB, Soler Artigas M, Gharib SA, Henry A, Manichaikul A, Ramasamy A, Loth DW, Imboden M, Koch B, McArdle WL, Smith AV, Smolonska J, Sood A, Tang W, Wilk JB, Zhai G, Zhao JH, Aschard H, Burkart KM, Curjuric I, Eijgelsheim M, Elliott P, Gu X, Harris TB, Janson C, Homuth G, Hysi PG, Liu JZ, Loehr LR, Lohman K, Loos RJF, Manning AK, Marciante KD, Obeidat Me, Postma DS, Aldrich MC, Brusselle GG, Chen T-h, Eiriksdottir G, Franceschini N, Heinrich J, Rotter JI, Wijmenga C, Williams OD, Bentley AR, Hofman A, Laurie CC, Lumley T, Morrison AC, Joubert BR, Rivadeneira F, Couper DJ, Kritchevsky SB, Liu Y, Wjst M, Wain LV, Vonk JM, Uitterlinden AG, Rochat T, Rich SS, Psaty BM, O’Connor GT, North KE, Mirel DB, Meibohm B, Launer LJ, Khaw K-T, Hartikainen A-L, Hammond CJ, Gläser S, Marchini J, Kraft P, Wareham NJ, Völzke H, Stricker BHC, Spector TD, Probst-Hensch NM, Jarvis D, Jarvelin M-R, Heckbert SR, Gudnason V, Boezen HM, Barr RG, Cassano PA, Strachan DP, Fornage M, Hall IP, Dupuis J, Tobin MD, London SJ. Genome-Wide Joint Meta-Analysis of SNP and SNP-by-Smoking Interaction Identifies Novel Loci for Pulmonary Function. PLoS Genet. 2012;8(12):e1003098. doi: 10.1371/journal.pgen.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, Zhao JH, Ramasamy A, Zhai G, Vitart V, Huffman JE, Igl W, Albrecht E, Deloukas P, Henderson J, Granell R, McArdle WL, Rudnicka AR, Wellcome Trust Case Control C. Barroso I, Loos RJ, Wareham NJ, Mustelin L, Rantanen T, Surakka I, Imboden M, Wichmann HE, Grkovic I, Jankovic S, Zgaga L, Hartikainen AL, Peltonen L, Gyllensten U, Johansson A, Zaboli G, Campbell H, Wild SH, Wilson JF, Glaser S, Homuth G, Volzke H, Mangino M, Soranzo N, Spector TD, Polasek O, Rudan I, Wright AF, Heliovaara M, Ripatti S, Pouta A, Naluai AT, Olin AC, Toren K, Cooper MN, James AL, Palmer LJ, Hingorani AD, Wannamethee SG, Whincup PH, Smith GD, Ebrahim S, McKeever TM, Pavord ID, MacLeod AK, Morris AD, Porteous DJ, Cooper C, Dennison E, Shaheen S, Karrasch S, Schnabel E, Schulz H, Grallert H, Bouatia-Naji N, Delplanque J, Froguel P, Blakey JD, Team NRS. Britton JR, Morris RW, Holloway JW, Lawlor DA, Hui J, Nyberg F, Jarvelin MR, Jackson C, Kahonen M, Kaprio J, Probst-Hensch NM, Koch B, Hayward C, Evans DM, Elliott P, Strachan DP, Hall IP, Tobin MD. Genome-wide association study identifies five loci associated with lung function. Nature Genetics. 2010;42(1):36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, Franceschini N, van Durme YM, Chen TH, Barr RG, Schabath MB, Couper DJ, Brusselle GG, Psaty BM, van Duijn CM, Rotter JI, Uitterlinden AG, Hofman A, Punjabi NM, Rivadeneira F, Morrison AC, Enright PL, North KE, Heckbert SR, Lumley T, Stricker BH, O’Connor GT, London SJ. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nature Genetics. 2010;42(1):45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilk JB. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, DeMeo DL, Hunninghake GM, Litonjua AA, Sparrow D, Lange C, Won S, Murphy JR, Beaty TH, Regan EA, Make BJ, Hokanson JE, Crapo JD, Kong X, Anderson WH, Tal-Singer R, Lomas DA, Bakke P, Gulsvik A, Pillai SG, Silverman EK. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42(3):200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soler Artigas M, Wain LV, Repapi E, Obeidat Me, Sayers I, Burton PR, Johnson T, Zhao JH, Albrecht E, Dominiczak AF, Kerr SM, Smith BH, Cadby G, Hui J, Palmer LJ, Hingorani AD, Wannamethee SG, Whincup PH, Ebrahim S, Smith GD, Barroso I, Loos RJF, Wareham NJ, Cooper C, Dennison E, Shaheen SO, Liu JZ, Marchini J, Health MRCNSo. Team DRS. Dahgam S, Naluai ÅT, Olin A-C, Karrasch S, Heinrich J, Schulz H, McKeever TM, Pavord ID, Heliövaara M, Ripatti S, Surakka I, Blakey JD, Kähönen M, Britton JR, Nyberg F, Holloway JW, Lawlor DA, Morris RW, James AL, Jackson CM, Hall IP, Tobin MD, Consortium tS Effect of Five Genetic Variants Associated with Lung Function on the Risk of Chronic Obstructive Lung Disease, and Their Joint Effects on Lung Function. American Journal of Respiratory and Critical Care Medicine. 2011;184(7):786–795. doi: 10.1164/rccm.201102-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castaldi PJ, Cho MH, Litonjua AA, Bakke P, Gulsvik A, Lomas DA, Anderson W, Beaty TH, Hokanson JE, Crapo JD, Laird N, Silverman EK, COPDGene ft. Investigators E The Association of Genome-Wide Significant Spirometric Loci with Chronic Obstructive Pulmonary Disease Susceptibility. American Journal of Respiratory Cell and Molecular Biology. 2011;45(6):1147–1153. doi: 10.1165/rcmb.2011-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel BD, Coxson HO, Pillai SG, Agusti AGN, Calverley PMA, Donner CF, Make BJ, Muller NL, Rennard SI, Vestbo J, Wouters EFM, Hiorns MP, Nakano Y, Camp PG, Nasute Fauerbach PV, Screaton NJ, Campbell EJ, Anderson WH, Pare PD, Levy RD, Lake SL, Silverman EK, Lomas DA, International COPD Genetics Network Airway Wall Thickening and Emphysema Show Independent Familial Aggregation in Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2008;178(5):500–505. doi: 10.1164/rccm.200801-059OC. [DOI] [PubMed] [Google Scholar]
- 18.Kong X, Cho MH, Anderson W, Coxson HO, Muller N, Washko G, Hoffman EA, Bakke P, Gulsvik A, Lomas DA, Silverman EK, Pillai SG, ECLIPSE Study NETT Investigators Genome-wide Association Study Identifies BICD1 as a Susceptibility Gene for Emphysema. Am J Respir Crit Care Med. 2011;183(1):43–49. doi: 10.1164/rccm.201004-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeMeo DL, Hersh CP, Hoffman EA, Litonjua AA, Lazarus R, Sparrow D, Benditt JO, Criner G, Make B, Martinez FJ, Scanlon PD, Sciurba FC, Utz JP, Reilly JJ, Silverman EK. Genetic Determinants of Emphysema Distribution in the National Emphysema Treatment Trial. Am J Respir Crit Care Med. 2007;176(1):42–48. doi: 10.1164/rccm.200612-1797OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hersh CP, Hansel NN, Barnes KC, Lomas DA, Pillai SG, Coxson HO, Mathias RA, Rafaels NM, Wise RA, Connett JE, Klanderman BJ, Jacobson FL, Gill R, Litonjua AA, Sparrow D, Reilly JJ, Silverman EK, Investigators I Transforming growth factor-beta receptor-3 is associated with pulmonary emphysema. American Journal of Respiratory Cell & Molecular Biology. 2009;41(3):324–331. doi: 10.1165/rcmb.2008-0427OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambrechts D, Buysschaert I, Zanen P, Coolen J, Lays N, Cuppens H, Groen HJM, Dewever W, van Klaveren RJ, Verschakelen J, Wijmenga C, Postma DS, Decramer M, Janssens W. The 15q24/25 Susceptibility Variant for Lung Cancer and Chronic Obstructive Pulmonary Disease Is Associated with Emphysema. Am J Respir Crit Care Med. 2010;181(5):486–493. doi: 10.1164/rccm.200909-1364OC. [DOI] [PubMed] [Google Scholar]
- 22.Kim DK, Hersh C, Washko G, Hokanson J, Lynch D, Newell J, Murphy J, Crapo J, Silverman E, Investigators tCG Epidemiology, radiology, and genetics of nicotine dependence in COPD. Respiratory Research. 2011;12(1):9. doi: 10.1186/1465-9921-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamada M, Ishii T, Ikeda S, Naka-Mieno M, Tanaka N, Arai T, Kumasaka T, Gemma A, Kida K, Muramatsu M, Sawabe M. Association of fucosyltransferase 8 (FUT8) polymorphism Thr267Lys with pulmonary emphysema. Journal of Human Genetics. 2011;56(12):857–860. doi: 10.1038/jhg.2011.118. [DOI] [PubMed] [Google Scholar]
- 24.Ishii T, Hagiwara K, Ikeda S, Arai T, Mieno MN, Kumasaka T, Muramatsu M, Sawabe M, Gemma A, Kida K. Association between genetic variations in surfactant protein d and emphysema, interstitial pneumonia, and lung cancer in a Japanese population. Copd: Journal of Chronic Obstructive Pulmonary Disease. 2012;9(4):409–416. doi: 10.3109/15412555.2012.676110. [DOI] [PubMed] [Google Scholar]
- 25.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SFA, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, DerOhannessian S, de Bakker PIW, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CWK, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, FitzGerald GA. Concept, Design and Implementation of a Cardiovascular Gene-Centric 50 K SNP Array for Large-Scale Genomic Association Studies. PLoS One. 2008;3(10):e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giordano RJ, Lahdenranta J, Zhen L, Chukwueke U, Petrache I, Langley RR, Fidler IJ, Pasqualini R, Tuder RM, Arap W. Targeted Induction of Lung Endothelial Cell Apoptosis Causes Emphysema-like Changes in the Mouse. Journal of Biological Chemistry. 2008;283(43):29447–29460. doi: 10.1074/jbc.M804595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clauss M, Voswinckel R, Rajashekhar G, Sigua NL, Fehrenbach H, Rush NI, Schweitzer KS, Yildirim AÖ, Kamocki K, Fisher AJ, Gu Y, Safadi B, Nikam S, Hubbard WC, Tuder RM, Twigg HL, Presson RG, Sethi S, Petrache I. Lung endothelial monocyte-activating protein 2 is a mediator of cigarette smoke-induced emphysema in mice. The Journal of Clinical Investigation. 2011;121(6):2470–2479. doi: 10.1172/JCI43881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barr RG, Bluemke DA, Ahmed FS, Carr JJ, Enright PL, Hoffman EA, Jiang R, Kawut SM, Kronmal RA, Lima JAC, Shahar E, Smith LJ, Watson KE. Percent Emphysema, Airflow Obstruction, and Impaired Left Ventricular Filling. New England Journal of Medicine. 2010;362(3):217–227. doi: 10.1056/NEJMoa0808836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barr RG, Ahmed FS, Carr JJ, Hoffman EA, Jiang R, Kawut SM, Watson K. Subclinical atherosclerosis, airflow obstruction and emphysema: the MESA Lung Study. European Respiratory Journal. 2012;39(4):846–854. doi: 10.1183/09031936.00165410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J-a, Montagnani M, Koh KK, Quon MJ. Reciprocal relationships between insulin resistance and endothelial dysfunction: molecular and pathophysiological mechanisms. Circulation. 2006;113(15):1888–1904. doi: 10.1161/CIRCULATIONAHA.105.563213. [DOI] [PubMed] [Google Scholar]
- 31.Kasahara Y, Tuder RM, Cool CD, Lynch DA, Flores SC, Voelkel NF. Endothelial cell death and decreased expression of vascular endothelial growth factor and vascular endothelial growth factor receptor 2 in emphysema. American Journal of Respiratory & Critical Care Medicine. 2001;163(3 Pt 1):737–744. doi: 10.1164/ajrccm.163.3.2002117. [DOI] [PubMed] [Google Scholar]
- 32.Santos S, Peinado VI, Ramirez J, Morales-Blanhir J, Bastos R, Roca J, Rodriguez-Roisin R, Barbera JA. Enhanced expression of vascular endothelial growth factor in pulmonary arteries of smokers and patients with moderate chronic obstructive pulmonary disease. American Journal of Respiratory & Critical Care Medicine. 2003;167(9):1250–1256. doi: 10.1164/rccm.200210-1233OC. [DOI] [PubMed] [Google Scholar]
- 33.Kanazawa H, Asai K, Hirata K, Yoshikawa J. Possible effects of vascular endothelial growth factor in the pathogenesis of chronic obstructive pulmonary disease. American Journal of Medicine. 2003;114(5):354–358. doi: 10.1016/s0002-9343(02)01562-0. [DOI] [PubMed] [Google Scholar]
- 34.Leibow AA. Pulmonary emphysema with special reference to vascular change. Am Rev Respir Dis. 1959;80:67–93. doi: 10.1164/arrd.1959.80.1P2.67. [DOI] [PubMed] [Google Scholar]
- 35.Barr RG, Mesia-Vela S, Austin JHM, Basner RC, Keller BM, Reeves AP, Shimbo D, Stevenson L. Impaired Flow-mediated Dilation Is Associated with Low Pulmonary Function and Emphysema in Ex-smokers: The Emphysema and Cancer Action Project (EMCAP) Study. Am J Respir Crit Care Med. 2007;176(12):1200–1207. doi: 10.1164/rccm.200707-980OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuder RM, Petrache I, Elias JA, Voelkel NF, Henson PM. Apoptosis and emphysema: the missing link. American Journal of Respiratory Cell & Molecular Biology. 2003;28(5):551–554. doi: 10.1165/rcmb.F269. comment. [DOI] [PubMed] [Google Scholar]
- 37.Tuder RM, Zhen L, Cho CY, Taraseviciene-Stewart L, Kasahara Y, Salvemini D, Voelkel NF, Flores SC. Oxidative stress and apoptosis interact and cause emphysema due to vascular endothelial growth factor receptor blockade. American Journal of Respiratory Cell & Molecular Biology. 2003;29(1):88–97. doi: 10.1165/rcmb.2002-0228OC. [DOI] [PubMed] [Google Scholar]
- 38.Petrache I, Natarajan V, Zhen L, Medler TR, Richter AT, Cho C, Hubbard WC, Berdyshev EV, Tuder RM. Ceramide upregulation causes pulmonary cell apoptosis and emphysema-like disease in mice. Nature Medicine. 2005;11(5):491–498. doi: 10.1038/nm1238. see comment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia P, Vadas MA, Rye K-A, Barter PJ, Gamble JR. High Density Lipoproteins (HDL) Interrupt the Sphingosine Kinase Signaling Pathway. Journal of Biological Chemistry. 1999;274(46):33143–33147. doi: 10.1074/jbc.274.46.33143. [DOI] [PubMed] [Google Scholar]
- 40.Wolfrum C, Poy MN, Stoffel M. Apolipoprotein M is required for prebeta-HDL formation and cholesterol efflux to HDL and protects against atherosclerosis. Nature Medicine. 2005;11(4):418–422. doi: 10.1038/nm1211. [DOI] [PubMed] [Google Scholar]
- 41.Christoffersen C, Jauhiainen M, Moser M, Porse B, Ehnholm C, Boesl M, Dahlbäck B, Nielsen LB. Effect of Apolipoprotein M on High Density Lipoprotein Metabolism and Atherosclerosis in Low Density Lipoprotein Receptor Knock-out Mice. Journal of Biological Chemistry. 2008;283(4):1839–1847. doi: 10.1074/jbc.M704576200. [DOI] [PubMed] [Google Scholar]
- 42.Christoffersen C, Obinata H, Kumaraswamy SB, Galvani S, Ahnström J, Sevvana M, Egerer-Sieber C, Muller YA, Hla T, Nielsen LB, Dahlbäck B. Endothelium-protective sphingosine-1-phosphate provided by HDL-associated apolipoprotein M. Proceedings of the National Academy of Sciences. 2011;108(23):9613–9618. doi: 10.1073/pnas.1103187108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musunuru K, Lettre G, Young T, Farlow DN, Pirruccello JP, Ejebe KG, Keating BJ, Yang Q, Chen M-H, Lapchyk N, Crenshaw A, Ziaugra L, Rachupka A, Benjamin EJ, Cupples LA, Fornage M, Fox ER, Heckbert SR, Hirschhorn JN, Newton-Cheh C, Nizzari MM, Paltoo DN, Papanicolaou GJ, Patel SR, Psaty BM, Rader DJ, Redline S, Rich SS, Rotter JI, Taylor HA, Jr, Tracy RP, Vasan RS, Wilson JG, Kathiresan S, Fabsitz RR, Boerwinkle E, Gabriel SB, NHLBI Candidate Gene Association Resource Candidate Gene Association Resource (CARe): Design, Methods, and Proof of Concept. Circ Cardiovasc Genet. 2010;3(3):267–275. doi: 10.1161/CIRCGENETICS.109.882696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacobs DR, Jr, Kronmal R, Liu K, Nelson JC, O’Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-ethnic study of atherosclerosis: objectives and design. American Journal of Epidemiology. 2002;156(9):871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 45.The ARIC investigators The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 46.Fried LP. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 47.Dawber TR, Kannel WB. The Framingham study. An epidemiological approach to coronary heart disease. Circulation. 1966;34:553–555. doi: 10.1161/01.cir.34.4.553. [DOI] [PubMed] [Google Scholar]
- 48.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4:518–525. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 49.Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR, Liu K, Savage PJ. Cardia: study design, recruitment, and some characteristics of the examined subjects. Journal of Clinical Epidemiology. 1988;41(11):1105–1116. doi: 10.1016/0895-4356(88)90080-7. [DOI] [PubMed] [Google Scholar]
- 50.Buxbaum SG, Elston RC, Tishler PV, Redline S. Genetics of the apnea hypopnea index in Caucasians and African Americans: I. Segregation analysis. Genetic Epidemiology. 2002;22(3):243–253. doi: 10.1002/gepi.0170. [DOI] [PubMed] [Google Scholar]
- 51.Taylor HA., Jr. The Jackson Heart Study: an overview. Ethnicity & Disease. 2005;15(4 Suppl 6):S6-1–3. [PubMed] [Google Scholar]
- 52.Carpenter MA, Crow R, Steffes M, Rock W, Heilbraun J, Evans G, Skelton T, Jensen R, Sarpong D. Laboratory, reading center, and coordinating center data management methods in the Jackson Heart Study. American Journal of the Medical Sciences. 2004;328(3):131–144. doi: 10.1097/00000441-200409000-00001. [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez J, Jiang R, Johnson WC, MacKenzie BA, Smith LJ, Barr RG. The Association of Pipe and Cigar Use With Cotinine Levels, Lung Function, and Airflow Obstruction. Annals of Internal Medicine. 2010;152(4):201–210. doi: 10.1059/0003-4819-152-4-201002160-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. American Journal of Respiratory & Critical Care Medicine. 1999;159(1):179–187. doi: 10.1164/ajrccm.159.1.9712108. see comment. [DOI] [PubMed] [Google Scholar]
- 55.Manichaikul A, Chen W-M, Williams K, Wong Q, Sale M, Pankow J, Tsai M, Rotter J, Rich S, Mychaleckyj J. Analysis of family- and population-based samples in cohort genome-wide association studies. Human Genetics. 2012;131(2):275–287. doi: 10.1007/s00439-011-1071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaufman JD, Adar SD, Allen RW, Barr RG, Budoff MJ, Burke GL, Casillas AM, Cohen MA, Curl CL, Daviglus ML, Diez Roux AV, Jacobs DR, Jr., Kronmal RA, Larson TV, Liu SL-J, Lumley T, Navas-Acien A, O’Leary DH, Rotter JI, Sampson PD, Sheppard L, Siscovick DS, Stein JH, Szpiro AA, Tracy RP. Prospective study of particulate air pollution exposures, subclinical atherosclerosis, and clinical cardiovascular disease: The Multi-Ethnic Study of Atherosclerosis and Air Pollution (MESA Air) American Journal of Epidemiology. 2012;176(9):825–837. doi: 10.1093/aje/kws169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffman EA, Jiang R, Baumhauer H, Brooks MA, Carr JJ, Detrano R, Reinhardt J, Rodriguez J, Stukovsky K, Wong ND, Barr RG. Reproducibility and Validity of Lung Density Measures from Cardiac CT Scans--The Multi-Ethnic Study of Atherosclerosis (MESA) Lung Study1. Academic Radiology. 2009;16(6):689–699. doi: 10.1016/j.acra.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai MY, Johnson C, Kao WHL, Sharrett AR, Arends VL, Kronmal R, Jenny NS, Jacobs DR, Jr, Arnett D, O’Leary D, Post W. Cholesteryl ester transfer protein genetic polymorphisms, HDL cholesterol, and subclinical cardiovascular disease in the Multi-Ethnic Study of Atherosclerosis. Atherosclerosis. 2008;200(2):359–367. doi: 10.1016/j.atherosclerosis.2007.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Purcell S. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen M-H, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26(4):580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price AL. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 62.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scandinavian Journal of Statistics. 1979;6(2):65–70. [Google Scholar]
- 64.Kappelle PJWH, Lambert G, Dahlbäck B, Nielsen LB, Dullaart RPF. Relationship of plasma apolipoprotein M with proprotein convertase subtilisin–kexin type 9 levels in non-diabetic subjects. Atherosclerosis. 2011;214(2):492–494. doi: 10.1016/j.atherosclerosis.2010.10.041. [DOI] [PubMed] [Google Scholar]
- 65.Hu HJ, Jin EH, Yim SH, Yang SY, Jung SH, Shin SH, Kim WU, Shim SC, Kim TG, Chung YJ. Common variants at the promoter region of the APOM confer a risk of rheumatoid arthritis. Experimental & Molecular Medicine. 2011;43(11):613–621. doi: 10.3858/emm.2011.43.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu N, Dahlbäck B. A Novel Human Apolipoprotein (apoM) Journal of Biological Chemistry. 1999;274(44):31286–31290. doi: 10.1074/jbc.274.44.31286. [DOI] [PubMed] [Google Scholar]
- 67.Nielsen LB, Christoffersen C, Ahnström J, Dahlbäck B. ApoM: gene regulation and effects on HDL metabolism. Trends in Endocrinology & Metabolism. 2009;20(2):66–71. doi: 10.1016/j.tem.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 68.Xia P, Gamble JR, Rye KA, Wang L, Hii CS, Cockerill P, Khew-Goodall Y, Bert AG, Barter PJ, Vadas MA. Tumor necrosis factor-alpha induces adhesion molecule expression through the sphingosine kinase pathway. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(24):14196–14201. doi: 10.1073/pnas.95.24.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karuna R, Park R, Othman A, Holleboom AG, Motazacker MM, Sutter I, Kuivenhoven JA, Rohrer L, Matile H, Hornemann T, Stoffel M, Rentsch KM, von Eckardstein A. Plasma levels of sphingosine-1-phosphate and apolipoprotein M in patients with monogenic disorders of HDL metabolism. Atherosclerosis. 2011;219(2):855–863. doi: 10.1016/j.atherosclerosis.2011.08.049. [DOI] [PubMed] [Google Scholar]
- 70.Diab KJ, Adamowicz JJ, Kamocki K, Rush NI, Garrison J, Gu Y, Schweitzer KS, Skobeleva A, Rajashekhar G, Hubbard WC, Berdyshev EV, Petrache I. Stimulation of Sphingosine 1-Phosphate Signaling as an Alveolar Cell Survival Strategy in Emphysema. Am J Respir Crit Care Med. 2010;181(4):344–352. doi: 10.1164/rccm.200906-0826OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karlsson H, Leanderson P, Tagesson C, Lindahl M. Lipoproteomics II: Mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. PROTEOMICS. 2005;5(5):1431–1445. doi: 10.1002/pmic.200401010. [DOI] [PubMed] [Google Scholar]
- 72.Ortiz-Muñoz G, Houard X, Martín-Ventura J-L, Ishida BY, Loyau S, Rossignol P, Moreno J-A, Kane JP, Chalkley RJ, Burlingame AL, Michel J-B, Meilhac O. HDL antielastase activity prevents smooth muscle cell anoikis, a potential new antiatherogenic property. The FASEB Journal. 2009;23(9):3129–3139. doi: 10.1096/fj.08-127928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Holla ØL, Laerdahl JK, Strøm TB, Tveten K, Cameron J, Berge KE, Leren TP. Removal of acidic residues of the prodomain of PCSK9 increases its activity towards the LDL receptor. Biochemical and Biophysical Research Communications. 2011;406(2):234–238. doi: 10.1016/j.bbrc.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 74.Abifadel M, Varret M, Rabes J-P, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, Derre A, Villeger L, Farnier M, Beucler I, Bruckert E, Chambaz J, Chanu B, Lecerf J-M, Luc G, Moulin P, Weissenbach J, Prat A, Krempf M, Junien C, Seidah NG, Boileau C. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34(2):154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 75.Abifadel M, Rabes J-P, Devillers M, Munnich A, Erlich D, Junien C, Varret M, Boileau C. Mutations and polymorphisms in the proprotein convertase subtilisin kexin 9 (PCSK9) gene in cholesterol metabolism and disease. Human Mutation. 2009;30(4):520–529. doi: 10.1002/humu.20882. [DOI] [PubMed] [Google Scholar]
- 76.Aung LHH, Yin R-X, Miao L, Hu X-J, Yan T-T, Cao X-L, Wu D-F, Li Q, Pan S-L, Wu J-Z. The proprotein convertase subtilisin/kexin type 9 gene E670G polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids in Health & Disease. 2011;10:5. doi: 10.1186/1476-511X-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cohen JC, Boerwinkle E, Mosley TH, Hobbs HH. Sequence Variations in PCSK9, Low LDL, and Protection against Coronary Heart Disease. New England Journal of Medicine. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 78.Sullivan D, Olsson AG, Scott R, Kim JB, Xue A, Gebski V, Wasserman SM, Stein EA. Effect of a monoclonal antibody to PCSK9 on low-density lipoprotein cholesterol levels in statin-intolerant patients: the GAUSS randomized trial. JAMA. 2012;308(23):2497–2506. doi: 10.1001/jama.2012.25790. [DOI] [PubMed] [Google Scholar]
- 79.Raal F, Scott R, Somaratne R, Bridges I, Li G, Wasserman SM, Stein EA. Low-density lipoprotein cholesterol-lowering effects of AMG 145, a monoclonal antibody to proprotein convertase subtilisin/kexin type 9 serine protease in patients with heterozygous familial hypercholesterolemia: the Reduction of LDL-C with PCSK9 Inhibition in Heterozygous Familial Hypercholesterolemia Disorder (RUTHERFORD) randomized trial. Circulation. 2012;126(20):2408–2417. doi: 10.1161/CIRCULATIONAHA.112.144055. [DOI] [PubMed] [Google Scholar]
- 80.Giugliano RP, Desai NR, Kohli P, Rogers WJ, Somaratne R, Huang F, Liu T, Mohanavelu S, Hoffman EB, McDonald ST, Abrahamsen TE, Wasserman SM, Scott R, Sabatine MS. Efficacy, safety, and tolerability of a monoclonal antibody to proprotein convertase subtilisin/kexin type 9 in combination with a statin in patients with hypercholesterolaemia (LAPLACE-TIMI 57): a randomised, placebo-controlled, dose-ranging, phase 2 study. The Lancet. 380(9858):2007–2017. doi: 10.1016/S0140-6736(12)61770-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roth EM, McKenney JM, Hanotin C, Asset G, Stein EA. Atorvastatin with or without an Antibody to PCSK9 in Primary Hypercholesterolemia. New England Journal of Medicine. 2012;367(20):1891–1900. doi: 10.1056/NEJMoa1201832. [DOI] [PubMed] [Google Scholar]
- 82.Dias CS, Shaywitz AJ, Wasserman SM, Smith BP, Gao B, Stolman DS, Crispino CP, Smirnakis KV, Emery MG, Colbert A, Gibbs JP, Retter MW, Cooke BP, Uy ST, Matson M, Stein EA. Effects of AMG 145 on Low-Density Lipoprotein Cholesterol Levels: Results From 2 Randomized, Double-Blind, Placebo-Controlled, Ascending-Dose Phase 1 Studies in Healthy Volunteers and Hypercholesterolemic Subjects on Statins. Journal of the American College of Cardiology. 2012;60(19):1888–1898. doi: 10.1016/j.jacc.2012.08.986. [DOI] [PubMed] [Google Scholar]
- 83.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Hólm H, Ding EL, Johnson T, Schunkert H, Samani NJ, Clarke R, Hopewell JC, Thompson JF, Li M, Thorleifsson G, Newton-Cheh C, Musunuru K, Pirruccello JP, Saleheen D, Chen L, Stewart AFR, Schillert A, Thorsteinsdottir U, Thorgeirsson G, Anand S, Engert JC, Morgan T, Spertus J, Stoll M, Berger K, Martinelli N, Girelli D, McKeown PP, Patterson CC, Epstein SE, Devaney J, Burnett M-S, Mooser V, Ripatti S, Surakka I, Nieminen MS, Sinisalo J, Lokki M-L, Havulinna A, de Faire U, Gigante B, Ingelsson E, Zeller T, Wild P, de Bakker PIW, Klungel OH, Maitland-van der Zee A-H, Peters BJM, de Boer A, Grobbee DE, Kamphuisen PW, Deneer VHM, Elbers CC, Onland-Moret NC, Hofker MH, Wijmenga C, Verschuren WMM, Boer JMA, van der Schouw YT, Rasheed A, Frossard P, Demissie S, Willer C, Do R, Ordovas JM, Abecasis GR, Boehnke M, Mohlke KL, Daly MJ, Guiducci C, Burtt NP, Surti A, Gonzalez E, Purcell S, Gabriel S, Marrugat J, Peden J, Erdmann J, Diemert P, Willenborg C, König IR, Fischer M, Hengstenberg C, Ziegler A, Buysschaert I, Lambrechts D, Van de Werf F, Fox KA, El Mokhtari NE, Rubin D, Schrezenmeir J, Schreiber S, Schäfer A, Danesh J, Blankenberg S, Roberts R, McPherson R, Watkins H, Hall AS, Overvad K, Rimm E, Boerwinkle E, Tybjaerg-Hansen A, Cupples LA, Reilly MP, Melander O, Mannucci PM, Ardissino D, Siscovick D, Elosua R, Stefansson K, O’Donnell CJ, Salomaa V, Rader DJ, Peltonen L, Schwartz SM, Altshuler D, Kathiresan S, Perola M. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. The Lancet. 2012 doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJP, Komajda M, Lopez-Sendon J, Mosca L, Tardif J-C, Waters DD, Shear CL, Revkin JH, Buhr KA, Fisher MR, Tall AR, Brewer B. Effects of Torcetrapib in Patients at High Risk for Coronary Events. New England Journal of Medicine. 2007;357(21):2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 85.Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, Chaitman BR, Holme IM, Kallend D, Leiter LA, Leitersdorf E, McMurray JJV, Mundl H, Nicholls SJ, Shah PK, Tardif J-C, Wright RS. Effects of Dalcetrapib in Patients with a Recent Acute Coronary Syndrome. New England Journal of Medicine. 2012;367(22):2089–2099. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 86.Ahnstrom J, Axler O, Jauhiainen M, Salomaa V, Havulinna AS, Ehnholm C, Frikke-Schmidt R, Tybjaerg-Hansen A, Dahlback B. Levels of apolipoprotein M are not associated with the risk of coronary heart disease in two independent case-control studies. Journal of Lipid Research. 2008;49(9):1912–1917. doi: 10.1194/jlr.M700471-JLR200. [DOI] [PubMed] [Google Scholar]
- 87.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Research. 2009;37:D32–36. doi: 10.1093/nar/gkn721. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rudd MF, Webb EL, Matakidou A, Sellick GS, Williams RD, Bridle H, Eisen T, Houlston RS, Consortium G Variants in the GH-IGF axis confer susceptibility to lung cancer. Genome Research. 2006;16(6):693–701. doi: 10.1101/gr.5120106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV, Spitz MR, Eisen T, Amos CI, Houlston RS. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40(12):1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tisi GM, Conrique A, Barrett-Connor E, Grundy SM. Increased high density lipoprotein cholesterol in obstructive pulmonary disease (predominant emphysematous type) Metabolism: Clinical & Experimental. 1981;30(4):340–346. doi: 10.1016/0026-0495(81)90113-x. [DOI] [PubMed] [Google Scholar]
- 91.Reed RM, Iacono A, DeFilippis A, Eberlein M, Girgis RE, Jones S. Advanced chronic obstructive pulmonary disease is associated with high levels of high-density lipoprotein cholesterol. The Journal of heart and lung transplantation: the official publication of the International Society for Heart Transplantation. 2011;30(6):674–678. doi: 10.1016/j.healun.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 92.Cirillo DJ, Agrawal Y, Cassano PA. Lipids and pulmonary function in the Third National Health and Nutrition Examination Survey. American Journal of Epidemiology. 2002;155(9):842–848. doi: 10.1093/aje/155.9.842. [DOI] [PubMed] [Google Scholar]
- 93.Ubhi BK, Riley JH, Shaw PA, Lomas DA, Tal-Singer R, MacNee W, Griffin JL, Connor SC. Metabolic profiling detects biomarkers of protein degradation in COPD patients. European Respiratory Journal. 2012;40(2):345–355. doi: 10.1183/09031936.00112411. [DOI] [PubMed] [Google Scholar]
- 94.Reed RM, Hashmi S, Eberlein M, Iacono A, Netzer G, DeFilippis A, Girgis RE, Toth PP, Scharf S, Jones S. Impact of lung transplantation on serum lipids in COPD. Respiratory Medicine. (0) doi: 10.1016/j.rmed.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 95.Lovasi GS, Diez Roux AV, Hoffman EA, Smith LJ, Jiang R, Carr JJ, Barr RG. Socioeconomic Status is Positively Associated with Percent Emphysema on CT Scan: The MESA Lung Study. Academic Radiology. 2011;18(2):199–204. doi: 10.1016/j.acra.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ashraf H, Lo P, Shaker SB, de Bruijne M, Dirksen A, Tønnesen P, Dahlbäck M, Pedersen JH. Short-term effect of changes in smoking behaviour on emphysema quantification by CT. Thorax. 2011;66(1):55–60. doi: 10.1136/thx.2009.132688. [DOI] [PubMed] [Google Scholar]
- 97.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Cohort and race specific quantile-quantile plots of association results for FEV1/FVC.
Cohort and race specific quantile-quantile results for all SNPs associated with FEV1/FVC. (A) Cohort specific genomic inflation factors calculated after controlling for 10 principal components analyses for ancestry for European-American participants. (B) Cohort specific genomic inflation factors calculated after controlling for 10 principal components analyses for ancestry for African-American participants.
Supplementary Figure 7 Linkage Disequilibrium Plots based on population samples.
D’ pairwise linkage disequilibrium (LD) plots of population data from CEU-Utah residents with Northern and Western ancestry from CEPH collection (A) and ASW-African ancestry in Southwest USA (B). LD plots created with Haploview, HapMap 3 release 2. Red: D’ = 1 (LOD ≥ 2.0); blue: D’ = 1 (LOD < 2.0); pink: D’ < 1 (LOD ≥ 2.0); white: D’ < 1 (LOD < 2.0).
APOM SNP (rs805297), BAG6 SNP (rs805301) and GPANK1 SNP (rs707974) are circled in green. In the CEU population (a) Block 2 is 11kb in length and includes rs805301 and rs805297. Although rs707974 is in Block 3 (1kb), the D’=1 (LOD ≥ 2.0) for rs707974 and rs805297. In the ASW population (b) Block 3 is 11 kb in length and includes rs805297 and rs707974. SNP rs805301 is in Block 2 (2 kb) and the D’=0.78 (LOD<2.0) for rs805301 and rs805297.
APOM SNP (rs805297) is in LD with rs707974 among African-Americans and European-Americans (r2=0.36 and r2=0.32, respectively) and it is in LD with rs805301 among European-Americans (r2=0.23) but not African-Americans (r2=0.06) using HapMap3 release2.
LOD = logarithm of odds.
Supplementary Figure 2 Quantile-quantile plots of meta-analysis association results for FEV1/FVC stratified by race and across all races.
Quantile-quantile results are for meta-analysis of cohorts for SNPs associated with FEV1/FVC stratified by race and combined across all races. Genomic inflation factors calculated after controlling for 10 principal components for ancestry.
Supplementary Figure 3 Regional association plots of top SNPs associated with FEV1/FVC stratified by race and combined across all races.
Regional association plots of top SNPs for FEV1/FVC in European-American participants (A-D), African-American participants (E-H) and combined analysis of European-American and African-American participants (I-M). The selected SNPs with the lowest p value are illustrated by the purple diamond. The correlations (r2) of surrounding SNPs in the region are indicated by the colors shown on the graph. A 500Kb flanking size was selected for all plots. Plots were generated using LocusZoom. The Genome builds/LD populations implemented were hg 18/HapMap Phase II CEU for European-American and combined European-American and African-American analyses. The hg/19 1000 Genomes Nov 2010 AFR for African-American participants.
Abbreviations: SNPs = single nucleotide polymorphisms, FEV1/FVC = ratio of forced expiratory volume in one second over forced vital capacity.
Supplementary Figure 4 Manhattan Plots ordered by chromosome position of association results for FEV1.
(A) Meta-analysis of 38,294 SNPs among 20,077 European-American participants. (B) Meta-analysis of 44,416 SNPs among 6,900 African-American participants. (C) Meta-analysis of SNPs among combined European-American & African-American participants.
Abbreviations: FEV1 = forced expiratory volume in one second.
Supplementary Figure 5 Quantile-quantile plots of meta-analysis association results for FEV1 stratified by race and across all races.
Quantile-quantile results are for meta-analysis of cohorts for SNPs associated with FEV1 stratified by race and combined across all races. Genomic inflation factors calculated after controlling for 10 principal components.
Supplementary Figure 6 Regional association plots of top SNPs associated with FEV1 stratified by race and combined across all races.
Regional association plots of top SNPs for FEV1 in European-American participants (A-E), African-American participants (F-I) and combined analysis of European-American and African-American participants (J-N). The selected SNPs with the lowest p value are illustrated by the purple diamond. The correlations (r2) of surrounding SNPs in the region are indicated by the colors shown on the graph. A 500Kb flanking size was selected for all plots. Plots were generated using LocusZoom. The Genome builds/LD populations implemented were hg 18/HapMap Phase II CEU for European-American and combined European-American and African-American analyses. The hg/19 1000 Genomes Nov 2010 AFR for African-American participants.
Abbreviations: SNPs = single nucleotide polymorphisms, FEV1/FVC = ratio of forced expiratory volume in one second over forced vital capacity.


