Abstract
Lipins are phosphatidic acid phosphatases involved in the biosynthesis of triacylglycerols and phospholipids. They are associated with the endoplasmic reticulum but can also travel into the nucleus and alter gene expression. Previous studies indicate lipins in solution form high molecular weight complexes, possibly tetramers. This study was undertaken to determine if lipins form complexes on membranes as well. Murine lipin 1b was applied to a supported bilayer of phosphatidylcholine, phosphatidylserine, and cholesterol and examined by atomic force microscopy (AFM) over time. Lipin on bare mica appeared as a symmetric particle with a volume consistent with the size of a monomer. On the bilayer lipin initially bound as asymmetric, curved particles which sometimes assembled into circular structures with an open center. Subsequently, lipin assemblies grew into large, symmetric particles with an average volume twelve times that of the monomer. Over time some of the lipin assemblies were removed from the bilayer by the AFM probe leaving behind “footprints” composed of complex patterns that may reflect the substructure of the lipin assemblies. The lipin complexes appeared very flat, with a diameter 20 times their height. The footprints had a similar diameter, providing confirmation of the extensive deformation of the protein under the AFM probe. The ability of lipin to form large complexes on membranes may have significant implications for the local concentrations of the product, diacylglycerol, formed during hydrolysis of phosphatidic acid and for cooperative hormonal regulation of lipin activity through phosphorylation of one or more monomers in the complexes.
Introduction
Lipins are highly conserved, dual-function proteins centrally involved in both lipid metabolism and the regulation of the expression of certain genes.1 They are phosphatidic acid phosphatases that generate diacylglycerol as an intermediate in the production of triacylglycerol and phospholipids. They are peripherally associated with the endoplasmic reticulum where they participate in lipid metabolism but also can travel to the nucleus and influence the activity of a number of transcription factors, thereby potentially linking lipid metabolism to gene expression. The lipins are subject to changes in their phosphorylation state in cells in response to insulin, catecholamines, or free fatty acids2 and therefore may play an important role in hormonal regulation of both lipid metabolism and gene expression.
While many other enzymes involved in triacylglycerol and phospholipid metabolism are integral membrane proteins, the ability of lipin to associate reversibly with membranes plays an essential mechanistic role in enabling and regulating its dual activities. Biophysical studies of soluble lipin indicate that the protein can form homomeric and heteromeric complexes, possibly tetramers, with other lipin isoforms both in solution and in the cytoplasm of cells.3 However, the nature of the protein-lipid complexes that lipin forms when interacting with membranes is not known. Because of the fundamental importance of the ability of lipin to interact reversibly with membranes, this study was undertaken to visualize the interaction of lipin with membranes in vitro by atomic force microscopy (AFM). The results provide novel insights into the interaction of lipin with itself on membranes to form unexpectedly large complexes. The nature of these complexes may significantly influence the mechanism of lipin action on membrane-bound substrates and the cooperative regulation of lipin by phosphorylation of monomers within the complexes.
Materials and Methods
Purification of recombinant lipin
FLAG-tagged murine lipin 1b was expressed from vector pRK5-FLAG-lipin 1b in HEK-293T cells.4 Cells were cultured in 15 cm plates in DMEM containing 5% fetal calf serum and Penicillin/Streptomycin (Gibco). Cells were transiently transfected with 30 μg of plasmid per 15 cm plate using Lipofectamine 2000 at a 2:1 ratio of DNA:Lipofectamine. Eighteen to 24 plates were used for each purification of lipin 1. Transfected HEK293T cells were harvested by centrifugation at 16,000 x g for 10 min, washed with ice cold phosphate-buffered saline (PBS), and either used directly or frozen at −80°C. Cells were lysed in Buffer A (150 mM NaCl, 20 mM Hepes, pH 7.2, 0.1% Brij 35) by passage 5 times through a 22 gauge needle, the lysates cleared by centrifugation at 16,000 g for 10 min, and the supernatant was incubated with anti-FLAG beads for 2-4 h at 4°C. Beads were isolated by centrifugation at 2,000 x g and the supernatant was removed. After washing, the slurry was incubated for 30 min in 100 mM NaCl, 1 mM MnCl2, 2 mM DTT, 50 mM Hepes, pH 7 and subjected to gentle agitation. The slurry was packed onto a small column, washed with Buffer A, and lipin was eluted by 5 successive additions of an equal volume of 0.5 mg/mL FLAG peptide (Lifetein) in 150 mM NaCl, 20 mM Hepes, pH 7.2. Elution fractions containing lipin were pooled and dialyzed three times against 150 mM NaCl, 20 mM Hepes, pH 7.2 and 1% glycerol. Purified lipin was quantitated using UV absorbance and comparison of bands of lipin and bovine serum albumin standards on Coomassie Blue-stained SDS-PAGE gels. The typical yield was 25 ug per 15 cm culture plate.
Preparation of lipin samples for dry imaging in air
100 μl of a 0.57 μg/ml solution of lipin in 150 mM NaCl, 50 mM HEPES-NaOH (pH7.4), 1 mM EGTA was applied to a 13 mm freshly cleaved mica disk glued to a 15 mm steel support disk. After 5 min the lipin solution was washed off with several mL of deionized water and the mica surface dried under a stream of argon before the disk was mounted in the AFM.
Preparation of lipin on supported lipid bilayers for imaging in buffer
Supported bilayers were prepared by the vesicle fusion technique.5-7 1,2-Dioleoyl-sn-glycero-3-phosphatidylcholine (PC), porcine brain L-α-phosphatidylserine (PS) with acyl chain composition 42% 18:0, 30% 18:1, 2% 20:4, 11% 22:6, 15 % unknown, and ovine cholesterol obtained from Avanti Polar Lipids as chloroform stocks were mixed in a ratio 2:1:1 by weight. The chloroform was evaporated under a stream of argon, and the lipids were rehydrated in deionized water to give a total lipid concentration of 2 mg/mL. The lipid mixture was vortexed to produce large multilamellar vesicles, from which small unilamellar vesicles were prepared by sonication with a probe sonicator for 1 min. For the formation of supported lipid bilayers, 50 μL of the vesicle suspension was mixed with 950 μL of 150 mM NaCl, 50 mM HEPES-NaOH (pH7.4), 2 mM CaCl2 and 100 μL of this suspension was then applied to a freshly cleaved mica disk. After 2 min the mica was washed with several mL of the same buffer to remove unbound vesicles. With this lipid composition the bilayers were anticipated to be in a liquid state, and no indications of phase separation were seen by AFM. Lipin was applied to the bilayer in 100 μL of the same buffer at a concentration of 0.57 μg/ml. After 5 min the lipin concentration was reduced by a brief wash with 2 mL of buffer and the sample was then imaged in the AFM under buffer. This final wash was necessary to reduce the free lipin concentration sufficiently that it did not adhere to the probe and interfere with imaging.
Atomic force microscopy
Samples were examined with a Veeco Multimode 8 atomic force microscope. Dry samples in air were imaged in Peak-Force tapping mode8 using SCANASYST-AIR probes (Veeco) with a spring constant of 0.4 N/m. Samples under fluid were imaged in tapping mode9 with Micromasch NSC19ALBS probes with a spring constant of 0.6 N/m. Samples were examined at 29 to 30 degrees C, the ambient temperature of the microscope head during operation. Images were acquired at a scan rate of 2 Hz with a resolution of 512 lines X 512 pixels per line. Image forces were kept to the minimum values compatible with stable imaging by maximizing the amplitude set point in standard tapping mode and minimizing the peak force set point in Peak-Force tapping mode (typically less than 0.1V). For images acquired in tapping mode both the height image, to give topographic information, and the amplitude error image, to give better edge definition, are shown.
Particle heights and diameters at half-height were determined from the average of measurements of two cross sections at approximately right angles to one another. Following the procedure of Schneider and colleagues10, molecular volumes were calculated by modeling the particles as spherical caps and using the formula
where h = the height of the particle and r = the radius of the particle at half height. The diameter and radius were taken at half height since previous empirical studies suggest this tends to compensate for the increase in apparent diameter due to the finite size of the probe when measuring the volumes of isolated proteins.10 Statistics on particle measurements are given as means +/− sample standard deviations.
Results
Lipin in air on mica
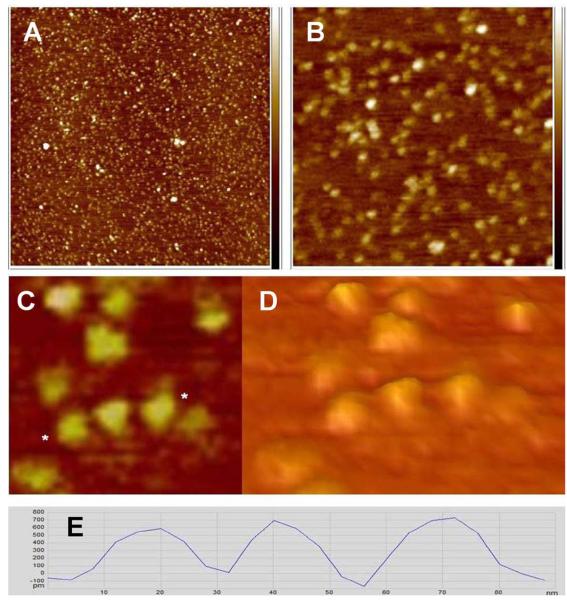
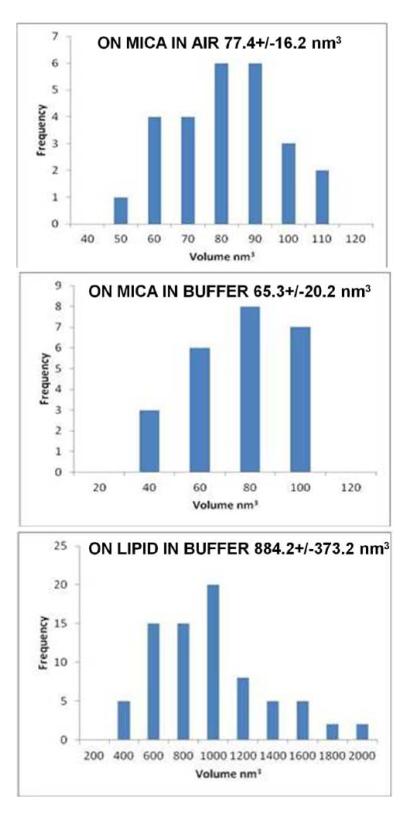
Lipin was initially imaged in air on a bare mica support. Single particles and clusters were observed as seen in Figure 1. Analysis of single particles indicated an average height of 0.676+/−.078 nm (n=26), diameter at half height of 17.0 +/− 1.7 nm and a calculated molecular volume (see Materials and Methods) of 77.4 +/− 16.2 nm3. The distribution of particle volumes are shown in the histogram in Figure 2. At higher magnification (Figure 1C and D) the individual particles had a lumpy appearance indicative of some substructure, but because of the different orientations of the particles on the mica a particular molecular shape could not be assigned to the particles.
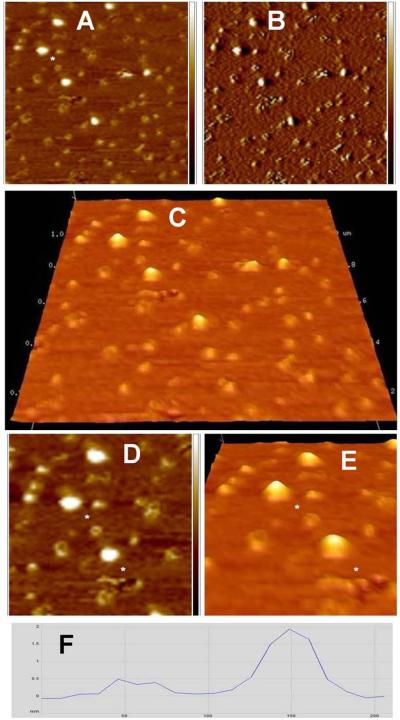
Figure 1.
Lipin imaged in air on mica. A, B, and C: Height images obtained in peak force tapping mode. A 2 × 2 μm, B 500 × 500 nm, C 130 × 130 nm. Height scale on the right in A and B is -1.5 to +1.5 nm. D: Three dimensional representation of the same area seen in C, viewed at an oblique angle. E: Cross section profile of the three particles between the asterisks in C. Vertical scale on the cross section, -100 to +800 pm, horizontal scale 0 to 90 nm.
Figure 2.
Histograms of the molecular volumes of lipin particles calculated as described in Materials and Methods. Top, lipin visualized on mica in air. Middle, lipin on mica in buffer. Bottom, lipin on the lipid bilayer in buffer.
Lipin in buffer on mica and on a supported bilayer
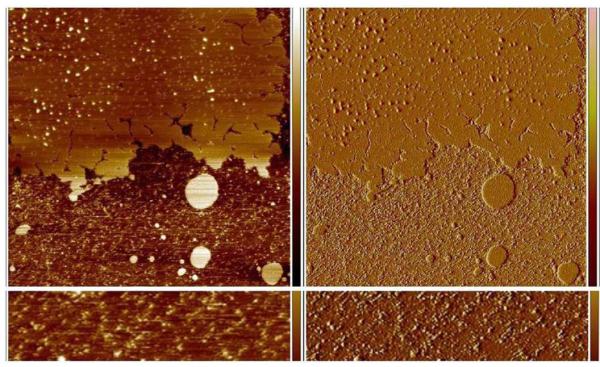
Lipin was visualized after applying to a supported lipid bilayer composed of 50% PC, 25%PS, 25% cholesterol by weight in a buffer of 150 mM NaCl, 50 mM HEPES-NaOH (pH7.4), 2 mM CaCl2. Because the bilayer was not complete it was possible to visualize lipin both on the bilayer and on the bare mica (Figure 3). Initially the lipin did not adhere tightly enough to the bare mica to be imaged and the particles appeared as streaks as they were pushed ahead of the probe during imaging (Figure 3, bottom). After 90 minutes the lipin adhered more tightly to the mica and could be imaged although the particles were somewhat extended in the direction of probe motion suggesting some movement under the action of the probe (Figure 4) and the particles did not have the lumpy appearance that was resolved when lipin was imaged in air. Nonetheless, the dimensions of the particles on mica under buffer were similar to those visualized in air: height, 0.603 +/− 0.082 nm (n=24); diameter 16.4 +/− 2.3 nm; and molecular volume 65.3 +/−20.2 nm3 (see histogram of volumes in Figure 2). This similarity in size of the “dry” lipin versus lipin in buffer suggests the protein remains hydrated on the mica surface during imaging in air, and has also been reported in comparisons of other proteins observed by AFM in air and in buffer.10
Figure 3.
Lipin imaged in buffer on a supported lipid bilayer and on mica. Top left panel, height image 5 × 5 μm, height scale on right – 2 to + 2 nm. Dark areas represent the mica substrate, lighter areas the lipid bilayer. Lipin appears as a variety of size particles on the lipid bilayer. Panel at the bottom left is a higher magnification view (0.5 × 2 μm) of a portion of the bare mica seen in the height image above. Lipin on the mica is not stable to imaging and is pushed by the probe creating the streaks at a 45 degree angle. Panels on the right are the amplitude error images of the same areas seen on the left, amplitude error scale −5 to +5 mv.
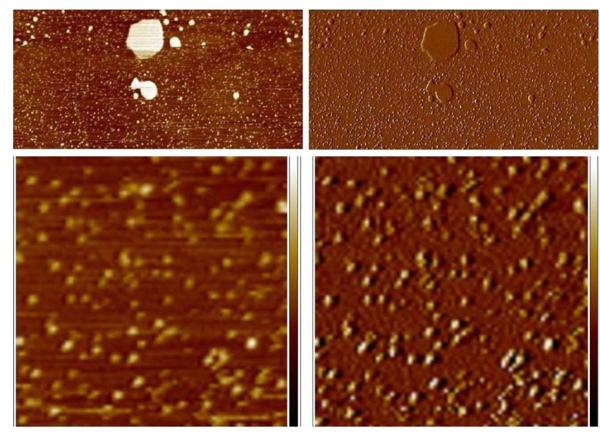
Figure 4.
After extended incubation in buffer, lipin adheres strongly enough to the mica to be visualized. An area of bare mica adjacent to the patch of bilayer seen in Figure 3 is shown at low magnification in the top panels (2.4 × 5 μm), left panel height image, right panel amplitude error image of the same region. The large bright objects in the height image are small patches of lipid bilayer, the small particles on the mica are lipin. The lower panels show a portion of the same field at higher magnification (750 nm × 750 nm), left panel height image, height scale −2 to + 2 nm, right panel corresponding amplitude error image, amplitude error scale −10 to +10 mv. The lipin particles move slightly toward the upper right under the force of the probe, distorting the appearance of the particles.
In contrast, when lipin bound to the lipid bilayer it assembled into larger particles. After 30 minutes a mixture of particle sizes was visualized (Figure 5). The smaller particles appeared as oblong, slightly curved structures, sometimes assembled into complete circles (Figure 5). The heights of these structures above the bilayer ranged from 0.2 to 0.5 nm (e.g., Figure 5F). Also present were larger, essentially globular structures. However, in some cases imaging of the same spot revealed that a larger particle had been apparently partially disrupted, leaving a ring-like structure behind.
Figure 5.
Higher magnification view of lipin bound to the supported lipid bilayer seen in Figure 3. A: Height image 1 × 1 μm, height scale – 1.5 nm to + 1.5 nm. B: Amplitude error image corresponding to the height image in A, amplitude error scale – 5 to + 5 mv. Note the coexistence of large, round lipin particles (appearing bright in the height image A) and smaller, lower, lipin particles that have a linear, curved, or near circular morphology. One example of a particle with circular morphology is seen to the lower right of the asterisk in panel A. This particle is seen at higher magnification in panels D and E. C: Three dimensional representation of the area seen in panels A and B. The vertical scale has been magnified 10-fold to aid in visualization of the topography. D and E: Higher magnification view of various size lipin aggregates from a portion of panel A. D: Height image 615 × 615 nm, height scale -1 to +1 nm. A three dimensional representation of the same area is shown in E with the vertical scale magnified 10 fold to aid in visualization of the topography. F: Cross section profile between the two asterisks in panels D and E goes through the small circular lipin particle and the adjacent large particle. Vertical scale on the cross section, 0 to 2 nm, horizontal scale 0 to 200 nm.
After 90 minutes, most of the particles on the bilayer were of the larger class (Figure 6), suggesting that the smaller structures were intermediates in the pathway to formation of the larger structures. The larger particles which became stabilized on the bilayer had a height of 1.63 +/− 0.24 nm (n=78), diameter at half-height of 36.2 +/− 5.9 nm, and molecular volume of 884.2 +/− 373.2 nm3 (see histogram of volumes in Figure 2).
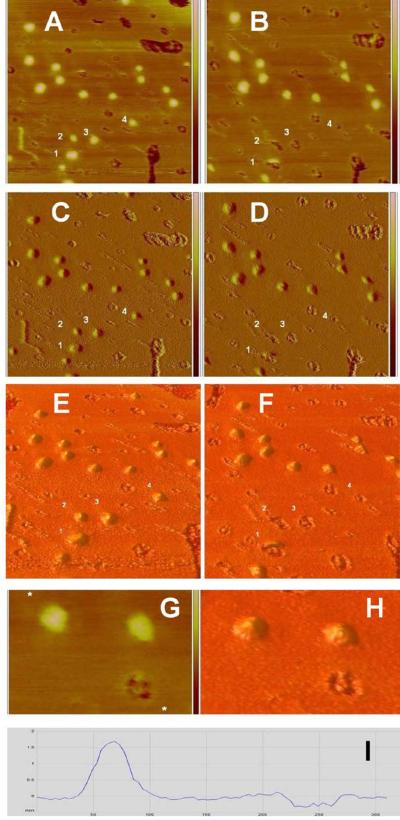
Figure 6.
Large lipin particles leave behind “footprints” when they are displaced by the AFM probe. Panels A through F: Lipin bound to the supported lipid bilayer is viewed in two scans 512 seconds apart. Left panels (A, C, and E) are from a scan in the downward direction taking 256 seconds. The height image is in panel A, 1 × 1 μm, height scale −2 to + 2 nm; the corresponding amplitude error image is in panel C, amplitude error scale -5 to +5 mv. Panel E shows a three dimensional representation of the area seen in panels A and C with the vertical scale magnified 10-fold to aid visualization of the topography. The panels on the right, B (height image), D (amplitude error image), and F (three dimensional representation), were captured at the same location during the next downward pass of the probe (after one upward pass). Lipin aggregates appear as bright objects in the height images. Depressions, holes, or footprints in the bilayer appear as dark areas. Note that the lipin particles labeled 1, 2, 3, and 4 in the left panels (particles are immediately to the lower right of the numbers) are replaced by corresponding lipin “footprints” labeled 1, 2, 3, 4 in the right panels. Particle 1 appears to have been sliced in half (panels B, D, and F) because the probe displaced the particle when it was passing near the center of the particle. The footprint left by particle 2 appears to be a doublet, possibly indicating the lipin particle rolled ahead of the probe and formed a second footprint before being fully displaced. Panels G, H, and I: Higher magnification view of 2 lipin aggregates and a lipin footprint on the supported bilayer. Height image (G) is zoomed from a region near the center of panel B, 220 × 330 nm, height scale is truncated; refer to full height scale −2 to + 2 nm in panel B. The corresponding three dimensional representation is in panel H. Note the complex pattern present in the single “footprint” in panels G and H. A cross section profile across one of the lipin particles and the lipin footprint (between the asterisks marked on the height image G) is shown in panel I, vertical scale −0.5 to +2 nm, horizontal scale 0 to 300 nm.
“Footprints” left in the bilayer by displacement of lipin particles
After the lipin particles on the bilayer were fairly stable (after 90 minutes), they were sometimes knocked off the bilayer by the AFM probe (Figure 6A-F). At this point in time they did not leave a smaller structure behind as they did when first forming on the bilayer. Instead a distinct “footprint” was left as a partial hole in the bilayer. In several instances the displacement occurred after the probe had made sufficient contact with the particle to generate an image of part of the particle, then, as the probe continued the particle was removed, resulting in an image of part of a particle and part of a footprint (Figure 6A-F). These footprints were complex, consisting of three or four adjacent holes (e.g., Figure 6G,H), possibly as a result of removal of parts of the membrane underlying the particle that were tightly bound to the protein multimers. The diameter of the footprints was typically close to but slightly greater than the apparent diameter of the protein before it was displaced (Table 1). The depth ranged up to 0.3 nm (e.g., Figure 6I), however, it is not clear whether the probe would have been able to reach the bottom of the holes because of the finite diameter of the probe.
Table 1.
Dimensions (nm) of lipin particles and their respective footprints on the supported bilayer
| Particlea | Particle Horizontal Width b |
Particle Vertical Width b |
Footprint Horizontal Width |
Footprint Vertical Width |
|---|---|---|---|---|
| #1 | 71.7c | 49.0 | 78.3 | n.d.d |
| #2 | 42.4 | 42.6 | 48.9 | n.d.e |
| #3 | 55.5 | 45.8 | 58.7 | 52.3 |
| #4 | 52.2 | 42.5 | 52.2 | 45.7 |
Particle numbers correspond to the particles as numbered in Figure 6.
Particle widths were measured at the base (full height) not the half height.
The profile of this particle suggests it is two poorly resolved lipin particles sitting adjacent to one another in the horizontal direction resulting in this large horizontal measurement.
Not determined because part of the footprint is occupied by the particle which was removed during imaging.
Not determined because this footprint appears as a doublet in the vertical direction possibly because the particle was pushed ahead of the probe during imaging and created two footprints.
Discussion
Lipin assembles into multimeric complexes on a lipid bilayer
Comparing the images of lipin bound directly to mica, either in air or under buffer, with lipin bound to a supported lipid bilayer it is apparent that lipin undergoes a process of assembly into larger complexes on the bilayer, and does not remain bound to the bilayer as single particles. The sizes of lipin particles on the bare mica and on the bilayer are summarized in Table 2, including the molecular volumes calculated from the AFM measurements by the method described in Materials and Methods. The transition to the larger size particle was monitored with the AFM and the process went through intermediate steps. The smallest particles initially seen bound to the bilayer were not as high as lipin bound to mica (0.2 to 0.5 nm on the bilayer versus 0.6 to 0.7 nm on mica) and had a more elongated shape. These intermediate structures were typically curved and often appeared to assemble to form nearly circular structures. For a period of 30 to 60 minutes these structures increased in size to form globular particles, although sometimes the larger particles were reduced in size and reverted to the circular shapes. The impression given by these images is that the initial assembly process is dynamic and reversible. It is possible that perturbation by the AFM probe was responsible for causing the disassembly of intermediate size particles before they achieved a more stable structural endpoint.
Table 2.
Average dimensions of lipin particles; comparison with hypothetical volumes based on molecular weight
| Imaging conditions | Ha, nm | Dhalf, nm | Vhalf, nm3 | Dfull, nm | Vfull, nm3 | Vc, nm3 |
|---|---|---|---|---|---|---|
| In air on mica | 0.676 | 17.0 | 77.4 | 24.02 | 153.4 | 193.4 (monomer) |
| In buffer on mica | 0.603 | 16.4 | 65.3 | 23.20 | 127.3 | 193.4 (monomer) |
| In buffer on bilayer | 1.63 | 36.2 | 884.2 | 51.14 | 1676.5 | 774 (tetramer) |
His the particle height. Dhalf is the particle diameter at half height. Vhalf is the calculated volume of the particle when modeled as a spherical cap using the diameter at half height in the calculation following the method of Schneider and colleagues.10 Dfull is the diameter at full height of the particle estimated from the relationship Dfull = √2 Dhalf (see Discussion). Vfull is the calculated volume of the particle using the diameter at full height in the calculation. Vc is the calculated volume of a lipin monomer or tetramer based on its molecular weight using the method described in the Discussion and reference 10.
Subsequent to this period of growth the larger particles that formed were stable but had a fairly broad distribution in sizes. The average size corresponded to a volume that was twelve fold greater than the initial size of independent lipin particles on mica (Table 2). This may be an underestimate if some of the lipin was buried in the bilayer because the height of the particles was measured relative to the surface of the bilayer. This average size would correspond to a dodecamer of lipin monomers. However, the breadth of the histogram of molecular volumes (Figure 2) suggests the calculated average size likely includes contributions from complexes of a range of sizes whose exact compositions could not be resolved. For example, in the case of particle number 1 in Figure 6A-F, a cross section revealed that this particle had a saddle at its peak and was twice as wide in one dimension as the average particle width. Therefore it was likely to have been a poorly resolved multiple particle. Such particles were excluded from the determination of the average size of particles when, as in this case, their morphology clearly indicated they were composed of two or more large particles. However, even after excluding these there remained a significant fraction of particles that were twice the average volume (see the histogram in Figure 2) that were retained in the analysis because no clear resolution onto multiple particles was evident from cross section analysis.
The lipid phosphatase catalytic domain of lipin is a member of the haloacid dehalogenase structural superfamily.11 A homologous dehalogenase domain in vertebrate cytidine monophosphate-sialic acid synthetase forms a tetramer which in turn drives tetramerization of the complete synthetase molecule.12 It is possible that the corresponding domain in lipin might drive the formation of a lipin tetramer. Indeed, gel filtration analysis of soluble lipin indicates a molecular size of about 600 kDa, possibly representing a tetramer or a higher order complex of the 100 kDa monomer.3 However, the gel filtration analysis may be misleading if lipin in solution is a highly asymmetric molecule. Our observations of lipin on mica suggest it is not asymmetric, and the molecular volumes we measured for lipin on mica are more consistent with a monomer than a tetramer (see Table 2). However, the smallest lipin particles seen initially on the bilayer during assembly of the larger complexes were distinctly asymmetric as described above. When interacting with a lipid bilayer the globular monomer may undergo a conformational change to form the extended molecules that were initially seen on the bilayer. These extended monomers may then assemble to form tetramers, which may also associate with one another on the bilayer to form octamers, dodecamers, or even higher order structures.
Such a self-assembly process may be important for the function of lipin on membranes. Cooperative interactions between membrane-bound lipin molecules may strengthen the interaction with the bilayer, resulting in a structure that cannot be removed from the membrane without damaging the membrane, as suggested by the appearance of the footprints when mature lipin complexes were displaced by the AFM probe. The assembly of lipin complexes on the membrane may also enable lipin to form high local concentrations of diacylglycerol from hydrolysis of its substrate, phosphatidic acid.
Lipin “footprints” in the bilayer provide insights into protein deformation during AFM imaging
Some of the observations of lipin obtained here may provide more general insights into the capabilities and limitations of the AFM for the study of proteins under physiological conditions. The image of an object obtained with the AFM is the result of analyzing the height of the probe as it passes over objects while maintaining a constant force of interaction with the object and the surrounding substrate. This height is in turn determined by the height of the object, but this may be altered by deformation of the object due to pressure from the probe and by movement of the object while the probe passes over it. There may also be differing forces of interaction between the probe and different materials due to charge differences or other physical-chemical interactions creating attraction or repulsion. In addition, there is a broadening of objects in lateral dimensions because of the finite size of the probe. All of these factors may need to be taken into account in relating an AFM image to the actual structure of the object being observed. As will be discussed below, the fortuitous formation of “footprints” in the bilayer by lipin provides an unusual, independent measurement of the physical size of lipin particles under observation that may be of assistance in interpreting the AFM images.
A number of previous AFM studies of the morphology of isolated proteins on solid supports have yielded greatly flattened molecular shapes as was seen here for lipin on mica or on the supported bilayer10,13-15 in spite of efforts to use the minimum imaging force that was compatible with stable imaging. Proteins in dense arrays, such as two dimensional crystals of annexin A5 on a supported bilayer, do not appear to be subject to such flattening, possibly because of lateral support from the protein lattice.16 The ratio of the diameter at half height to the height of lipin particles was 25(in air) to 27 (in buffer) for lipin on mica and 22 for the larger lipin particles seen on the supported bilayer. As is a common practice in AFM studies of this nature, in order to aid in visualization of the features of the lipin particles in the three dimensional representations the vertical axis has been expanded 10 to 20 fold so this flattening is not so apparent in the figures. This twenty-fold flattening of the lipin particles is even greater than the ratio of tenfold observed for a series of proteins studied by Schneider and colleagues10 who proposed the method for calculating molecular volumes from AFM measurements that was applied in this study (see Materials and Methods). In spite of this obvious distortion of the shape of what are likely globular proteins, Schneider and colleagues found that the molecular volumes of a series of test proteins observed with tapping mode AFM and modeled as spherical caps, were close to the volumes expected on the basis of molecular weight and a typical density for proteins.10 The best correlation between such measured and calculated volumes was obtained by using the diameter of the spherical cap at half -height rather than full-height in the formula for a spherical cap. Although this was a somewhat ad hoc modification of the calculation it was justified as helping to correct for tip broadening – the artifactual increase in observed size due to the finite dimensions of the AFM probe tip. Measuring the diameter at half height also made the measurement of the diameter operationally easier to obtain since the profiles of the particles visualized in the AFM typically do not have a sharply defined bottom.
In order to estimate the contribution tip broadening might have made to the measurements in our experiments we examined the morphology of the Micromasch NSC19 probes used. This analysis was performed by imaging the rough, non-deformable surface of a polycrystalline titanium “roughness sample” provided for this purpose by the AFM manufacturer. The “tip estimation” software provided by the manufacturer analyzes each peak in the topographic data from the roughness sample and generates a model of the tip based on the principle that no data in the image can have a slope steeper than the slope of the tip. An image of the tip model generated this way is shown in Figure 7A. A cross section of this image (Figure 7B) suggests the tip can be modeled as a hemisphere of radius 8.5 nm. To appreciate the potential tip broadening effect from this finite size, we consider the possible size of a lipin tetramer with subunits of mass 101,789 Da. Using the method of Schneider and colleagues10 we can calculate an expected molecular volume, Vc, for the protein from the formula
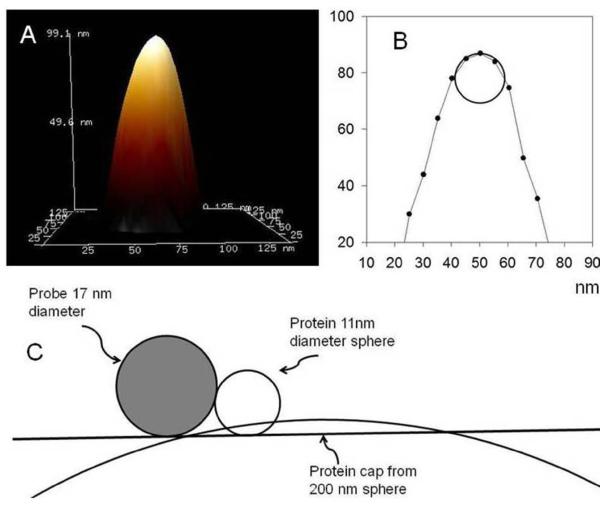
Figure 7.
Modeling of the NSC-19 AFM probe used in this study. Panel A: A three dimensional representation of the probe tip determined by characterization of the image made with the probe of a non-deformable tip calibration sample (see Discussion). Panel B: Cross section of the probe tip illustrated in Panel A, showing the tip modeled as a 8.5 nm radius sphere. Panel C: Schematic illustration of the model probe approaching a spherical protein of diameter 11 nm and a protein cap of the same volume which has a diameter to height ratio of 20, similar to the images of lipin seen in this study.
where M0 is the molecular weight, N0 is Avogadro’s number, and V1 and V2 are the partial specific volumes of the protein (0.74 cm3g-1 ) and water (1 cm3g-1), respectively. d is the extent of protein hydration (0.4 mol H2O/mol protein). For the lipin tetramer the volume calculated this way is 774 nm3 and a sphere of this volume has a diameter of 11 nm (or 16 nm for a lipin dodecamer). Figure 7C illustrates that the probe broadening for a sphere of this size would add approximately 8 nm to the radius of the sphere, therefore increasing the observed diameter of a rigid sphere from 11 nm to 27 nm (or to 32 nm for a dodecamer). The average observed diameter of the lipin particles on the bilayer was 36.2 nm at half height and 51.1 nm at full height (see derivation of diameter at full height below). Therefore tip broadening alone would not appear to be sufficient to explain the large diameter of the lipin particles on the bilayer, measured at full height, if they are globular tetramers or dodecamers. This suggests that the large diameter of the particles is likely partly due, in addition, to deformation of the particles due to pressure from the AFM probe.
Additional evidence that the lipin complexes were flattened during imaging comes from the close correlation of the size of the “footprints” left by lipin removed by the probe and the size of the particles seen before they were removed. The dimensions of the four particles seen displaced in Figure 6 are compared with the dimensions of their footprints in Table 1. All of the particles are on the order of 40 to 60 nm wide (measured at their base or “full height”, not the half-height as was used in the volume calculations), and the footprints correlate very well with the different size particles, although the footprints are a few nm larger in some cases.
If the lipin particles remained partially or fully flattened between passes of the probe, the amount of probe broadening due to the tip geometry would have been much less than the 8 nm on each side of the protein as suggested above for a rigid sphere model of the protein. Figure 7C shows how the probe would approach a flattened lipin particle of the same volume as the spherical particle. This construction suggests probe broadening would not have been greater than about 1 nm when the probe approaches the flattened particle. Probe broadening should also affect the images of the footprints, although in this case the lower topology of the holes would cause the apparent diameter of the hole to be reduced rather than increased as occurs with a particle projecting above the surface. The actual depth of the footprints is uncertain because they generally contained “islands” of material so it is not certain the probe, with a 17 nm diameter could have reached the bottom of the holes. The measured depth was only on the order of 0.3 nm so, again, one would expect the diameters of the holes to be underestimated by no more than one nm.
However, it is not actually known if the proteins remain flattened in between acquisition of the images or if they are deformed only when the probe is passing over them. When a particle was seen to be displaced during the instant it was being imaged, as in the case of particle number 1 in Figure 6 (and several others not shown), only a partial particle is seen, although the remaining part of the footprint is still present. This might imply that the particle was flattened in a previous scan and was then peeled off like a pancake, revealing the remainder of the footprint. On the other hand if the membrane had been damaged to form a footprint underneath the protein during the previous pass of the probe, the full footprint could still be there even if the protein recovered a globular shape between images. It is therefore unclear whether the actual protein particle is the flat object recorded or is a more dynamic structure being squeezed out under each tap of the probe during imaging. Nonetheless, the footprints may provide an accurate record of either the dimensions of the static flat object the protein has become or the extent of the periodic dynamic extrusion of the bulk of the protein during imaging.
One argument that favors the interpretation that the proteins must to some extent recover their shape in between images is that the assembly of larger complexes was seen to progress even during continuous scanning of the sample. It is difficult to imagine that the assembly of flattened disk-like proteins would follow the same pathway, to the same endpoint, as undisturbed globular proteins. When areas of the membrane adjacent to the area being monitored were examined at the end of the time course, the same types of fully assembled lipin particles were seen, although these had developed in the absence of perturbation by the probe. Whether this assembly occurred at the same rate as with the perturbed particles is not known. Nonetheless, it appears the proteins are remarkably resilient and have the ability to recover from extreme deformation and continue to the same final assembly endpoint.
The degree of apparent flattening of proteins observed in the AFM is affected by probe sharpness
AFM images of the cytoplasmic domain of synaptotagmin on mica or bound to a lipid bilayer obtained by Shahin and colleagues7 are also flattened with a height that is about one tenth the diameter at half height. This is similar to the degree of flattening seen by Schneider and colleagues for a series of unrelated proteins bound to mica.10 As described above, we found the degree of flattening of lipin to be twice as great – i.e. the ratio of the diameter to at half height to the height was over 20. It is possible that this difference is due to a greater sharpness of the probes we used compared to the probes used in the earlier studies since a sharper probe might enter the protein to a greater depth. Schneider and colleagues did not specify the sharpness of the probes they used. Shahin and colleagues used Veeco DNP-S probes. We used the tip calibration software to analyze the sharpness of a DNP-S probe and found it has a radius of 22 nm compared to the 8.5 nm of the Micromasch NSC-19 probes used here. Both of these measurements compare well with the tip size specifications from the respective manufacturers. It was previously reported that in a direct comparison of the height of a supported lipid bilayer measured by AFM using the DNP-S probe versus a Micromasch NSC-18 probe the bilayer height was found to be 4 nm with the DNP-S probe and 2.5 nm with the Micromasch NSC-18 probe.17 This suggests the Micromasch probe was partially penetrating into the bilayer. The NSC-18 probe has a stiffer spring constant (3.5 N/m) than the DNP-S probe (0.58 N/m) and it was speculated previously that this difference might underlie the greater penetration of the bilayer by this probe.17 However, the NSC-19 probe has a similar spring constant (0.63 N/m) to the DNP-S probe and, in the present study, we found that the height of the supported bilayer (e.g. in Figure 3) was also measured as 2.5 nm with the NSC-19 probe. We measured the radius of the NSC-18 probe and found it was 8.2 nm, comparable to the NSC-19 probe (radius 8.5 nm). Therefore, we conclude that the probe sharpness is likely to have been the most important factor in determining the degree of penetration of the probe into the bilayer in these studies. It is possible that this difference in sharpness also explains the apparent two fold greater penetration of globular proteins with the NSC-19 probe versus the DNP-S probe. The ScanAsyst probe used to measure the height of the lipin particles in air (Figure 1) also gave a ratio of diameter at half height to height of lipin greater than 20. We determined the radius of the tip of this probe to be 10.6 nm, also comparable to the NSC-19 probe, reinforcing the conclusion that the probe sharpness is an important determinant of probe penetration and the measured height of isolated proteins. In a recent study discussed further below, Neaves and colleagues18 also concluded that probe sharpness as well as spring constant contributed significantly to probe penetration in a study of a series of isolated proteins supported on mica.
Calculation of molecular volumes using the full diameter of the spherical cap model
The measured molecular volume of the lipin monomer on mica was 77.4 nm3 in air and 65.3 nm3 in buffer (Table 2) when the diameter of the particle at half height was used to calculate the volume. This does not correlate well with the volume of 193.4 nm3 expected on the basis of the molecular weight of lipin and a typical protein density. Since tip broadening in our experiments may not have contributed to as great an extent to the measurement of molecular volume as in previous studies, we considered that it might be better to calculate the volume of the particles based on the full diameter of the spherical cap model. However, it is difficult to clearly define the bottom of the cap when viewed in cross section (e.g. in Figure 6I). Therefore we have analyzed how to calculate the diameter at full height given a measurement of the diameter at half height, which is easier to determine unequivocally. If the diameter at half height of a spherical cap is Dhalf, then the diameter at full height of the same cap, Dfull, can be obtained from the formula
where Hfull is the measured full height. A derivation of this formula is provided in the legend to Figure 8. For very shallow spherical caps, as with the AFM images of lipin obtained here, this reduces to a limit of
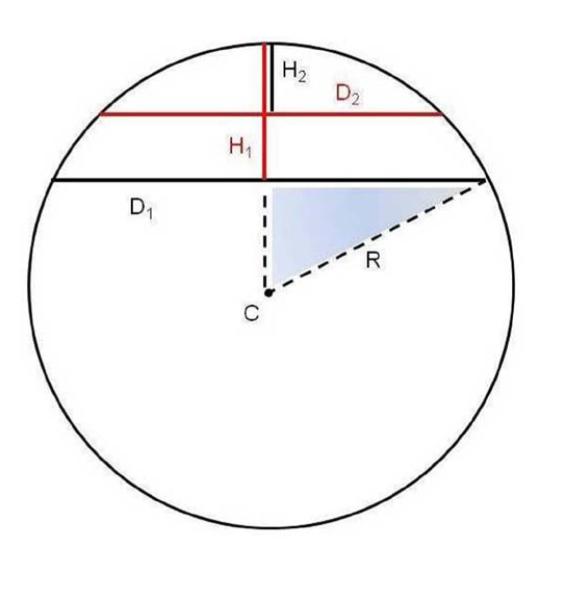
Figure 8.
Calculation of the full diameter of lipin particles from the diameter at half height when the particles are modeled as spherical caps. The sphere shown has a radius of R and center at point C. Two spherical caps are shown with height and diameter of H1 and D1, and H2 and D2, respectively. H1 = 2 H2 (full height = 2 × half height). We wish to determine D1, the diameter of the cap at full height, in terms of D2, the diameter of the cap at half height, and H1, the full height of the cap.
For example, for lipin the ratio of the value of the diameter at half-height to the value of the full height was over 20. For a ratio of 20, an exact calculation gives
And for a ratio of 10, similar to the observations of Schneider and colleagues10,
These coefficients may be compared to the square root of 2 which is 1.4142. We suggest is a useful approximation for AFM studies of this nature, if the observed morphology of the protein can be reasonably modeled as a spherical cap.
Using the diameter at full height when calculating the molecular volumes of the lipin particles yields a better correlation between the measured volume of the lipin monomer on mica (153.4 nm3 in air and 127.3 nm3 in buffer) and the calculated volume of a monomer based on molecular weight and protein density (193.4 nm3), as summarized in Table 2. However, the ratio of the volumes of the lipin complexes on the bilayer to the particles on mica is independent of the method of calculation and remains 12 fold (Table 2).
Interestingly , Neaves and colleagues18 recently came to a similar conclusion that using the diameter of the spherical cap model at the base gave a better correlation between the measured and calculated volumes of proteins measured by AFM. In their study they were also using probes that were both sharper than the ones used by Schneider and colleagues10 and had a much higher spring constant (42 N/m versus 0.12 N/m).18 Both the greater sharpness and the higher spring constant of the probes used by Neaves and colleagues may have contributed to greater penetration of the proteins during imaging, necessitating a similar modification of the method of calculating the molecular volumes of the proteins under study.
Lipin “footprints” provide additional information about the nature of the lipin-bilayer interaction
What do the footprints left in the bilayer by lipin represent? Presumably some of the lipid bilayer has been displaced or removed as a result of interaction with lipin prior to or during imaging. When the lipin complex is displaced it may take some lipid with it, leaving defects in the membrane. In this case the complexity of the footprints, usually having 3 or 4 depressions in a single footprint, may reflect the complexity of the lipin particle that might have several sites that interact most strongly with the bilayer.
Alternatively, lipids may have been squeezed out from underneath the lipin particle when it was flattened instead of being removed with the lipin. In this case the complexity of the footprints could also reflect the complexity of the lipin multimers as firmer parts of the multimers might have been pressed into the bilayer to form the patterns in the footprint.
Another possibility is that the association of the lipin with the bilayer promoted the demixing of some of the components of the bilayer (phosphatidylcholine, phosphatidylserine, and cholesterol) creating domains of different composition that allowed the probe to enter the bilayer more deeply. On the one hand, it seems unlikely such domains would have been as stable as they were found to be: they did not dissipate over multiple scans. On the other hand, since the imaging was done in the presence of 2 mM Ca2+, possibly interactions between Ca2+ and phosphatidylserine could have provided some stability to such domains.
Footprints possibly similar in nature to the ones seen here with lipin were observed by Shahin and colleagues in their study of the interaction of the cytoplasmic domain of synaptotagmin with a supported lipid bilayer.7 When synaptotagmin complexes were displaced from the bilayer by the AFM probe stable “indentations” of depth 1.8 nm and of diameters similar to those of the protein complexes before their ejection were left behind. However, in the case of synaptotagmin, the indentations did not have an apparent substructure as was seen here with the lipin footprints.
Conclusion
In spite of the uncertainties inherent in determining the exact molecular volumes of lipin complexes by AFM, and the significant deformation of the complexes that occurred during imaging, the conclusion that lipin assembles on membranes to form particles that on average are 12 fold larger than the lipin monomer seems firm. This result is likely to be of considerable importance in understanding the regulation of lipin as well as the nature of the deposition of the product, diacylglycerol, in the membrane during complex lipid synthesis.
Several aspects of the lipin assembly process observed here warrant more detailed investigation. Electron microscopy studies of lipin monomers and higher order complexes will be essential to determine unequivocally their morphology, molecular content, and organization. A previously identified polybasic domain of lipin 1 is responsible for the binding of the protein to phosphatidic acid and is subject to negative regulation by phosphorylation.19,20 AFM studies of lipin 1 with mutation or deletion of the polybasic domain, or of lipin 1 in different phosphorylation states, could be used to determine the role of this domain in the formation of lipin complexes and lipin footprints on bilayers. The possible effects of lipid composition, particularly the inclusion of the substrate, phosphatidic acid, and product, diacylglycerol, in the bilayer could likewise be determined. It would also be of interest to assess whether heterocomplexes of different lipin isoforms might assemble by different pathways or to different final states since multiple lipin isoforms are co-expressed in cells.21-23 The AFM technology might also be used to address whether lipin may co-assemble with other enzymes in the triacylglycerol or phospholipid synthetic pathways to form efficient lipid-modifying protein assemblies amenable to coordinated regulation by phosphorylation or other modifications.
Acknowledgments
Funding Source Statement:
This study was supported by grants from The National Institutes of Health (S10-RR026490) and the American Diabetes Association (ADA 1-11-JF-21).
Abbreviations
- PC
1,2 dioleyl-sn-glycero-3-phosphocholine
- PS
L-α-phosphatidylserine from porcine brain
References
- 1.Harris TE, Finck BN. Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol Metab. 2011;22:226–233. doi: 10.1016/j.tem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris TE, Huffman TA, Chi A, Shabanowitz J, Hunt DF, Kumar A, Lawrence JC. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J Biol Chem. 2007;282:277–286. doi: 10.1074/jbc.M609537200. [DOI] [PubMed] [Google Scholar]
- 3.Liu GH, Qu J, Carmack AE, Kim HB, Chen C, Ren H, Morris AJ, Finck BN, Harris TE. Lipin proteins form homo- and hetero-oligomers. Biochem J. 2010;432:65–76. doi: 10.1042/BJ20100584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mou J, Yang J, Shao Z. Tris(hydroxymethyl)aminomethane (C4H11NO3) induced a ripple phase in supported unilamellar phospholipid bilayers. Biochemistry. 1994;33:4439–4443. doi: 10.1021/bi00181a001. [DOI] [PubMed] [Google Scholar]
- 6.Brian AA, McConnell HM. Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proc Natl Acad Sci U S A. 1984;81:6159–6163. doi: 10.1073/pnas.81.19.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shahin V, Datta D, Hui E, Henderson RM, Chapman ER, Edwardson JM. Synaptotagmin perturbs the structure of phospholipid bilayers. Biochemistry. 2008;47:2143–2152. doi: 10.1021/bi701879g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alsteens D, Dupres V, Yunus S, Latgé JP, Heinisch JJ, Dufrêne YF. High-resolution imaging of chemical and biological sites on living cells using peak force tapping atomic force microscopy. Langmuir. 2012;28:16738–16744. doi: 10.1021/la303891j. [DOI] [PubMed] [Google Scholar]
- 9.Hansma P, Cleveland J, Radmacher M, Walters D, Hillner P, Bezanilla M, Fritz M, Vie D, Hansma H, Prater C, Massie J, Fukunaga L, Gurley J, Elings V. Tapping mode atomic-force microscopy in liquids. Applied Physics Letters. 1994;64:1738–1740. [Google Scholar]
- 10.Schneider SW, Lärmer J, Henderson RM, Oberleithner H. Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflugers Arch. 1998;435:362–367. doi: 10.1007/s004240050524. [DOI] [PubMed] [Google Scholar]
- 11.Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oschlies M, Dickmanns A, Haselhorst T, Schaper W, Stummeyer K, Tiralongo J, Weinhold B, Gerardy-Schahn R, von Itzstein M, Ficner R, Münster-Kühnel AK. A C-terminal phosphatase module conserved in vertebrate CMP-sialic acid synthetases provides a tetramerization interface for the physiologically active enzyme. J Mol Biol. 2009;393:83–97. doi: 10.1016/j.jmb.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Barrera NP, Betts J, You H, Henderson RM, Martin IL, Dunn SM, Edwardson JM. Atomic force microscopy reveals the stoichiometry and subunit arrangement of the alpha4beta3delta GABA(A) receptor. Mol Pharmacol. 2008;73:960–967. doi: 10.1124/mol.107.042481. [DOI] [PubMed] [Google Scholar]
- 14.Oatley P, Stewart AP, Sandford R, Edwardson JM. Atomic force microscopy imaging reveals the domain structure of polycystin-1. Biochemistry. 2012;51:2879–2888. doi: 10.1021/bi300134b. [DOI] [PubMed] [Google Scholar]
- 15.Stewart AP, Egressy K, Lim A, Edwardson JM. AFM imaging reveals the tetrameric structure of the TRPM8 channel. Biochem Biophys Res Commun. 2010;394:383–386. doi: 10.1016/j.bbrc.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 16.Reviakine I, Bergsma-Schutter W, Mazeres-Dubut C, Govorukhina N, Brisson A. Surface topography of the p3 and p6 annexin V crystal forms determined by atomic force microscopy. Journal of Structural Biology. 2000;131:234–239. doi: 10.1006/jsbi.2000.4286. [DOI] [PubMed] [Google Scholar]
- 17.Creutz CE, Edwardson JM. Organization and synergistic binding of copine I and annexin A1 on supported lipid bilayers observed by atomic force microscopy. Biochim Biophys Acta. 2009;1788:1950–1961. doi: 10.1016/j.bbamem.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Neaves KJ, Cooper LP, White JH, Carnally SM, Dryden DT, Edwardson JM, Henderson RM. Atomic force microscopy of the EcoKI Type I DNA restriction enzyme bound to DNA shows enzyme dimerization and DNA looping. Nucleic Acids Res. 2009;37:2053–2063. doi: 10.1093/nar/gkp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren H, Federico L, Huang H, Sunkara M, Drennan T, Frohman MA, Smyth SS, Morris AJ. A phosphatidic acid binding/nuclear localization motif determines lipin1 function in lipid metabolism and adipogenesis. Mol Biol Cell. 2010;21:3171–3181. doi: 10.1091/mbc.E10-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eaton JM, Mullins GR, Brindley DN, Harris TE. Phosphorylation of lipin 1 and charge on the phosphatidic acid head group control its phosphatidic acid phosphatase activity and membrane association. J Biol Chem. 2013;288:9933–9945. doi: 10.1074/jbc.M112.441493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donkor J, Sariahmetoglu M, Dewald J, Brindley DN, Reue K. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J Biol Chem. 2007;282:3450–3457. doi: 10.1074/jbc.M610745200. [DOI] [PubMed] [Google Scholar]
- 22.Grimsey N, Han GS, O'Hara L, Rochford JJ, Carman GM, Siniossoglou S. Temporal and spatial regulation of the phosphatidate phosphatases lipin 1 and 2. J Biol Chem. 2008;283:29166–29174. doi: 10.1074/jbc.M804278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gropler MC, Harris TE, Hall AM, Wolins NE, Gross RW, Han X, Chen Z, Finck BN. Lipin 2 is a liver-enriched phosphatidate phosphohydrolase enzyme that is dynamically regulated by fasting and obesity in mice. J Biol Chem. 2009;284:6763–6772. doi: 10.1074/jbc.M807882200. [DOI] [PMC free article] [PubMed] [Google Scholar]