Abstract
The long-term survival of plasma cells is entirely dependent on signals derived from their environment. These extrinsic factors presumably induce and sustain the expression of antiapoptotic proteins of the Bcl-2 family. It is uncertain whether there is specificity among Bcl-2 family members in the survival of plasma cells and whether their expression is linked to specific extrinsic factors. We found here that deletion of the gene encoding the antiapoptotic protein Mcl-1 in plasma cells resulted in rapid depletion of this population in vivo. Furthermore, we found that the receptor BCMA was needed to establish high expression of Mcl-1 in bone marrow but not spleen plasma cells and that establishing this survival pathway preceded the component of plasma cell differentiation that depends on the transcriptional repressor Blimp-1. Our results identify a critical role for Mcl-1 in the maintenance of plasma cells.
High-affinity neutralizing antibodies protect the body against foreign organisms and their toxic products. These antigen-specific antibodies are secreted by plasma cells, which arise by differentiation from activated B lymphocytes. The completion of plasma cell differentiation, rather than its initiation, depends on the expression of the transcriptional repressor Blimp-1 (refs. 1–3); indeed, subsets of plasma cells can be identified on the basis of high and intermediate Blimp-1 expression, as has been shown through the use of a green fluorescent protein (GFP) reporter4. Most long-lived plasma cells are produced in response to protein-containing antigens in transient structures called ‘germinal centers’, in which B lymphocytes undergo extensive proliferation and maturation. Throughout rounds of proliferation, a fraction of B cells undergo differentiation and leave the germinal centers as either plasma cells or memory B cells. Both of these cell types have the capacity for longevity; plasma cells migrate to specialized niches5, whereas memory B cells recirculate between lymphoid organs6,7. Germinal center–derived plasma cells seem to persist mainly in the bone marrow. The emigration of plasma cells from secondary lymphoid organs is mediated by the induction of sphingosine 1-phosphate receptor type 1 in plasma cells8, whereas expression of the chemokine receptor CXCR4 promotes the recruitment of plasma cells into bone marrow niches and contributes to the positioning of plasma cells near bone marrow stromal cells expressing the chemokine CXCL12 (ref. 9). Most plasma cells in the bone marrow have a mature plasma cell phenotype, as indicated by low expression of major histocompatibility complex class II and the B cell–associated markers B220 and CD19 but high Blimp-1 expression10. In addition to giving rise to germinal center–derived plasma cells, activated B cells can give rise to plasma cells in extrafollicular foci in the spleen10. These antibody-secreting cells are generated soon after immunization or infection, have lower expression of Blimp-1 than do long-lived plasma cells in the spleen and bone marrow4 and are generally short-lived, dying by apoptosis in situ11.
In niches that promote the survival of plasma cells, plasma cells receive signals from local stromal cells that are crucial for their long-term survival. Proposed mediators of survival include ligands for the receptor BCMA (APRIL (a proliferation-inducing ligand) and BAFF (a B cell–activation factor))12,13, the cytokines interleukin 4 (IL-4), IL-5 and IL-6, tumor-necrosis factor14,15, ligands of the activation and memory marker CD44, costimulatory molecules CD80 and CD86 (ref. 16) and CXCL12 (ref. 17). Many of these promote the expression of antiapoptotic proteins of the Bcl-2 family12,18–21, but the in vivo expression of Bcl-2 family members and their relevance, individually and collectively, in the survival of long-lived plasma cells in the whole animal remain to be elucidated.
Treatment of mice with the Bcl-2 homology domain mimetic ABT-737, a compound that binds in Biacore studies with high affinity to Bcl-2 and the Bcl-2 family members Bcl-w and Bcl-xL (but not to Mcl-1t or A1), inhibits the appearance of plasma cells in the bone marrow during the treatment period but does not affect the maintenance of pre-existing plasma cells22. Notably, in contrast to those affinity measurements made with Biacore, other studies have shown that Bcl-2 (but not Bcl-xL or Bcl-w) is the main target of ABT-737 in the lymphoid lineage in vivo23. Together those observations suggest that newly formed plasma cells depend at least transiently on Bcl-2 expression but that such dependence on Bcl-2 may be lost in long-lived plasma cells. Additionally, Cre recombinase–mediated deletion of the gene encoding Bcl-XL (Bcl2l1; called ‘Bclx’ here) in B cells expressing activation-induced cytidine deaminase leads to a significant decrease in the abundance of of antigen-specific plasma cells in the bone marrow at 14 d after immunization, but not after 21 d (ref. 24). This suggests that Bcl-xL may also support the survival of recently generated plasma cells. In contrast to the described roles of Bcl-2 and Bcl-xL in newly formed plasma cells, it is unknown which survival molecule(s) mediate(s) the survival of long-lived plasma cells in their niche.
Although plasma cells have a crucial role in maintaining lifelong humoral immunity, and the relatively long life span of plasma cells is essential for this function, the underlying molecular mechanism of their long-term survival in vivo is unknown. To address this issue, we examined the expression of prosurvival molecules of the Bcl-2 family and examined their function in vivo. We identified Mcl-1 as being essential for the survival of plasma cells and found that signaling through the receptor BCMA facilitated high expression of Mcl-1 in bone marrow plasma cells. This insight may aid the development of new therapeutic strategies for the manipulation of long-lived plasma cells in autoimmune diseases and plasma cell–derived cancers, such as multiple myeloma.
RESULTS
Expression of plasma cell prosurvival Bcl-2 proteins
We examined the expression of various members of the Bcl-2 family in plasma cells isolated from the spleen and bone marrow of adult mice, and compared that to their expression in naive and germinal center B cells. Transcripts encoding Bcl-2 (Bcl2), Bcl-w (Bcl2l2) and Mcl-1 (Mcl1) were all detectable in plasma cells (Fig. 1a), whereas, as reported25, expression of the Bcl-2 family member A1 (encoded by Bcl2a1a) was strongly repressed (Fig. 1a). Expression of prosurvival Bcl-2 family members in the Blimp-1hi and Blimp-1int plasma cell subsets, identified through the use of Blimp-1–GFP reporter mice (in which sequence encoding GFP is expressed from the gene encoding Blimp-1 (Prdm1GFP)4), was essentially unchanged from that in the unseparated plasma cell population (Supplementary Fig. 1). The greater abundance of both Bcl2 mRNA and Mcl1 mRNA in plasma cells than in naive B cells was reflected in the greater abundance of their proteins, as determined by immunoblot analysis of extracts of plasma cells isolated from spleen and bone marrow (Fig. 1b and Supplementary Fig. 2).
Figure 1.
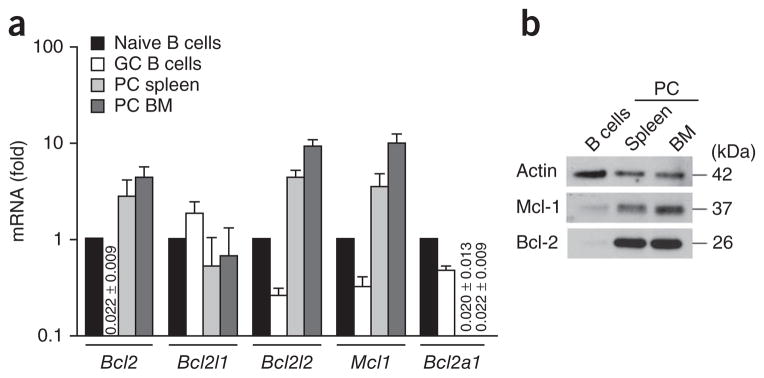
Expression of prosurvival members of the Bcl-2 family in plasma cells. (a) Quantitative PCR analysis of mRNA encoding members of the Bcl-2 family in CD19+PNA+ germinal center (GC) B cells sorted from spleen and B220−CD138+ plasma cells (PC) sorted from spleen or bone marrow (BM) of wild-type mice, normalized to expression of the housekeeping gene Hprt and presented relative to expression in naive (B220+) B cells, set as 1. Numbers in graph indicate expression <0.1 (± s.e.m.). (b) Immunoblot analysis of Mcl-1 and Bcl-2 in naive B cells and plasma cells isolated as in a. Data are representative of four independent sorts (a; mean and s.e.m.) or three experiments (b).
Published experiments with ABT-737 have ruled out a substantial role for Bcl-2 in the survival of existing plasma cells22,23. However, the low but detectable expression of Bcl2l1 (Fig. 1a) could have supported survival of long-lived plasma cells. Therefore, we examined the effect of conditional deletion of Bcl2l1 in existing plasma cells in vivo. For this, we reconstituted lethally irradiated Ly5.1+ recipient mice with a mixture of μMT bone marrow (which can generate all hematopoietic cells except B cells) and bone marrow in which all B cells carried the Rosa26-CreERT2 allele (and thus expressed a tamoxifen-sensitive variant of Cre recombinase from the ubiquitous Rosa26 locus; called ‘CreERT2’ here) and were either Bcl2l1+/+ or had loxP-flanked Bcl2l1 alleles (Bcl2l1fl/fl). We immunized those bone marrow chimeras with nitrophenyl coupled to keyhole limpet hemocyanin (NP-KLH) in the adjuvant alum and treated them with two doses of tamoxifen (by oral gavage) 14 d later. To measure Bcl2l1 expression after tamoxifen treatment, we isolated plasma cells from the bone marrow 2 d after the start of treatment and detected a lower abundance of Bcl2l1 transcripts (Supplementary Fig. 3a). Because Bcl-xL protein is expected to be relatively stable, we assessed the consequences of the deletion of Bcl2l1 on plasma cell frequency 4 d after the start of tamoxifen treatment, which was 18 d after immunization. We observed no significant difference between Bcl2l1+/+CreERT2 and Bcl2l1fl/flCreERT2 mice in percentage or absolute number of either total or antigen-specific plasma cells in the spleen or bone marrow (P > 0.05; Supplementary Fig. 3b,c). These data indicated that expression of Bcl-xL was not crucial for the survival of existing plasma cells.
BCMA regulates bone marrow plasma cell Mcl-1 expression
Next we investigated several extracellular factors and signals available in bone marrow niches for their ability to elicit the expression of Bcl-2, Bcl-xL, Bcl-w and Mcl-1 in plasma cells. Ligand-receptor interactions, including IL-6–IL-6 receptor, CD80- and/or CD86–CD28 and APRIL-BCMA can promote the survival of plasma cells in vivo26. We analyzed the frequency of plasma cells in mice deficient in the gene encoding IL-6 (Il6−/−) or BCMA (Tnfrsf17−/−; called ‘Bcma−/−’ here) or deficient in the genes encoding CD80 and CD86 (Cd80−/−Cd86−/−). As we expected, we found that the lack of any of these three pathways in mice led to fewer plasma cells than those in wild-type mice; this difference was significant in the bone marrow compartment but not in the spleen (Fig. 2a,b).
Figure 2.
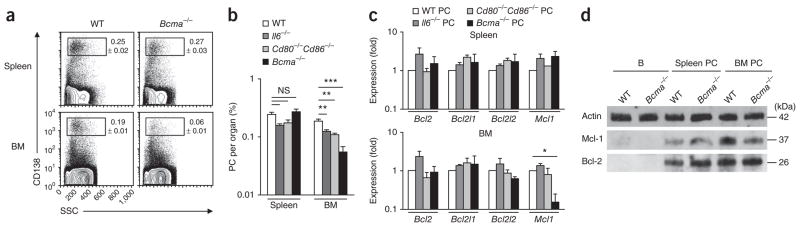
BCMA regulates Mcl-1 expression in bone marrow plasma cells. (a) Flow cytometry of cells from the spleen or bone marrow of wild-type (WT) or Bcma−/− mice, gated on B220− cells. Numbers adjacent to outlined areas indicate percent CD138+ (plasma) cells (± s.e.m.). SSC, side scatter. (b) Frequency of plasma cells in the spleen and bone marrow of wild-type, Il6−/−, Cd80−/−Cd86−/− and Bcma−/− mice (n = 3–4 per group), gated as in a. (c) Expression of genes encoding prosurvival members of the Bcl-2 family in plasma cells (B220−CD138+) isolated from the spleen or bone marrow of wild-type, Il6−/−, Cd80−/−Cd86−/− and Bcma−/− mice (n = 4 group; pooled cells), presented relative to that of wild-type plasma cells, set as 1. (d) Immunoblot analysis of Mcl-1 and Bcl-2 in B cells (B) and plasma cells (B220−CD138+) sorted from the spleen or bone marrow wild-type and Bcma−/− mice (n = 4–6 per genotype; pooled cells). NS, not significant; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (Student’s t-test). Data are representative of two experiments (a,c,d) or two experiments with similar results (b; mean and s.e.m. in b,c).
To determine whether those three distinct pathways contributed to the induction of prosurvival members of the Bcl-2 family, we isolated plasma cells from the spleen and bone marrow of control (wild-type), Il6−/−, Cd80−/−Cd86−/− and Bcma−/− mice and measured the mRNA enoding various prosurvival members of the Bcl-2 family. We observed no significant difference among genotypes in such expression in plasma cells isolated from the spleen (Fig. 2c). However, plasma cells from the bone marrow of Bcma−/− mice showed abnormally lower expression of both Mcl1 mRNA (Fig. 2c) and Mcl-1 protein (Fig. 2d and Supplementary Fig. 4). Although we observed no compensatory upregulation of the expression of other prosurvival members of the Bcl-2 family (Fig. 2c), we found significantly lower expression of the gene encoding Bim in plasma cells from the bone marrow of Bcma−/− mice (Supplementary Fig. 5). Thus, of the three different mutant mouse strains we examined, Bcma−/− mice had the fewest total bone marrow plasma cells relative to those in wild-type mice (30% as many; Fig. 2a,b). This was in agreement with existing literature showing a considerably fewer antigen-specific plasma cells (17% as many)12 and total plasma cells (less than 50% as many)27 in the bone marrow, but not the spleen, of Bcma−/− mice than in that of wild-type mice. Together these data showed that higher expression of the prosurvival protein Mcl-1 was influenced by expression of BCMA on plasma cells residing in the bone marrow compartment.
Bcma induction is Blimp-1 independent
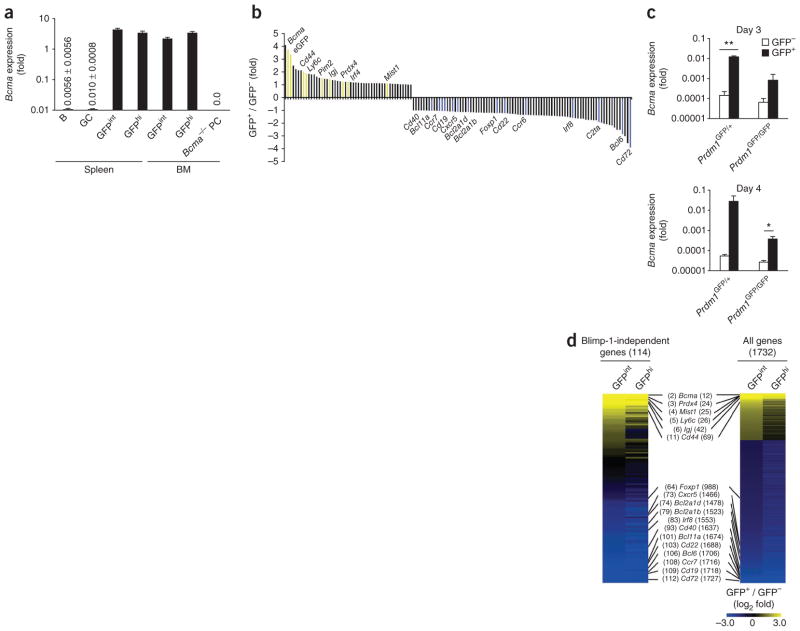
BCMA, a member of the tumor-necrosis factor receptor family, has high expression in mouse and human plasma cells but is absent from naive B cells, germinal center B cells and memory B cells13,28. We sought to measure BCMA relative to the changes in Blimp-1 expression that occur during the differentiation of activated B cells into plasma cells. As functional antibodies to mouse BCMA are lacking at present, we assessed the expression of Bcma mRNA by quantitative real-time PCR. We detected large amounts of Bcma mRNA in Blimp-1int plasma blasts and Blimp-1hi plasma cells in the spleen and bone marrow of Blimp-1–GFP reporter mice3 (Prdm1GFP/+; Fig. 3a). As we expected, we found that Bcma mRNA was absent from naive B cells and germinal center B cells in these mice (Fig. 3a). These results indicated that induction of BCMA was part of a transcriptional program specifically induced after plasma cell differentiation. However, when we cultured Blimp-1-deficient (Prdm1GFP/GFP) B cells3 under conditions of plasma cell differentiation, we observed considerable induction of Bcma expression in activated GFP+ B cells; that is, B cells with an active Prdm1 locus that were arrested in the earliest stage of plasma cell differentiation3 (Fig. 3b,c). This observation indicated that initiation of Bcma transcription was Blimp-1 independent. Moreover, genome-wide microarray analysis showed that Bcma was among the genes most induced in Blimp-1int plasma blasts in the spleen of Blimp-1–GFP reporter mice, relative to expression in naive B cells (Fig. 3d). Collectively, these data indicated that initiation of the BCMA-mediated plasma cell survival pathway was independent of the Blimp-1-dependent component of plasma cell differentiation and that it had a role in regulating Mcl-1 expression in plasma cells.
Figure 3.
The BCMA pathway is regulated independent of Blimp-1. (a) Bcma expression in naive B cells (B), germinal center B cells (GC), Blimp-1int (GFPint) plasmablasts and Blimp-1hi (GFPhi) plasma cells isolated from spleen and bone marrow of Prdm1GFP/+ mice (n = 3–4 per group), presented relative to Hprt expression (numbers in graphs as in Fig. 1). Bcma−/− plasma cells serve as a negative control. (b) Genome-wide oligonucleotide array analysis of gene expression in Blimp-1-deficient (Prdm1GFP/GFP) B cells cultured for 4 d, sorted as GFP− and GFP+ cells, presented as expression in GFP+ cells relative to expression in GFP− cells and ranked according to change in expression; colors indicate genes with known differences in expression in plasma cells (yellow, induced; blue, repressed)). (c) Quantitative PCR analysis of Bcma expression in Blimp-1–GFP (Prdm1GFP/+) B cells and Blimp-1-deficient (Prdm1GFP/GFP) B cells (GFP− or GFP+), assessed on days 3 and 4 of culture and presented relative to Hprt expression. (d) Genome-wide oligonucleotide array analysis of naive B cells, GFPint plasmablasts and GFPhi plasma cells isolated from the spleen of Prdm1GFP/+ mice, showing genes with a difference in expression of more than twofold relative to their expression in naive B cells (right; ranked (numbers in parentheses) according to change in expression in GFPint cells versus naive B cells) and the subset of genes regulated independently of Blimp-1 (left; as in b; ranked as at right). *P ≤ 0.05 and **P ≤ 0.01 (Student’s t-test). Data are representative of one experiment (a), two experiments (b,d) or three experiments (d; mean and s.e.m. in a,c).
Mcl-1 promotes plasma cell survival in vitro
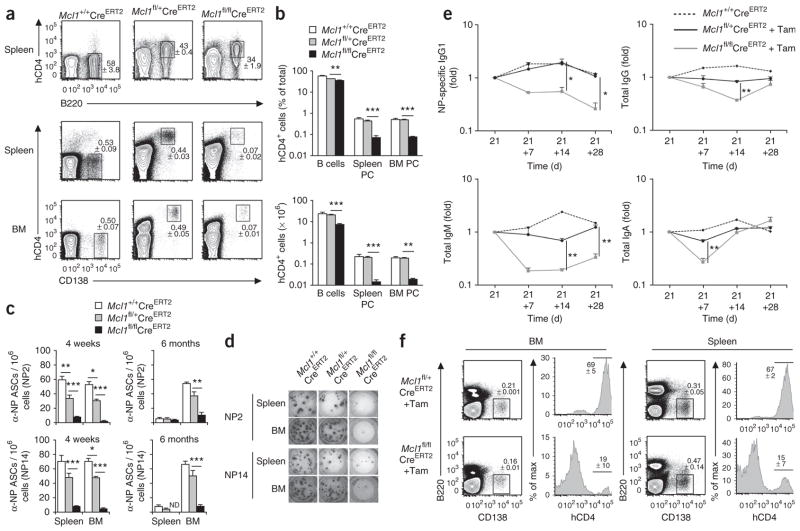
The higher expression of Mcl-1 in normal plasma cells and the lower Mcl-1 expression and plasma cell numbers in the absence of BCMA indicated that Mcl-1 might be an essential mediator of plasma cell survival. Indeed, plasma cells in the whole mouse are resistant to ABT-737, an apoptosis-inducing antagonist of Bcl-2, Bcl-xL and Bcl-w, which suggests that these three prosurvival proteins are dispensable for the survival of plasma cells22. To examine the requirement for Mcl-1 in the survival of plasma cells, we investigated the effect of conditional deletion of Mcl1 in plasma cells. We generated Mcl1fl/fl CreERT2 mice, in which, after Cre-mediated deletion of Mcl1, human CD4 was expressed on the cell surface under the control of Mcl1 regulatory elements24,29. Surface expression of human CD4 was therefore a reporter of Mcl1 deletion and an indicator of Mcl1 transcription. First, we used human CD4 expression as an indicator of Mcl1 transcription. We cultured B cells from the spleen of Mcl1fl/+CreERT2 mice under plasma cell–differentiating conditions30. We added the BCMA ligand APRIL to the culture medium on day 5 and induced tamoxifen-mediated deletion of the loxP-flanked Mcl1 allele 20 h before analysis on day 6 or 7 of culture. APRIL-mediated signaling resulted in a significant increase in the expression of human CD4 on plasma blasts over the 20-hour time period after tamoxifen addition (Fig. 4a,b). That result established a connection between APRIL and the induced transcription of Mcl1. Next we isolated B cells from Mcl1fl/+CreERT2 or Mcl1fl/flCreERT2 mice and cultured the cells under differentiation conditions. Again, we induced tamoxifen-mediated deletion of the loxP-flanked Mcl1 allele 20 h before analysis on day 6 or 7 of culture, and this time we measured the effect on cell survival. Analysis of Mcl-1 by immunoblot analysis showed efficient deletion of Mcl1 after in vitro treatment with tamoxifen (Supplementary Fig. 6). The absolute number of plasma blasts after treatment was lower on both days 6 and 7 of culture. Cells expressing the plasma cell marker CD138 were affected significantly more by Mcl1 deletion than were undifferentiated B cell blasts on day 7 (Fig. 4c,d). We concluded from these experiments that plasma blasts had both higher expression of Mcl-1 than that od B cell blasts and greater reliance on Mcl-1 for their survival.
Figure 4.
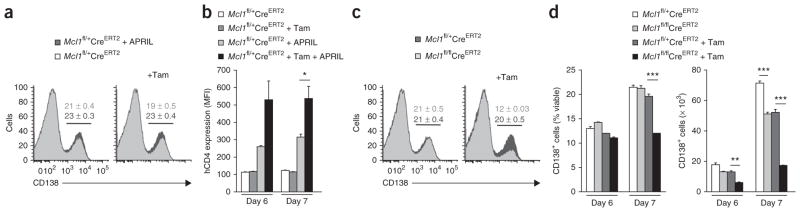
Dependence on Mcl-1 in in vitro cultured plasmablasts. (a) Flow cytometry analysis of the cell surface expression of CD138 (gated on viable cells) on activated Mcl1fl/+CreERT2 B cells cultured for 7 d in plasma cell–differentiating conditions, without tamoxifen (left) or with the addition of tamoxifen 20 h before analysis (Tam), with (+ APRIL) or without the addition of APRIL on day 5 of culture. (b) Mean fluorescence intensity (MFI) of human CD4 (hCD4) on day 6 or 7 of culture (as in a) of Mcl1fl/+CreERT2 and Mcl1fl/flCreERT2 cells, gated on CD138+ plasmablasts as in a. (c) Flow cytometry of activated Mcl1fl/+CreERT2 and Mcl1fl/flCreERT2 B cells cultured in plasma cell–differentiating conditions, with or without tamoxifen (as in a) added on day 7 of culture. (d) Frequency and absolute number of CD138+ Mcl1fl/+CreERT2 or Mcl1fl/flCreERT2 plasmablasts on day 6 or 7 of culture with or without tamoxifen (as in a), gated as in a. Numbers above bracketed lines (a,c) indicate percent CD138+ cells (± s.e.m.).*P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001 (Student’s t-test). Data are representative of two experiments (a–c), or two experiments with similar results (d; mean and s.e.m. of three samples per group in b,d).
Mcl-1-dependent survival of plasma cells in vivo
Our in vitro experiments suggested that Mcl-1 might be an important mediator of plasma cell survival in vivo. To examine the effect of Mcl1 deletion on existing plasma cells in vivo, we created mice through hematopoietic reconstitution after lethal irradiation such that all B cells carried the Rosa26-CreERT2 allele and were Mcl1+/+, Mcl1fl/+ or Mcl1fl/fl. We immunized reconstituted mice with NP-KLH in alum and subsequently treated the mice with two doses of tamoxifen (by oral gavage) either 4 weeks later (Fig. 5a–d) or 26 weeks later (Fig. 5c). We analyzed spleen and bone marrow 2 d after the start of tamoxifen administration. Mcl1 was deleted in existing plasma cells residing in these tissues, as judged by the cell-surface expression of human CD4 (Fig. 5a). The percentage and absolute number of total plasma cells (Fig. 5a,b) and antigen-specific plasma cells (Fig. 5c,d) were significantly lower after deletion of both alleles of Mcl1 than in wild-type mice. This difference was considerably greater than that of naive B cells (either immature or mature) and similar to the difference seen with germinal center B cells24 (Fig. 5a,b and data not shown). Deletion of one allele of Mcl1 did not affect the percentage or absolute number of total plasma cells (Fig. 5b) but did lead to fewer NP-specific plasma cells 4 weeks after immunization (Fig. 5c). To examine the effect of B cell–specific deletion of Mcl1 on serum immunoglobulin titers, we immunized additional Mcl1fl/flCreERT2 mixed–bone marrow chimeras with NP-KLH in alum, treated them with tamoxifen 21 and 22 d later and sampled their serum at three time points thereafter. As expected, we observed a decrease in NP-specific immunoglobulin G1 (IgG1) over time after deletion of both alleles of Mcl1 (Fig. 5e). Whereas the rate of loss of IgG1 antibody to NP in the first 5 d after tamoxifen treatment matched that expected from the 8-day half-life of serum IgG1 in mice31, the titer at the end of the experiment was higher than expected, possibly owing to the incomplete depletion of NP-specific plasma cells (Fig. 5c). We observed also a decrease in the serum titers of total IgG, IgM and IgA that was temporary in all cases (Fig. 5e). The magnitude and rate of the decrease reflected the serum half-life of the isotypes in that IgM, with a serum half-life of 2 d (ref. 31), decreased 80% within 5 d of treatment, whereas IgG, with a half-life of 8 d, decreased 30% (Fig. 5e). At 28 d after tamoxifen treatment, the final time point of this experiment, we counted plasma cells from spleen and bone marrow by flow cytometry. In contrast to our earlier findings (Fig. 5a), we observed abundant human CD4–negative Mcl1fl/flCreERT2 plasma cells (Fig. 5f), which were probably plasma cells differentiated from B cells that had escaped the inducible deletion of Mcl1. We presumed that these cells accounted for the rebound in concentrations of total IgG, IgM and IgA after treatment with tamoxifen.
Figure 5.
Mcl-1 expression is required for the survival of plasma cells. (a) Flow cytometry of cells from the spleen and bone marrow of mixed–bone marrow chimeras after inducible deletion of Mcl1. Numbers adjacent to outlined areas indicate percent (±s.e.m.) human CD4–positive B220+ cells (B cells with deletion of Mcl1; top row) or B220−CD138+ (plasma) cells (bottom row). (b) Frequency and absolute number of plasma cells and B cells after Mcl1 deletion, gated as in a. (c,d) Enzyme-linked immunospot assay of total (NP14) or high-affinity (NP2) NP-specific antibody–secreting cells (α-NP ASCs) after deletion of Mcl1, 4 weeks or 6 months after immunization of mice (n = 4 per group) with NP-KLH, shown as frequency of ASCs (c) or images of assays (d). ND, not detected. (e) Enzyme-linked immunosorbent assay of NP-specific IgG1, total IgG, total IgM and total IgA in blood after inducible deletion of Mcl1, assessed on day 21 after immunization of mice and 7 d (+7), 14 d (+14) or 28 d (+28) thereafter, (n = 3 mice per group) and presented relative to titers on day 21. P values, compared with Mcl1fl/+CreERT2 + tamoxifen. (f) Flow cytometry of cells from the spleen or bone marrow of mice reconstituted with Mcl1fl/+CreERT2 or Mcl1fl/flCreERT2 bone marrow, assessed 28 d after deletion of Mcl1 as in e. Numbers adjacent to outlined areas indicate percent (±s.e.m.) B220−CD138+ (plasma) cells; numbers above bracketed lines indicate percent cells expressing human CD4, gated on plasma cells. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (Student’s t-test). Data are representative of three experiments (a), three experiments with four mice per group (b), three pooled experiments (4 weeks) or one experiment (6 months; c,d) or one experiment (e,f; mean and s.e.m. in b,c,e).
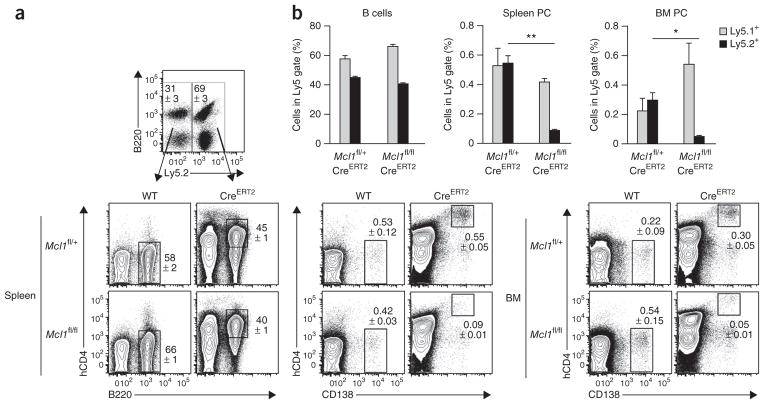
To rule out possible effects of the lower abundance of naive B cells on the maintenance of plasma cells, we generated mixed–bone marrow chimeras in which ~50% of the hematopoietic cells were wild-type (Ly5.1+) and ~50% were either Mcl1fl/+CreERT2 or Mcl1fl/fl CreERT2 (Ly5.2+). We immunized those mice with NP-KLH in alum as described above and treated them with tamoxifen on days 26 and 27, followed by analysis 1 d later. Control Ly5.1+ B cells and plasma cells were unaffected by the tamoxifen treatment, but the proportion and number of Ly5.2+ human CD4–positive Mcl1fl/flCreERT2 plasma cells in both the spleen and bone marrow was significantly lower (Fig. 6). Thus, the requirement for continued expression of Mcl1 in the survival of plasma cells was cell intrinsic.
Figure 6.
Dependence of plasma cells on Mcl-1 is cell intrinsic. (a) Flow cytometry of plasma cells (CD138+) in mixed chimeras after inducible deletion of Mcl1 (Ly5.1+, wild type (WT); Ly5.2+, Mcl1fl/+CreERT2 or Mcl1fl/flCreERT2 (CreERT2)). Numbers adjacent to outlined areas indicate percent cells in each. (b) Frequency of cells in populations gated as in a. Ly5.1+ (wild type) cells are defined as B220+ B cells or CD138+ plasma cells; cells with a loxP-flanked Mcl1 allele are Ly5.2+ and hCD4+B220+ B cells or hCD4+CD138+ plasma cells. *P ≤ 0.01 and **P ≤ 0.001 (Student’s t-test). Data are representative of three experiments (mean and s.e.m. of four mice per group in b).
DISCUSSION
The lifespan of plasma cells is believed to reach years in mice and decades in humans, and they provide protective antibodies for a lifetime in some cases32–34. To achieve such longevity, plasma cells must locate themselves in specialized niches composed of cells that provide the signals necessary to maintain plasma cell viability35. Niches in the bone marrow are the best characterized, but long-lived plasma cells are also present in the red pulp of the spleen and mucosa-associated lymphoid tissues26. Bone marrow niches contain monocytes, eosinophils, megakaryocytes, CXCL12+ reticular stromal cells, basophils, osteoclasts and osteoblasts26. Collectively, these cells provide attractants, anchoring molecules and survival factors that recruit, retain and then sustain bone marrow plasma cells. Among the sustaining molecules are BAFF and APRIL (members of the tumor-necrosis factor superfamily), the cytokine IL-6, and the coreceptor CD28 ligands CD80 and CD86. Determining how these factors mediate the survival of plasma cells, although crucial for understanding the persistence of immunity, has been hampered by lack of knowledge about what molecular mechanism they activate, individually or combined, in the plasma cell. We reasoned that identifying the survival mechanism of plasma cells should help determine the factor(s) responsible for activating that mechanism and the signaling pathways involved. The data presented here have shown that sustained expression of the Bcl-2 family member Mcl-1 exemplifies how niche-derived survival factors mediate the persistence of plasma cells and have further shown that ligands of BCMA mediate this effect in the bone marrow.
BCMA is important for plasma cell survival in vivo, as immunized mice deficient in Bcma have abnormally low numbers of antigen- specific plasma cells in their bone marrow12. APRIL, a ligand for BCMA, is produced by bone marrow–resident eosinophils36, basophils15 and monocytes or macrophages37 in mice, and by neutrophils38 in humans, and these cell types promote the survival and accumulation of plasma cells. As with immunized Bcma−/− mice, the bone marrow of APRIL-deficient mice supports considerably fewer antigen-specific plasma blasts and plasma cells than does wild-type bone marrow, albeit after transfer rather than with de novo generation20. Our results have confirmed the importance of the APRIL-BCMA axis in plasma cell survival in the bone marrow and indicate that this process requires transcriptional induction of Mcl1. How does this model account for plasma cells present in the bone marrow of Bcma−/− mice, albeit at lower frequency and with lower expression of Mcl-1 than that of wild-type mice? This presumably reflects the expression pattern of genes encoding survival molecules that is due to factors acting in addition to BCMA. This suggests that BCMA is positioned to sustain high Mcl-1 expression. The low Bim expression found in Bcma−/− bone marrow plasma cells, although necessary for survival in this case, could reflect the usual expression of Bim among some newly generated plasma cells. Such plasma cells with intrinsically low Bim expression might be infrequent; this could explain the difficulty in establishing new specificities into the Bcma−/− bone marrow plasma cell population12.
Notably, we found that that BCMA deficiency affected bone marrow plasma cells much more than spleen plasma cells in both frequency and expression of genes encoding survival molecules. The Bcma−/− bone marrow plasma cell population reflects survival mediated by signals derived from receptors other than BCMA. Splenic plasma cells were sustained at near normal frequency in BCMA-deficient mice and had normal expression of Mcl-1. Thus, the spleen must contain factors that can substitute for BCMA signaling to induce Mcl-1. Plasma cells in spleen and bone marrow continue to express mRNA encoding the transmembrane activator TACI, albeit at lower amounts than its expression by B cell blasts. TACI binds APRIL and BAFF with different affinities, and these ligands might be present in different amounts and/or configurations in spleen and bone marrow, thereby mediating different outcomes through the same receptor. Alternatively, a ligand-receptor system may be available to plasma cells in the spleen that is not available in bone marrow; this system may be composed of some combination of already defined factors or be previously unknown. Whichever is true, analyzing the signals that mediate high expression of Mcl-1 in the persisting long-lived splenic plasma cell will help resolve these issues.
Our data have shown that BCMA-mediated expression of Mcl-1 constitutes a plasma cell–survival pathway that arises independently of the Blimp-1-controlled plasma cell–differentiation pathway. B cells induced to differentiate without Blimp-1 progress to a ‘pre-plasmablast’ stage at which cells secrete small amounts of immunoglobulin, upregulate the transcription factor XBP-1 and downregulate transcription factor Pax5 (ref. 3). We propose that these cells represent the earliest stage of plasma cell differentiation, one that is by definition Blimp-1 independent3. The findings that induction of BCMA expression occurred independently of Blimp-1, that BCMA is encoded by a plasma cell–specific gene and that BCMA was crucial for the survival of fully differentiated cells at the terminus of this pathway support the proposal that Blimp-1 consolidates and amplifies plasma cell differentiation rather than being the ‘master regulator’ of the process. Indeed, the term “master regulator” may be better reserved for the factors that trigger immunoglobulin secretion and initiate the plasma cell–survival program, such as the transcription factor IRF4. Upregulation of IRF4 expression precedes the induction of Blimp-1 expression39, and both Bcma and Prdm1 have been identified as part of a plasma cell–differentiation program induced by IRF4 (ref. 40).
Plasma cells accumulate ‘preferentially’ in specific niches in the bone marrow, and it has been suggested that a wide variety of cell types and ligand-receptor pairs contribute to their longevity41. However, loss of many of these ligand-receptor pairs on their own diminishes the survival of plasma cells only marginally in vivo12,14,16,26,27. We therefore postulate that many (perhaps all) of these signaling pathways converge after the transcriptional induction of Mcl1 and that their combined action allows the high expression of Mcl-1 in plasma cells. Here we found that the maintenance of Mcl-1 expression in plasma cells was crucial for their survival, as loss of Mcl1 led to rapid disappearance of this cell population. Moreover, we found that signaling through BCMA promoted transcription of Mcl1 and thus was important for the long-term maintenance of bone marrow plasma cells. Both BCMA42,43 and Mcl-1 (ref. 44) have high expression in plasma cell–derived cancerous multiple myeloma cells and their expression seems to be correlated with tumor cell persistence. Furthermore, two recent clinical phase 2 trials showed that the APRIL- and BAFF-inhibitor atacicept significantly decreased both the number of plasma cells and the amount of immunoglobulins in rheumatoid arthritis patients45,46. This highlights the potential clinical importance of interfering with the survival pathways that promote Mcl-1 expression in plasma cells.
ONLINE METHODS
Mice, antigens and immunization
Mcl1fl mice24, Bcl2l1flox mice24, Rosa26- CreERT2 mice24 (Taconic Artemis), Prdm1GFP/+ mice3, Il6−/− mice47, Cd80−/−Cd86−/− mice48 and μMT-deficient mice49 were bred and maintained at the animal facilities of the Walter and Eliza Hall Institute of Medical Research and were derived from C57BL/6 embryonic stem cells or from 129SV-derived embryonic stem cells but backcrossed to the C57BL/6 background for more than ten generations. All animal procedures were approved by The Walter and Eliza Hall Institute Animal Ethics Committee. Prdm1GFP/GFP mice were produced from fetal liver–derived stem cells as described3. Bcma−/− mice were on a C57BL/6-and-C3H/He mixed genetic background. Bone marrow–reconstituted mice were generated by the following protocols. Lethally irradiated (two doses of 5.5 Gy, 3 h apart) C57BL/6 (Ly5.1+) mice were reconstituted with either 80% B cell–deficient (μMT-deficient) bone marrow plus 20% (Ly5.2+) Mcl1+/+CreERT2, Mcl1fl/+CreERT2 or Mcl1fl/flCreERT2 bone marrow or 50% C57BL/6 (Ly5.1+) bone marrow plus 50% Mcl1fl/+CreERT2 or Mcl1fl/flCreERT2 bone marrow (each C57BL/6 Ly5.2+). Immunization was a single intraperitoneal injection of 100 μg NP-KLH at a ratio of 21:1 (NPH/KLH) and precipitated onto alum as described24. Estrogen receptor–mediated deletion of loxP-flanked Mcl1 alleles was done by oral gavage of tamoxifen (5 mg (Sigma) in 83 μl of a solution of 90% peanut oil (Sigma) and 10% ethanol) on two successive days before analysis after immunization. Deletion of the loxP-flanked Mcl1 allele(s) was monitored by human CD4 expression24.
Antibodies, flow cytometry and cell sorting
Single-cell suspensions were stained with the following antibodies to surface molecules: anti-B220 (RA3-6B2; produced in-house), anti-CD138 (281.2; BD Biosciences), anti-FcγR (2.4G2; produced in-house), anti-CD45.2 (104; BD Biosciences) and anti–human CD4 (OKT4; produced in-house). Peanut agglutinin coupled to fluorescein isothiocyanate was used for detection of germinal center B cells. NP binding was detected as described24. Stained cells were analyzed on a FACSCantoII (BD Biosciences). Naive B cells (B220+), germinal center B cells (B220+PNA+) and plasma cells (B220−CD138+) were sorted from splenocytes and bone marrow cells with a Moflow cytometer (BD Biosciences) to a purity of 98%. Enrichment for plasma cells was achieved by magnetic-activated cell separation beads coated with anti-CD138 (130-092-530; Miltenyi) before sorting by flow cytometry.
RNA and cDNA preparation and quantitative PCR
RNA was prepared from cells with an RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen), then cDNA was prepared from total RNA with reverse-transcription reagents (Applied Biosystems). Primers for measurement of RNA expression (designed by and obtained from Geneworks) were as follows: Bcl2 (forward 5′-GAGCGTCAACAGGGAGATG-3′, reverse 5′-CAGAGACAGCCAGGAGAAATC-3′), Bcl2l1 (forward 5′-GGAAAGC GTAGACAAGGAGATG-3′, reverse 5′-CCCGTAGAGATCCACAAAAG TG-3′), Bcl2l2 (forward 5′-CGTCTTGTGGCATTCTTTGTC-3′, reverse 5′-TCCCCGTATAGAGCTGTGAA-3′), Bcl2a1 (forward 5′-TGAATAACA CAGGAGAATGGATACG-3′, reverse 5′-GAAATGCCAAGTGCTGATAA CC-3′), Bcl2l11 (forward 5′-GAGTTGTGACAAGTCAACACAAACC-3′, reverse 5′-GAAGATAAAGCGTAACAGTTGTAAGATAACC-3′), Bbc3 (forward 5′-ACCTCAACGCGCAGTAC-3′, reverse 5′-GAGATTGTAC ATGACCCTCCAG-3′), Bid (forward 5′-TCTGAAAGTCAGGAAGAAA TCATCC-3′, reverse 5′-GGTTAATAAAGTTGACAGTCGTGTGG-3′), Pmaip1 (forward 5′-ACTGTGGTTCTGGCGCAGAT-3′, reverse 5′-TT GAGCACACTCGTCCTTCAA-3′), Hprt (forward 5′-GGGGGCTATAAG TTCTTTGC, reverse 5′-TCCAACACTTCGAGAGGTCC) and Bcma (forward 5′-GTCACTTGCGATGTTCCAAC-3′, reverse 5′-TACGTCCC TTTCACTGAACTG-3′). The Mcl1 primer assay (Quantitect, mM_Mcl1_1_ SG) was from Qiagen.
Enzyme-linked immunospot assay and enzyme-linked immunosorbent assay
The frequency of NP-specific antibody-secreting cells and total antibody-secreting cells was determined as described24. Enzyme-linked immunosorbent assays of anti-NP and immunoglobulin isotypes were done as described24.
Immunoblot
Cells were lysed for 30 min on ice in lysis buffer containing 1% NP-40. Cell lysates (equivalent to 100,000 cells per lane) were separated by size on precast 4–12% SDS gradient gels (NuPage, Invitrogen). Membranes were probed overnight at 4 °C with the following antibodies: rat anti–mouse Mcl-1 (19C4-15; prepared in-house), hamster anti–mouse Bcl-2 (3F11; BD Biosciences) and anti-actin (loading control; I-19; Santa Cruz). Primary antibodies were visualized by incubation with horseradish peroxidase–conjugated species-specific secondary antibodies to IgG (NA9350 (GE Healthcare) and 6060-05 (Southern Biotechnology).
In vitro generation of plasmablasts
For some experiments (Fig. 3b and Supplementary Fig. 2), immunomagnetically selected naive B cells (Miltenyi) were cultured for 4 d with soluble CD40L (ref. 50), IL-4 (10 ng/ml; Peprotech) and IL-5 (5 ng/ml; Peprotech) in RPMI medium supplemented with 5% FCS, β-mercaptoethanol and antibiotics. For other experiments (Fig. 4), immuno-magnetically selected naive B cells (Miltenyi) were cultured for 4 d on irradiated 3T3 feeder cells expressing CD40L on their cell surface in RPMI medium supplemented with 5% FCS and IL-4 (10 ng/ml; Peprotech). Then, B cells were isolated and cultured with newly irradiated 3T3 feeder cells expressing CD40L in RPMI medium supplemented with 5% FCS, IL-21 (50 ng/ml; Peprotech) and IL-5 (5 ng/ml; Peprotech) as described30. MegaAPRIL (200 ng/ml; Alexis) was added at day 5 of culture. Where needed, 4-hydroxytamoxifen (1 mM; Sigma) was added to the culture supernatant 20 h before analysis.
Microarray analysis
CD138+GFPint and CD138+GFPhi plasma cell populations from the spleen of naive Prdm1GFP/+ mice were sorted by flow cytometry. RNA was purified with RNeasy Micro kits (Qiagen) and microarrays were done with Illumina oligonucleotide arrays at the Australian Genome Research Facility (Melbourne and Brisbane). For analysis of Blimp-1-independent transcriptional changes, Prdm1GFP/GFP B cells were cultured for 4 d with IL-4 (10 ng/ml; Peprotech), IL-5 (5 ng/ml; Peprotech) and CD40L (ref. 50) as described above and GFP− and GFP+ cell populations were isolated by flow cytometry for RNA extraction.
Statistical analysis
Statistical analysis was done with GraphPad Prism software (GraphPad Software). A Student’s t-test with two-tailed distributions for two samples with equal variance was used. P values of less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank the facilities of our respective institutes, particularly those responsible for animal husbandry and flow cytometry; and C. Wellard, S. Chevrier, D. Segal, D. Huang, S. Heinzel, J. Marchingo, S. Willis and P. Bouillet for assistance. Supported by the Australia National Health and Medical Research Council (1021374 to I.V.; 356202 to D.M.T. and S.L.N.; 461221 to A.S.; 637326 to S.P.G.; 516786 to K.F.; and the Independent Research Institute Infrastructure Support Scheme), the Multiple Myeloma Research Foundation USA (V.P.), the European Molecular Biology Organization (V.P.), the US National Institutes of Health (AI093722 to L.D.E.) and Victorian State Government Operational Infrastructure Support.
Footnotes
Accession codes. GEO: microarray data, GSE43215.
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
V.P., I.V. and D.M.T. designed the research; V.P., I.V., J.W., S.P.G., M.L., C.M.C. and K.F. did experiments and contributed to interpretation and discussion; L.D.E., F.M., A.S. and S.L.N. contributed to the design of experiments, interpretation of results, and drafting the manuscript; V.P. and I.V. analyzed data and prepared figures; and V.P., I.V. and D.M.T. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Shaffer AL, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro-Shelef M, et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 3.Kallies A, et al. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 2007;26:555–566. doi: 10.1016/j.immuni.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Kallies A, et al. Plasma cell ontogeny defined by quantitative changes in Blimp-1 expression. J Exp Med. 2004;200:967–977. doi: 10.1084/jem.20040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sze DM, Toellner KM, Garcia de Vinuesa C, Taylor DR, MacLennan IC. Intrinsic constraint on plasmablast growth and extrinsic limits of plasma cell survival. J Exp Med. 2000;192:813–821. doi: 10.1084/jem.192.6.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarlinton D. B-cell memory: are subsets necessary? Nat Rev Immunol. 2006;6:785–790. doi: 10.1038/nri1938. [DOI] [PubMed] [Google Scholar]
- 7.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 8.Kabashima K, et al. Plasma cell S1P1 expression determines secondary lymphoid organ retention versus bone marrow tropism. J Exp Med. 2006;203:2683–2690. doi: 10.1084/jem.20061289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarlinton D, Radbruch A, Hiepe F, Dorner T. Plasma cell differentiation and survival. Curr Opin Immunol. 2008;20:162–169. doi: 10.1016/j.coi.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 10.Oracki SA, Walker JA, Hibbs ML, Corcoran LM, Tarlinton DM. Plasma cell development and survival. Immunol Rev. 2010;237:140–159. doi: 10.1111/j.1600-065X.2010.00940.x. [DOI] [PubMed] [Google Scholar]
- 11.Smith KG, Hewitson TD, Nossal GJ, Tarlinton DM. The phenotype and fate of the antibody-forming cells of the splenic foci. Eur J Immunol. 1996;26:444–448. doi: 10.1002/eji.1830260226. [DOI] [PubMed] [Google Scholar]
- 12.O’Connor BP, et al. BCMA is essential for the survival of long-lived bone marrow plasma cells. J Exp Med. 2004;199:91–98. doi: 10.1084/jem.20031330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson MJ, et al. Cutting edge: the dependence of plasma cells and independence of memory B cells on BAFF and APRIL. J Immunol. 2008;180:3655–3659. doi: 10.4049/jimmunol.180.6.3655. [DOI] [PubMed] [Google Scholar]
- 14.Cassese G, et al. Plasma cell survival is mediated by synergistic effects of cytokines and adhesion-dependent signals. J Immunol. 2003;171:1684–1690. doi: 10.4049/jimmunol.171.4.1684. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez Gomez M, et al. Basophils support the survival of plasma cells in mice. J Immunol. 2010;185:7180–7185. doi: 10.4049/jimmunol.1002319. [DOI] [PubMed] [Google Scholar]
- 16.Rozanski CH, et al. Sustained antibody responses depend on CD28 function in bone marrow-resident plasma cells. J Exp Med. 2011;208:1435–1446. doi: 10.1084/jem.20110040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokoyoda K, Egawa T, Sugiyama T, Choi BI, Nagasawa T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 2004;20:707–718. doi: 10.1016/j.immuni.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Le Gouill S, Podar K, Harousseau JL, Anderson KC. Mcl-1 regulation and its role in multiple myeloma. Cell Cycle. 2004;3:1259–1262. doi: 10.4161/cc.3.10.1196. [DOI] [PubMed] [Google Scholar]
- 19.Potter M. Neoplastic development in plasma cells. Immunol Rev. 2003;194:177–195. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- 20.Belnoue E, et al. APRIL is critical for plasmablast survival in the bone marrow and poorly expressed by early-life bone marrow stromal cells. Blood. 2008;111:2755–2764. doi: 10.1182/blood-2007-09-110858. [DOI] [PubMed] [Google Scholar]
- 21.Moreaux J, et al. BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone. Blood. 2004;103:3148–3157. doi: 10.1182/blood-2003-06-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrington EM, et al. BH3 mimetics antagonizing restricted prosurvival Bcl-2 proteins represent another class of selective immune modulatory drugs. Proc Natl Acad Sci USA. 2010;107:10967–10971. doi: 10.1073/pnas.1005256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mérino D, et al. Bcl-2, Bcl-x(L), and Bcl-w are not equivalent targets of ABT-737 and navitoclax (ABT-263) in lymphoid and leukemic cells. Blood. 2012;119:5807–5816. doi: 10.1182/blood-2011-12-400929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vikstrom I, et al. Mcl-1 is essential for germinal center formation and B cell memory. Science. 2010;330:1095–1099. doi: 10.1126/science.1191793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarte K, et al. The Bcl-2 family member Bfl-1/A1 is strongly repressed in normal and malignant plasma cells but is a potent anti-apoptotic factor for myeloma cells. Br J Haematol. 2004;125:373–382. doi: 10.1111/j.1365-2141.2004.04908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tangye SG. Staying alive: regulation of plasma cell survival. Trends Immunol. 2011;32:595–602. doi: 10.1016/j.it.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Jiang C, Loo WM, Greenley EJ, Tung KS, Erickson LD. B cell maturation antigen deficiency exacerbates lymphoproliferation and autoimmunity in murine lupus. J Immunol. 2011;186:6136–6147. doi: 10.4049/jimmunol.1001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Good KL, Avery DT, Tangye SG. Resting human memory B cells are intrinsically programmed for enhanced survival and responsiveness to diverse stimuli compared to naive B cells. J Immunol. 2009;182:890–901. doi: 10.4049/jimmunol.182.2.890. [DOI] [PubMed] [Google Scholar]
- 29.Glaser SP, et al. Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev. 2012;26:120–125. doi: 10.1101/gad.182980.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nojima T, et al. In vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun. 2011;2:465. doi: 10.1038/ncomms1475. [DOI] [PubMed] [Google Scholar]
- 31.Vieira P, Rajewsky K. The half-lives of serum immunoglobulins in adult mice. Eur J Immunol. 1988;18:313–316. doi: 10.1002/eji.1830180221. [DOI] [PubMed] [Google Scholar]
- 32.Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med. 2007;357:1903–1915. doi: 10.1056/NEJMoa066092. [DOI] [PubMed] [Google Scholar]
- 33.Hammarlund E, et al. Duration of antiviral immunity after smallpox vaccination. Nat Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 34.Crotty S, et al. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J Immunol. 2003;171:4969–4973. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- 35.Radbruch A, et al. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006;6:741–750. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- 36.Chu VT, et al. Eosinophils are required for the maintenance of plasma cells in the bone marrow. Nat Immunol. 2011;12:151–159. doi: 10.1038/ni.1981. [DOI] [PubMed] [Google Scholar]
- 37.Mohr E, et al. Dendritic cells and monocyte/macrophages that create the IL-6/APRIL-rich lymph node microenvironments where plasmablasts mature. J Immunol. 2009;182:2113–2123. doi: 10.4049/jimmunol.0802771. [DOI] [PubMed] [Google Scholar]
- 38.Huard B, et al. APRIL secreted by neutrophils binds to heparan sulfate proteoglycans to create plasma cell niches in human mucosa. J Clin Invest. 2008;118:2887–2895. doi: 10.1172/JCI33760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sciammas R, et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Shaffer AL, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manz RA, Hauser AE, Hiepe F, Radbruch A. Maintenance of serum antibody levels. Annu Rev Immunol. 2005;23:367–386. doi: 10.1146/annurev.immunol.23.021704.115723. [DOI] [PubMed] [Google Scholar]
- 42.Novak AJ, et al. Expression of BCMA, TACI, and BAFF-R in multiple myeloma: a mechanism for growth and survival. Blood. 2004;103:689–694. doi: 10.1182/blood-2003-06-2043. [DOI] [PubMed] [Google Scholar]
- 43.Tangye SG, Bryant VL, Cuss AK, Good KL. BAFF, APRIL and human B cell disorders. Semin Immunol. 2006;18:305–317. doi: 10.1016/j.smim.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Wuillème-Toumi S, et al. Mcl-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia. 2005;19:1248–1252. doi: 10.1038/sj.leu.2403784. [DOI] [PubMed] [Google Scholar]
- 45.van Vollenhoven RF, Kinnman N, Vincent E, Wax S, Bathon J. Atacicept in patients with rheumatoid arthritis and an inadequate response to methotrexate: results of a phase II, randomized, placebo-controlled trial. Arthritis Rheum. 2011;63:1782–1792. doi: 10.1002/art.30372. [DOI] [PubMed] [Google Scholar]
- 46.Genovese MC, Kinnman N, de La Bourdonnaye G, Pena Rossi C, Tak PP. Atacicept in patients with rheumatoid arthritis and an inadequate response to tumor necrosis factor antagonist therapy: results of a phase II, randomized, placebo-controlled, dose-finding trial. Arthritis Rheum. 2011;63:1793–1803. doi: 10.1002/art.30373. [DOI] [PubMed] [Google Scholar]
- 47.Kopf M, et al. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature. 1994;368:339–342. doi: 10.1038/368339a0. [DOI] [PubMed] [Google Scholar]
- 48.Borriello F, et al. B7–1 and B7–2 have overlapping, critical roles in immunoglobulin class switching and germinal center formation. Immunity. 1997;6:303–313. doi: 10.1016/s1074-7613(00)80333-7. [DOI] [PubMed] [Google Scholar]
- 49.Kitamura D, Roes J, Kuhn R, Rajewsky KA. B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 50.Janas ML, Hodgkin P, Hibbs M, Tarlinton D. Genetic evidence for Lyn as a negative regulator of IL-4 signaling. J Immunol. 1999;163:4192–4198. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.