Abstract
The activity of cyclin-dependent kinase 5 (Cdk5) depends on the association with one of its activators, p35 and p39, which are prominently expressed in the nervous system. Studies on the repertoire of protein substrates for Cdk5 have implicated the involvement of Cdk5 in neuronal migration and synaptic plasticity. Our recent analysis of the sequence of signal transducer and activator of transcription (STAT)3, a key transcription factor, reveals the presence of potential Cdk5 phosphorylation site. We report here that the Cdk5/p35 complex associates with STAT3 and phosphorylates STAT3 on the Ser-727 residue in vitro and in vivo. Intriguingly, whereas the Ser phosphorylation of STAT3 can be detected in embryonic and postnatal brain and muscle of wild-type mice, it is essentially absent from those of Cdk5-deficient embryos. In addition, treatment of cultured myotubes with neuregulin enhances the Ser phosphorylation of STAT3 and transcription of STAT3 target genes, such as c-fos and junB, in a Cdk5-dependent manner. Both the DNA-binding activity of STAT3 and the transcription of specific target genes, such as fibronectin, are reduced in Cdk5-deficient muscle. Taken together, these results reveal a physiological role of Cdk5 in regulating STAT3 phosphorylation and modulating its transcriptional activity.
Cyclin-dependent kinase 5 (Cdk5) has been implicated in various aspects of neural development (1). Distinct from other members of the Cdk family, Cdk5 is not involved in cell-cycle progression. The activation of Cdk5 does not require binding to cyclins, but rather, association with one of its regulatory subunits, p35 and p39 (2-4). The essentially exclusive expression of its regulators largely restricts the kinase activity of Cdk5 to the CNS (2-4). Recent studies, however, reveal the intriguing functions of Cdk5 outside the CNS, such as in muscle (5-8).
Analysis of the Cdk5 and p35 null mice shows that Cdk5 activity plays a pivotal role in neuronal migration, axon path-finding, and laminar organization in brain (9, 10). In addition to these studies, the identification of the repertoire of protein substrates for Cdk5 implicates its involvement in the regulation of neurite outgrowth, axon guidance, membrane transport, and synaptogenesis, as well as modulation of the efficacy of signaling pathways (1, 11, 12). We have previously identified ErbB receptors, which mediate actions of neuregulin (NRG), to be the substrates for Cdk5 (5). Inhibition of Cdk5 activity in myotube culture attenuates the NRG-stimulated ErbB receptor phosphorylation and down-regulates the synapse-specific gene expression (5). More recently, it was reported that transcription factors such as p53 and myocyte enhancer factor-2 can be phosphorylated and regulated by active Cdk5 (13, 14).
One family of transcription factor, signal transducer and activator of transcription (STAT), is known to play important physiological roles in embryogenesis, development, hematopoiesis, and immune response. STAT proteins are activated by cytokine receptors and growth factor receptors (15), as well as nonreceptor tyrosine kinases (16). Stimulation of the receptors leads to the Tyr phosphorylation and activation of Janus kinases that recruit cytoplasmic STAT proteins to the receptor complex and phosphorylate STATs on specific Tyr residues. Tyr-phosphorylated STATs then form homo- or heterodimers and translocate into the nucleus, where they interact with specific DNA-response elements and regulate the expression of target genes, including the activating protein 1 family of transcription factors such as c-fos and junB (16-18). In addition to Tyr phosphorylation, STAT proteins are also phosphorylated on Ser residue, in response to a variety of extracellular stimuli such as growth factors or environmental stress (17). The precise function of Ser phosphorylation on STAT proteins, however, is controversial. It has been suggested in some studies that Ser phosphorylation is required for STATs to achieve its maximal Tyr phosphorylation and transcriptional activity (19, 20), whereas in other studies, Ser phosphorylation has been shown to inhibit Tyr phosphorylation of STATs (21-23). In addition, it has also been reported that Ser phosphorylation has no effect on Tyr phosphorylation or DNA binding (24, 25).
The effect of Ser phosphorylation of STAT3 depends on the extracellular stimulus, cell type, and the activation status of the cell in question (26). It is noteworthy that the Ser-727 (S727) residue of STAT3 comprises a consensus phosphorylation site for mitogen-activated protein kinase (MAPK) and members of the Cdk family. Whereas extensive studies have shown that phosphorylation of STAT3 on S727 by MAPKs can regulate the activity of STAT3 (23, 27), the phosphorylation of STAT3 by Cdks has not been explored. In this study, we show that STAT3 is a substrate for Cdk5. The S727 phosphorylation of STAT3 is essentially absent in embryonic muscle and brain tissues of Cdk5 null mice, suggesting that Cdk5 is likely to be essential in modulating STAT3 phosphorylation in vivo. In addition, Cdk5 mediates NRG-stimulated Ser phosphorylation of STAT3 and gene transcription in cultured myotubes. More importantly, both the DNA-binding activity of STAT3 and specific target gene transcription are reduced in Cdk5-deficient muscle in vivo. Taken together, this report provides, to our knowledge, the first demonstration that Cdk5 plays an important physiological role in the regulation of STAT3 phosphorylation and transcriptional activity.
Materials and Methods
Plasmids and Abs. The GST-STAT3 fusion construct was generated by subcloning the transactivation domain of rat STAT3 (amino acids 660-770) into pGEX-6P-1 (Amersham Biosciences). STAT3 mutant (Ser-727 to Ala-727, STAT3M) was generated by PCR using complementary primers containing the mutations and was subcloned into pGEX-6P-1. Polyclonal Abs specific for Cdk5 (C8), p35 (C19), STAT1, and STAT3 were purchased from Santa Cruz Biotechnology. Abs specific for the phosphorylated forms of STAT3 were purchased from Cell Signaling Technology (Beverly, MA), phosphorylated Ser (P-Ser) STAT1 from Upstate Biotechnology (Lake Placid, NY), and mouse mAbs of STAT1 and STAT3 from Zymed and Transduction Laboratories (Lexington, KY), respectively. STAT3-inhibitory peptide, a STAT3 Src homology 2 domain-binding phosphopeptide that inhibits the formation of STAT dimer (28), was purchased from Calbiochem.
Animals, Cell Culture, and Transfection. Cdk5 and p35 knockout mice were kindly provided by A.B. Kulkarni, (National Institutes of Health, Bethesda), T. Curran, (St. Jude's Children's Research Hospital, Memphis, TN), and L.H. Tsai, (Harvard Medical School, Boston), respectively. Mice of different stages were collected and genotyped as described (9, 10). Primary muscle culture was prepared from wild-type and Cdk5 mutant embryos [embryonic day (E)18] as described (29).
Mouse C2C12 cells were maintained and differentiated as described (30). Differentiated C2C12 myotube culture was pretreated with roscovitine (Ros; 10 or 40 μM), AG490 (100 μM), or STAT3-inhibitory peptide (100 μM) before NRG treatment (3 nM). Primary muscle cultures were prepared from wild-type and Cdk5 mutant embryos and were treated with NRG for 30 min. COS-7 cells were cultured in DMEM (Invitrogen) supplemented with 10% heat-inactivated FBS plus antibiotics. COS-7 cells were transfected with p35, Cdk5, or STAT3 plasmids by using Lipofectamine plus (Invitrogen). C2C12 cells were transfected with dominant-negative (dn)Cdk5 construct by using SuperFect (Qiagen). Twenty-four hours after transfection, cells were switched to 2% horse serum for 2 days before the collection of cell lysates.
RNA Extraction and Northern Blot Analysis. Muscle tissues were obtained from the hind limbs of wild-type and Cdk5 mutant embryos (E18). Total RNAs of C2C12 myotubes and muscle tissues were prepared by guanidinium thiocyanate extraction and the RNeasy minikit (Qiagen, Valencia, CA), respectively. Northern blot analysis was performed as described (30).
Protein Extraction, Immunoprecipitation, and Western Blot Analysis.
COS-7 cells, brain, and muscle tissues were extracted in lysis buffer supplemented with various protease inhibitors, and Western blot analysis was performed as described (5). Nuclear and cytoplasmic fractions of C2C12 myotubes and knockout tissues were prepared by using the CelLytic NuCLEAR extraction kit (Sigma).
Fusion Protein and Pull-Down Assay. GST-p25, GST-Cdk5, and GST-STAT3 was expressed in the Escherichia coli (BL21 strain) and purified by using a glutathione-Sepharose 4B column following the instructions of the manufacturer (Amersham Biosciences). Pull-down studies were performed as described (31). For coimmunoprecipitation studies, 0.5-1 mg of lysates of COS-7 cells and C2C12 myotubes were incubated with the corresponding Ab (2 μg) at 4°C overnight and were then incubated with 40 μl of protein G-Sepharose at 4°C for 1 h. The samples were washed with lysis buffer and resuspended in SDS sample buffer. Immunoprecipitated proteins were detected by Western blot analysis.
In Vitro Phosphorylation Assay. A total of 200 ng of recombinant STAT3 and STAT3 mutant proteins was used as the substrates for reconstituted Cdk5/p35 or Cdk5/p25 in the in vitro assay. The kinase assay was performed at 30°C for 30 min in kinase buffer containing 1 μCi (1 Ci = 37 GBq) of [γ-32P] ATP as described (31). The phosphorylated proteins were separated by SDS/10% PAGE and were visualized by autoradiography.
DNA-Binding Assay. For protein-DNA-binding analysis, the nuclear and cytoplasmic extract of C2C12 myotubes or muscle was prepared as described (32), and incubated with 10 μg of sis-inducible elements agarose beads (Santa Cruz Biotechnology) for 30 min at room temperature. After incubation, the beads were washed extensively with lysis buffer. The sis-inducible elements bead-precipitated proteins were separated by SDS/PAGE and were then transferred to nitrocellulose membrane for immunoblotting with STAT3 Ab (Transduction Laboratories).
Promoter Luciferase Assay. Primary chick muscle cells were seeded in 24-well plates. Twenty-four hours after seeding, cells were transfected with 10 μg of plasmid DNA by using the calcium phosphate coprecipitation method. Transfection mixtures consisted of a mixture of pSTAT3-TA-Luc, pGL3-c-fos, or pGAS reporter together with p35 or the following cDNA constructs (dnCdk5, STAT3, or STAT3M) and β-galactosidase (βgal)pCMV constructs (Clontech). Luciferase assay was performed by using a kit purchased from Promega and β-gal activity was measured using a luminescent β-gal enzyme kit (Clontech). Luciferase activity was normalized against β-gal activity to correct for the variations in transfection efficiency.
Results
During our search for potential substrates that can be phosphorylated by Cdk5, we have identified the consensus sequence for Cdk5 phosphorylation, SPRT, in the region flanking S727 of STAT3. To examine whether S727 of STAT3 is a target for Cdk5, a fusion protein encoding the C-terminal transactivation domain of STAT3 was subjected to kinase assay by using reconstituted Cdk5/p35. We found that STAT3 was phosphorylated by Cdk5/p35 in vitro in a concentration-dependent manner (Fig. 1A). To examine whether Cdk5 phosphorylated STAT3 on S727, a STAT3 mutant (STAT3M) with the Ser residue substituted by Ala was generated and subjected to in vitro kinase assay. Unlike wild-type STAT3, STAT3M could not be phosphorylated by reconstituted Cdk5/p35 or Cdk5/p25 (Fig. 1B). To demonstrate that active Cdk5 phosphorylates STAT3 in mammalian cells, we examined the effect of overexpressing p35 on the phosphorylation of endogenous STAT3 in COS-7 cells. Overexpression of p35 alone in COS-7 cells was sufficient to enhance the Ser phosphorylation of endogenous STAT3, whereas cotransfection with dnCdk5 and p35 attenuated the P-Ser STAT3 (Fig. 1C). The effect of Cdk5/p35 on P-Ser STAT3 was specific, because it did not affect the Tyr phosphorylation of STAT3 on Y705 residue (Fig. 1C) nor Ser phosphorylation of STAT1 (data not shown). Similarly, when STAT3 and p35 or Cdk5/p35 were overexpressed in COS-7 cells, P-Ser STAT3 was significantly induced, whereas phosphorylated Tyr (P-Tyr) STAT3 was unaffected (Fig. 1C).
Fig. 1.
Phosphorylation of STAT3 by active Cdk5 in vitro and in vivo. (A) Fusion protein of STAT3 C-terminal fragment (amino acids 660-770) was phosphorylated by recombinant Cdk5/p35 in vitro. Cdk5/p35 recombinant protein (0-1 μg per reaction as indicated) was added to the reaction mix. (B) Single mutation of STAT3 at Ser-727 (S727A) completely abolished the in vitro phosphorylation of STAT3 by Cdk5/p35 and Cdk5/p25. STAT3, wild-type STAT3 fragment; STAT3M, S727A mutant. (C) COS-7 cells were transfected with p35, dnCdk5/p35 or Cdk5/p35 expression constructs with (Right) or without STAT3 (Left). The ratio of DNAs used for transfection was indicated at the left at the top. V, mock transfected control. Total cell lysates were subjected to Western blot analysis by using P-Ser and P-Tyr STAT3-specific Abs. Total STAT3 and p35 served as controls. (D) p25 binds STAT3 in vitro. STAT3 overexpressed in COS-7 cells was pulled down by purified GST-p25 protein (1-5 μg), but not GST (5 μg). A weak interaction between STAT3 and GST-Cdk5 (5 μg) was also detected. (E) The C-terminal fragment of STAT3 (amino acids 660-770, 5 μg) interacts with p35 overexpressed in COS-7 cells. STAT3 and STAT3M showed similar affinity in their binding to p35. (F) Cdk5/p35 protein complex in E18 brain lysate was pulled down by GST-STAT3. (G) Association of p35 with STAT3 in COS-7 cells. The lysate from p35-overexpressing COS-7 cells was immunoprecipitated with p35 Ab and immunoblotted with Abs specific for STAT3 (Upper) or p35 (Lower). The rabbit normal IgG was used as a negative control. (H) Association of p35 with STAT3 in C2C12 myotubes and cortical neurons cultured for 21 days. Lysates of C2C12 myotubes or cortical neurons were immunoprecipitated by using p35 Ab and were immunoblotted with STAT3 Ab.
To further verify whether STAT3 binds to p35 or Cdk5, an in vitro pull-down assay was performed. Cell lysate of COS-7 cells overexpressing STAT3 was incubated with GST, GST-p25, or GST-Cdk5. We found that GST-p25 interacted with STAT3 in a dosage-dependent manner, whereas weak association between GST-Cdk5 and STAT3 was also detected (Fig. 1D). Similar GST pull-down experiments using GST-STAT3 demonstrated that the C-terminal transactivation domain of STAT3 could bind to p35 overexpressed in COS-7 cells (Fig. 1E). Like wild-type STAT3, STAT3M also interacted with p35, suggesting that the association of STAT3 with p35 did not depend on the Ser phosphorylation. In addition, the interaction of STAT3 with both p35 and Cdk5 in brain lysate was detected. Because p35 normally exists in a complex with Cdk5 in vivo, this finding suggests that STAT3 associates with the Cdk5/p35 protein complex in vivo (Fig. 1F and ref. 33). The interaction of STAT3 and Cdk5/p35 complex was further confirmed by coimmunoprecipitation analysis. Interaction between p35 and STAT3 was observed in COS-7 cells overexpressing p35 and STAT3, cultured C2C12 myotubes, and primary cortical neurons (Fig. 1 G and H). These results demonstrated that STAT3 interacts with Cdk5/p35 in vitro as well as in vivo.
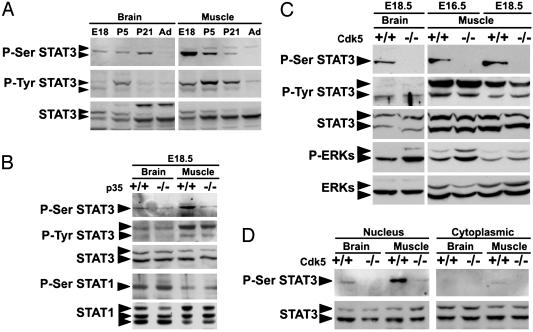
The temporal profile of STAT3 expression in brain and muscle was examined. Whereas STAT3 protein was expressed in rat brain and muscle throughout development, the phosphorylation of STAT3 on S727 and Y705 was predominantly detected in embryonic and postnatal brain and muscle (Fig. 2A). To further examine the regulation of Ser phosphorylation of STAT3 by Cdk5 in vivo, Western blot analysis of brain and muscle lysates prepared from Cdk5 and p35 null mice was performed. Intriguingly, whereas the Ser phosphorylation of STAT3 could be detected in E18.5 brain and muscle of wild-type mice, it was attenuated in p35 null mice (Fig. 2B). The P-Ser STAT1 remained unchanged in p35-deficient muscle, suggesting that the phosphorylation of STAT3 at S727 by Cdk5 in vivo is specific. Similar to the observation with p35 mutant embryos, P-Ser STAT3 was essentially absent in the brain and muscle of Cdk5-deficient embryos (Fig. 2C). In contrast, phosphorylated extracellular-regulated kinases (P-ERKs) were not down-regulated in brain or muscle lacking Cdk5, suggesting that the effect of Cdk5 on STAT3 in vivo did not depend on ERK activation. The increase of P-ERKs observed in Cdk5 mutant embryos is consistent with a previous report (34) that phosphorylation of MEK1 by Cdk5 reduced its activity. Similar to the observation in p35 mutant embryos, P-Ser STAT1 remained unchanged in Cdk5-deficient muscle (data not shown). The subcellular distribution of phosphorylated and total STAT proteins in brain and muscle of E18.5 wild-type and Cdk5 mutant mice was examined. Whereas P-Ser STAT3 was predominantly concentrated in the nuclear fraction of wild-type brain and muscle, it was drastically reduced in the nuclear fraction of Cdk5-deficient tissues at E18.5 (Fig. 2D).
Fig. 2.
Attenuation of Ser phosphorylation of STAT3 in brain and muscle of mice lacking Cdk5 activity. (A) Developmental expression of phosphorylated and total STAT3 in rat brain and muscle. Western blot analysis of P-Ser STAT3, P-Tyr STAT3, and total STAT3 in rat brain and muscle are shown. P5, postnatal day 5; Ad, adult. (B) Western blot analysis of P-Ser and P-Tyr STAT3 and total STAT3 in brain and muscle of p35 wild-type (+/+) and mutant (-/-) embryos at E18.5. P-Ser STAT1, and total STAT1 served as control. (C) Western blot analysis of P-Ser STAT3, P-Tyr STAT3, and P-ERKs in brain and muscle of wild-type (+/+) and Cdk5 mutant (-/-) embryos at E16.5 and E18.5. Total STAT3 and ERK proteins served as control. (D) P-Ser and total STAT3 in the nuclear and cytoplasmic fractions in brain and muscle of wild-type (+/+) and Cdk5 mutant (-/-) embryos at E18.5.
To further understand the role of Cdk5 in regulating the Ser phosphorylation of STAT3 in muscle, we examined whether NRG treatment of C2C12 myotubes could stimulate P-Ser STAT3 and whether Cdk5 could be involved in such regulation. NRG is a nerve-derived factor that regulates the transcription of a subset of synapse-specific genes in neurons or muscle (35). Consistent with previous report (36), incubation of C2C12 myotubes with NRG induced the mRNA expression of c-fos, a STAT3-responsive gene (Fig. 3A). Blockade of Cdk5 activity by a specific Cdk5 inhibitor, Ros (5, 37), or of STAT3 activity by specific STAT3-inhibitory peptide or Janus kinase 2 inhibitor AG490 attenuated the mRNA expression of c-fos in NRG-treated myotubes (Fig. 3A). This finding suggested that NRG-induced transcription of c-fos depends on the activation of Cdk5 and STAT3. The addition of NRG to cultured C2C12 myotubes increased the phosphorylation of STAT3 on S727 residue within 5 min, which was sustained up to at least 30 min (Fig. 3B). Whereas P-Tyr STAT3 was not affected by NRG treatment, stimulation of ERK phosphorylation was observed (Fig. 3B). To investigate whether the NRG-mediated STAT3 phosphorylation depends on Cdk5 activity, we examined the effect of Ros on the NRG-induced Ser phosphorylation of STAT3. We have previously demonstrated that the activities of Cdks such as Cdk2 are very low in myotubes and that the inhibition of Cdk5 kinase activity by Ros treatment in myotubes is specific (5). Preincubation of C2C12 myotubes with Ros abolished the NRG-induced Ser phosphorylation of STAT3 (Fig. 3C). A similar inhibitory effect on NRG-induced Ser phosphorylation of STAT3 was observed when the dnCdk5 expression construct was transiently transfected into C2C12 myotubes (Fig. 3D). The enhanced Ser phosphorylation of STAT3 was detected in both nuclear and cytoplasmic fractions of C2C12 myotubes after NRG treatment (Fig. 3E). Preincubation with Ros inhibited P-Ser STAT3, but not P-ERKs (Fig. 3E), suggesting that the Ser phosphorylation of STAT3 in NRG-treated C2C12 myotubes was mediated by Cdk5. Moreover, the expression of p35 in the nuclear fraction of C2C12 myotubes was increased after 30 min of NRG treatment, whereas Cdk5 protein expression remained unchanged (Fig. 3E).
Fig. 3.
Inhibition of Cdk5 activity abolished the NRG-induced Ser phosphorylation of STAT3. (A) Blockade of STAT3 activity and Cdk5 activity attenuated the NRG-induced c-fos transcription. Northern blot analysis of c-fos transcript after treatment of C2C12 myotubes with NRG (+) for 30 min. The myotubes were pretreated with different inhibitors: 40 μM Ros, 100 μM AG490 for 4 h, or 100 μM STAT3-inhibitory peptide (STAT3 peptide) for 24 h as indicated. (B) Cultured C2C12 myotubes were treated with NRG for 0-30 min. Total cell lysates were subjected to Western blot analysis using Abs specific for P-Ser STAT3, P-Tyr STAT3, and total STAT3 (Upper). Induction of P-ERKs served as positive control for NRG treatment (Lower). (C) Inhibition of Cdk5 activity with Ros abolished the NRG-induced P-Ser STAT3. -, untreated; +, NRG-treated for 15 min. DMSO served as control. (D) Expression of dnCdk5 in C2C12 myotubes attenuated the NRG-induced P-Ser STAT3. (E) Cultured C2C12 myotubes were treated with NRG for 0-30 min with or without Ros pretreatment. Lysates of nuclear (Left) or cytoplasmic (Right) fractions were prepared and were immunoblotted with specific Abs as depicted.
To investigate whether active Cdk5 regulates the transcriptional activity of STAT3, the DNA-binding activity of STAT3 in NRG-treated myotubes was examined in the presence or absence of Ros. We found that the addition of NRG to C2C12 myotubes increased STAT3 DNA-binding activity in the nuclear extract, and such an increase could be inhibited by Ros (Fig. 4A). To confirm whether Cdk5 is involved in regulating NRG-induced gene transcription, we examined the ability of NRG to induce the transcription of immediate early genes in Cdk5-deficient muscle. Primary myotube cultures were prepared from wild-type and Cdk5-deficient embryos (E18) and treated with NRG. The mRNA expression of c-fos and junB was increased in muscle culture prepared from wild-type embryos after NRG treatment. These two immediate early genes are well known targets for STAT3 in other cellular contexts, and the transcription of c-fos and junB was reported to be induced in myotube culture after NRG treatment (17, 18, 36). The NRG-induced increase of these two transcripts was attenuated in cultured myotubes isolated from Cdk5-deficient embryos (Fig. 4B).
Fig. 4.
Cdk5 regulated the STAT3 DNA-binding and transcriptional activity in vitro and in vivo. (A) Blockade of Cdk5 activity by Ros inhibited the NRG-induced increase in DNA binding of STAT3 in C2C12 myotubes. (Lower) Total STAT3. (Right) DNA-binding activity of STAT3 α and β was quantitated and expressed as a ratio of that in control myotubes pretreated with DMSO. Each value represented the mean ± SEM. of three representative experiments; *, P < 0.005. (B) Northern blot analysis of c-fos and junB in primary muscle cultures prepared from E18.5 wild-type (+/+) or Cdk5 null (-/-) embryos with (+) or without (-) NRG treatment. (Upper) c-fos. (Lower) junB. (C) DNA binding of STAT3 in E18.5 Cdk5 wild-type (+/+) and mutant (-/-) muscle (Left) and quantitation (Right). DNA-binding activity of STAT3 α and β was quantitated and expressed as a ratio of that in wild-type muscle. Each value represented the mean ± SEM. of three representative experiments; *, P < 0.005. (D) Northern blot analysis of fibronectin and muscle creatine kinase (MCK) in wild-type (+/+) and Cdk5-deficient (-/-) muscle of E18.5 embryos. MCK served as an equal loading control. (E) Overexpression of p35 increased the transcriptional activity of STAT3 in primary chick myotubes in a dose-dependent manner. Primary chick myotubes were transiently transfected with a STAT3 (pSTAT3-TA-Luc) or STAT1 (pGAS-TA-Luc) reporter gene construct and an internal control plasmid (β-gal-pCMV) with or without p35 or dnCdk5. The ratio of DNAs used for transfection was as depicted on the x axis. Luciferase activity was measured and normalized against the β-gal activity in the samples. Promoter activity was expressed as the ratio of luciferase activity in cells transfected with different combinations of p35 and dnCdk5 relative to that transfected with the empty vector. The data represented the mean ± SEM, n = 5. (F) Overexpression of p35 increased the transcriptional activity of c-fos. c-fos promoter-luciferase assay was performed as described in E with pSTAT3-TA-Luc (pSTAT3) as a positive control. Preincubation with Ros reduced Cdk5/p35-mediated transcriptional activity of pSTAT3 or c-fos. Overexpression of p35 could not increase the luciferase reporter activity under the control of STAT1 enhancer-responsive element (pGAS). The data represented mean ± SEM, n = 3. (G) The increase of STAT3 transcriptional activity depends on Ser-727 phosphorylation of STAT3. Primary muscle cultures were transfected with indicated expression constructs and luciferase assay was performed as in E; mean ± SEM, n = 3.
The STAT3 DNA-binding activity in vivo was also reduced in the nuclear extract of E18.5 Cdk5-deficient muscle when compared with that of wild type (≈40%; Fig. 4C). We have examined the transcription of a STAT3 target gene, fibronectin, in Cdk5-deficient muscle (38). It is noteworthy that fibronectin could be up-regulated by NRG in cultured myotubes (30). We found that the in vivo mRNA expression of fibronectin was reduced in E18 Cdk5-deficient muscle compared with that of wild type (Fig. 4D). In addition to modulating the transcriptional activity of STAT3 in NRG signaling, enhanced Cdk5 activity could increase the transcriptional activity of STAT3. Overexpression of p35 in primary chick muscle culture increased STAT3 transcriptional activity in a dose-dependent manner (Fig. 4E). When dnCdk5 was cotransfected with p35, the p35-increased STAT3 promoter activity was inhibited (Fig. 4E). On the other hand, Cdk5 did not regulate the STAT1-enhancer response element, pGAS-reporter activity, suggesting that the ability of Cdk5 to regulate the transcriptional activity of STAT3 was specific (Fig. 4F). Active Cdk5 also enhanced the transcriptional activity of a STAT3-regulated gene, c-fos (Fig. 4F). Treatment of cultured myotubes with Ros abolished the induction of STAT3 transcriptional activity by Cdk5/p35 (Fig. 4F). To further examine whether the ability of Cdk5/p35 to increase STAT3 transcriptional activity depended on S727 phosphorylation of STAT3, STAT3 or STAT3M was cotransfected with p35 and STAT3 promoter-reporter into primary muscle culture. The induction of STAT3 transcriptional activity by Cdk5/p35 was not observed in the presence of STAT3M (Fig. 4G).
Discussion
In the present study, we provide evidence that STAT3, a key transcription factor, is a substrate of Cdk5/p35. Our findings demonstrate that STAT3 is phosphorylated by active Cdk5 on a Ser residue, S727, both in vitro and in COS-7 cells overexpressing p35. This proline-directed Ser phosphorylation site comprises the consensus sequence for Cdk5 phosphorylation. The Ser phosphorylation of STAT3 in COS-7 cells is attenuated upon inhibition of Cdk5 activity. Furthermore, we found that incubation of C2C12 cells with NRG, previously shown to elevate Cdk5 activity (5), enhances the Ser phosphorylation of STAT3 in a Cdk5-dependent manner. Affinity pull-down assay and coimmunoprecipitation analysis reveal high-affinity association between STAT3 and Cdk5/p35. More importantly, the Ser phosphorylation of STAT3 can be detected in embryonic and postnatal brain and muscle of wild-type mice, but is essentially absent from those of Cdk5-deficient mice. Together with the observations that both DNA binding and transcriptional activity of STAT3 is reduced in myotube culture upon Cdk5 inhibition and also in Cdk5-deficient muscle, our results reveal a function of Cdk5 in the regulation of STAT3 in brain and muscle during development.
The regulation of STAT3 by Cdk5 appears to be highly specific. Although STAT1, STAT3, and STAT4 share a conserved stretch of amino acids (LPMSP) containing the proline-directed Ser residue (39), we found that the phosphorylation of STAT1 is not markedly reduced in Cdk5- or p35-deficient mouse brain and muscle. Whereas MAP kinase (ERK) has previously been reported to regulate the Ser phosphorylation of STAT3 in a variety of cellular systems (40), the endogenous ERK phosphorylation remains high in the Cdk5- and p35-deficient brain and muscle (this study and ref. 34). Thus, it is unlikely that the Ser phosphorylation of STAT3 observed in embryonic and postnatal mouse brain and muscle is mediated by ERKs. Similarly, the NRG-stimulated STAT3 Ser phosphorylation cannot be attributed to ERKs. Whereas preincubation of the NRG-treated myotube culture with Ros inhibits the increase in Ser phosphorylation of STAT3, ERK phosphorylation is not reduced. In addition to ERKs, p38 has been suggested to phosphorylate STAT3 on specific Ser residues (41). However, it has been reported that NRG treatment does not increase p38 activity in cultured myotubes (36). Our finding on the ability of Cdk5 to mediate the NRG-induced STAT3 Ser phosphorylation and specific endogenous genes provides a mechanism for NRG to regulate gene transcription in muscle. Recent studies from our laboratory indicate that NRG can also induce Ser phosphorylation of STAT3 in cultured cortical neurons (A.K.Y.F., Y.-P.N., and N.Y.I., unpublished observation). Work is in progress to investigate whether Cdk5 similarly mediate the NRG-induced STAT3 Ser phosphorylation in the CNS.
Both Ser and Tyr phosphorylation of STAT3 have been suggested to participate in modulating its transcriptional activity (42). Whereas the STAT transcriptional activity depends on Tyr phosphorylation, Ser phosphorylation appears to play a more subtle and supplementary role. The Ser phosphorylation of STAT3 by Cdk5 might affect STAT3 signaling by altering its Tyr phosphorylation, DNA binding, subcellular localization or finally, transcriptional activity (19, 20, 42). The modulatory effects of P-Ser STAT3 on gene transcription may also be through its effect on the interaction of STAT3 with coactivators such as p300 (20). In addition to regulating gene transcription, STAT3 has been suggested to operate as an adaptor protein to couple PI3K to cytokine receptors (43). In a similar fashion, through STAT3 Ser phosphorylation and specific association between STAT3 and Cdk5/p35 complex, the transcription factor may be shuttled between the membrane and nuclear fraction to modulate other protein-protein interactions. Whereas Ser phosphorylation of STATs has been extensively studied, precisely how the protein phosphorylation exerts its regulatory action on the transcriptional activity of STAT3 is still controversial (19, 20). The concentration of P-Ser STAT3 in the nucleus in vivo suggests the importance of Ser phosphorylation of STAT3 in nuclear functions. However, it is noteworthy that the subcellular distribution of P-Ser STAT3 varies depending on different cellular contexts; i.e., whereas comparable distribution of P-Ser STAT3 is found in cytosol and nucleus of C2C12 myotubes, P-Ser STAT3 is concentrated in the cytosol compartment in PC12 cells (A.K.Y.F., Y.-P.N., and N.Y.I., unpublished data).
Consistent with our finding on the ability of Cdk5 to modulate the transcriptional activity of STAT3, we provide evidence that both STAT3 DNA binding activity and transcription of STAT3 target genes, such as fibronectin, are attenuated in muscle of Cdk5-deficient embryos. Fibronectin has been reported to be prominently expressed in embryonic myotubes and is involved in cellular adhesion (44). The precise functional role of Cdk5-mediated fibronectin expression during the development of the neuromuscular junction, however, remains to be elucidated. On the other hand, microarray analysis of the gene expression profiling in Cdk5 mutant muscle also reveals the down-regulation of a number of transcripts, including potential STAT3 target genes (A.K.Y.F. and N.Y.I., unpublished observations). It was recently reported that the increase in Cdk5 kinase activity in response to neuronal insults regulates specific transcription factors such as p53 and myocyte enhancer factor-2 during neuronal apoptosis (13, 14). These results suggest that some of the genes that are down-regulated in the Cdk5-deficient muscle may be regulatory targets of transcription factors other than STAT3. Furthermore, the target genes for STAT3 vary among different cellular systems. For example, it has been reported that STAT3 may be involved in regulating acute-phase response gene in liver cells but modulating the transcription of a different set of genes in neurons (24). Future work is warranted to further identify and characterize the STAT3 target genes that are regulated by Cdk5 activity in brain and muscle.
Acknowledgments
We thank Drs. Ashok Kulkarni and Tom Curran for the Cdk5 null mice; Dr. Li-Huei Tsai for the p35 null mice; Dr. Zilong Wen for the STAT3 constructs; Dr. Shin-ichi Hisanaga for the recombinant Cdk5/p35 protein; Dr. Zhenguo Wu for helpful discussions; and Dr. Fanny C. F. Ip, Ms. Pu Shan Au, Ms. Hiu Yin Choi, and Ms. Janet Cheung for excellent technical assistance. This work was supported by Research Grants Council of Hong Kong Grants HKUST6091/01M, HKUST 6103/00M, and HKUST 2/99C, and the Area of Excellence Scheme Grant AoE/B-15/01. N.Y.I. was a recipient of the Croucher Foundation Senior Research Fellowship.
Abbreviations: Cdk5, cyclin-dependent kinase 5; dn, dominant-negative; NRG, neuregulin; STAT, signal transducer and activator of transcription; MAPK, mitogen-activated protein kinase; En, embryonic day n; β-gal, β-galactosidase; ERK, extracellular-regulated kinase; P-ERK, phosphorylated ERK; P-Ser, phosphorylated Ser; P-Tyr, phosphorylated Tyr; Ros, roscovitine.
References
- 1.Dhavan, R. & Tsai, L.-H. (2001) Nat. Rev. Mol. Cell Biol. 2, 749-759. [DOI] [PubMed] [Google Scholar]
- 2.Lew, J., Huang, Q. Q., Qi, Z., Winkfein, R. J., Aebersold, R., Hunt, T. & Wang, J. H. (1994) Nature 371, 423-426. [DOI] [PubMed] [Google Scholar]
- 3.Tsai, L.-H., Delalle, I., Caviness, V. S., Jr., Chae, T. & Harlow, E. (1994) Nature 371, 419-423. [DOI] [PubMed] [Google Scholar]
- 4.Tang, D., Yeung, J., Lee, K. Y., Matsushita, M., Matsui, H., Tomizawa, K., Hatase, O. & Wang, J. H. (1995) J. Biol. Chem. 270, 26897-26903. [DOI] [PubMed] [Google Scholar]
- 5.Fu, A. K., Fu, W.-Y., Cheung, J., Tsim, K. W., Ip, F. C., Wang, J. H. & Ip, N. Y. (2001) Nat. Neurosci. 4, 374-381. [DOI] [PubMed] [Google Scholar]
- 6.Lilja, L., Yang, S. N., Webb, D. L., Juntti-Berggren, L., Berggren, P. O. & Bark, C. (2001) J. Biol. Chem. 276, 34199-34205. [DOI] [PubMed] [Google Scholar]
- 7.Philpott, A., Porro, E. B., Kirschner, M. W. & Tsai, L.-H. (1997) Genes Dev. 11, 1409-1421. [DOI] [PubMed] [Google Scholar]
- 8.Lazaro, J. B., Kitzmann, M., Poul, M. A., Vandromme, M., Lamb, N. J. & Fernandez, A. (1997) J. Cell Sci. 110, 1251-1260. [DOI] [PubMed] [Google Scholar]
- 9.Ohshima, T., Ward, J. M., Huh, C. G., Longenecker, G., Veeranna, Pant, H. C., Brady, R. O., Martin, L. J. & Kulkarni, A. B. (1996) Proc. Natl. Acad. Sci. USA 93, 11173-11178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chae, T., Kwon, Y. T., Bronson, R., Dikkes, P., Li, E. & Tsai, L.-H. (1997) Neuron 18, 29-42. [DOI] [PubMed] [Google Scholar]
- 11.Lai, K. O. & Ip, N. Y. (2003) Trends Genet. 19, 395-402. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, K. & Ip, N. Y. NeuroSignals 12, 180-190.14673204
- 13.Zhang, J., Krishnamurthy, P. K. & Johnson, G. V. (2002) J. Neurochem. 81, 307-313. [DOI] [PubMed] [Google Scholar]
- 14.Gong, X., Tang, X., Wiedmann, M., Wang, X., Peng, J., Zheng, D., Blair, L. A., Marshall, J. & Mao, Z. (2003) Neuron 38, 33-46. [DOI] [PubMed] [Google Scholar]
- 15.Vignais, M. L., Sadowski, H. B., Watling, D., Rogers, N. C. & Gilman, M. (1996) Mol. Cell. Biol. 16, 1759-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy, D. E. & Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651-662. [DOI] [PubMed] [Google Scholar]
- 17.Coffer, P., Lutticken, C., van Puijenbroek, A., Klop-de Jonge, M., Horn, F. & Kruijer, W. (1995) Oncogene 10, 985-994. [PubMed] [Google Scholar]
- 18.Jenab, S. & Morris, P. L. (1996) Endocrinology 137, 4638-4743. [DOI] [PubMed] [Google Scholar]
- 19.Wen, Z., Zhong, Z. & Darnell, J. E., Jr. (1995) Cell 82, 241-250. [DOI] [PubMed] [Google Scholar]
- 20.Schuringa, J. J., Schepers, H., Vellenga, E. & Kruijer, W. (2001) FEBS Lett. 495, 71-76. [DOI] [PubMed] [Google Scholar]
- 21.Chung, J., Uchida, E., Grammer, T. C. & Blenis, J. (1997) Mol. Cell. Biol. 17, 6508-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain, N., Zhang, T., Fong, S. L., Lim, C. P. & Cao, X. (1998) Oncogene 17, 3157-3167. [DOI] [PubMed] [Google Scholar]
- 23.Lim, C. P. & Cao, X. (1999) J. Biol. Chem. 274, 31055-31061. [DOI] [PubMed] [Google Scholar]
- 24.Boulton, T. G., Zhong, Z., Wen, Z., Darnell, J. E., Jr., Stahl, N. & Yancopoulos, G. D. (1995) Proc. Natl. Acad. Sci. USA 92, 6915-6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng, J. & Cantrell, D. (1997) J. Biol. Chem. 272, 24542-24549. [DOI] [PubMed] [Google Scholar]
- 26.Decker, T. & Kovarik, P. (2000) Oncogene 19, 2628-2637. [DOI] [PubMed] [Google Scholar]
- 27.Lim, C. P. & Cao, X. (2001) J. Biol. Chem. 276, 21004-21011. [DOI] [PubMed] [Google Scholar]
- 28.Turkson, J., Ryan, D., Kim, J. S., Zhang, Y., Chen, Z., Haura, E., Laudano, A., Sebti, S., Hamilton, A. D. & Jove, R. (2001) J. Biol. Chem. 276, 45443-45455. [DOI] [PubMed] [Google Scholar]
- 29.Ip, F. C., Glass, D. G., Gies, D. R., Cheung, J., Lai, K. O., Fu, A. K., Yancopoulos, G. D. & Ip, N. Y. (2000) Mol. Cell. Neurosci. 16, 661-673. [DOI] [PubMed] [Google Scholar]
- 30.Fu, A. K., Cheung, W. M., Ip, F. C. & Ip, N. Y. (1999) Mol. Cell. Neurosci. 14, 241-253. [DOI] [PubMed] [Google Scholar]
- 31.Cheng, K., Li, Z., Fu, W.-Y., Wang, J. H., Fu, A. K. & Ip, N. Y. (2002) J. Biol. Chem. 277, 31988-31993. [DOI] [PubMed] [Google Scholar]
- 32.Chan, J. K., Sun, L., Yang, X. J., Zhu, G. & Wu, Z. (2003) J. Biol. Chem. 278, 23515-23521. [DOI] [PubMed] [Google Scholar]
- 33.Lee, K. Y., Rosales, J. L., Tang, D. & Wang, J. H. (1996) J. Biol. Chem. 271, 1538-1543. [DOI] [PubMed] [Google Scholar]
- 34.Sharma, P., Veeranna Sharma, M., Amin, N. D., Sihag, R. K., Grant, P., Ahn, N., Kulkarni, A. B. & Pant, H. C. (2002) J. Biol. Chem. 277, 528-534. [DOI] [PubMed] [Google Scholar]
- 35.Buonanno, A. & Fischbach, G. D. (2001) Curr. Opin. Neurobiol. 11, 287-296. [DOI] [PubMed] [Google Scholar]
- 36.Si, J., Wang, Q. & Mei, L. (1999) J. Neurosci. 19, 8498-8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meijer, L., Borgne, A., Mulner, O. Chong, J. P., Blow, J. J., Inagaki, N., Inagaki, M., Delcros, J. G. & Moulinoux, J. P. (1997) Eur. J. Biochem. 243, 527-536. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, F., Li, C., Halfter, H. & Liu, J. (2003) Oncogene 22, 894-905. [DOI] [PubMed] [Google Scholar]
- 39.Darnell, J. E., Jr. (1997) Science 277, 1630-1635. [DOI] [PubMed] [Google Scholar]
- 40.Kuroki, M. & O'Flaherty, J. T. (1999) Biochem. J. 341, 691-696. [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, B., Bhattacharjee, A., Roy, B., Xu, H. M., Anthony, D., Frank, D. A., Feldman, G. M. & Cathcart, M. K. (2003) Mol. Cell. Biol. 23, 3918-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aznar, S., Valeron, P. F., del Rincon, S. V., Perez, L. F., Perona, R. & Lacal, J. C. (2001) Mol. Biol. Cell 12, 3282-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeffer, L. M., Mullersman, J. E., Pfeffer, S. R., Murti, A., Shi, W. & Yang, C. H. (1997) Science 276, 1418-1420. [DOI] [PubMed] [Google Scholar]
- 44.Sanes, J. R., Schachner, M. & Covault, J. (1986) J. Cell Biol. 102, 420-431. [DOI] [PMC free article] [PubMed] [Google Scholar]