Summary
Anthrax toxin is an A/B bacterial protein toxin which is composed of the enzymatically active Lethal Factor (LF) and/or Oedema Factor (EF) bound to Protective Antigen 63 (PA63) which functions as both the receptor binding and transmembrane domains. Once the toxin binds to its cell surface receptors it is internalized into the cell and traffics through Rab5- and Rab7-associated endosomal vesicles. Following acidification of the vesicle lumen, PA63 undergoes a dynamic change forming a beta-barrel that inserts into and forms a pore through the endosomal membrane. It is widely recognized that LF, and the related fusion protein LFnDTA, must be completely denatured in order to transit through the PA63 formed pore and enter the eukaryotic cell cytosol. We demonstrate by protease protection assays that the molecular chaperone GRP78 mediates the unfolding of LFnDTA and LF at neutral pH and thereby converts these proteins from a trypsin resistant to sensitive conformation. We have used immuno-electron microscopy and gold-labeled antibodies to demonstrate that both GRP78 and GRP94 chaperones are present in the lumen of endosomal vesicles. Finally, we have used siRNA to demonstrate that knock down of GRP78 results in the emergence of resistance to anthrax lethal toxin and edema toxin action.
Keywords: unfoldase, anthrax toxin, siRNA, anthrax toxin entry
Introduction
Upon binding of Protective Antigen 83 (PA83) to its cell surfaces receptors, AntxR1 (tumor endothelial marker-8) and AntxR2 (capillary morphogenesis protein 2) (Bradley et al., 2001; Scobie et al., 2003), it is cleaved by the endoproteinase furin and oligomerizes into its heptameric prepore form PA63(7). Anthrax toxin is then assembled on the cell surface by the binding of up to three molecules of either Lethal Factor (LF) or Oedema Factor (EF), or any combination of the two (Pimental et al., 2004). Anthrax toxin is internalized into the cell in clathrin coated pits (Abrami et al., 2003) and enters an early endosomal vesicle compartment. Acidification of the vesicle lumen by vesicular (v)ATPase is known to trigger a dynamic change in heptameric PA63(7) leading toward the formation of a β-barrel that inserts into and forms a pore through the endosomal membrane (Miller et al., 1999; Finkelstein, 1994). While anthrax toxin is known to traffic through Rab5 positive early endosomes, optimal delivery of LF has been reported to be from a Rab7-associated late endosomal compartment (Abrami et al., 2004). At least the N-terminal portions of LF and EF bound by PA63(7) appear to be denatured in the low pH environment of the endosomal lumen and threaded into the PA63(7) pore (Krantz et al., 2004). Wesche et al. (1998) found that the introduction of unique disulfide bonds into LFnDTA, a fusion protein composed of the N-terminal 254 amino acids of LF genetically fused to the catalytic fragment of diphtheria toxin (DTA), prevented cytosolic delivery, thereby providing evidence that LFnDTA must completely unfold in order to pass through the PA63(7) trans-endosomal membrane pore.
Recently, we reported that both anthrax toxin and diphtheria toxin share a requirement for coatomer protein I (COPI) complex for efficient translocation of their respective catalytic domains from the lumen of toxin pre-loaded endosomal vesicles to the external milieu in vitro (Tamayo et al., 2008; Ratts et al., 2005). Since the presence of COPI complex was required but not by itself sufficient for in vitro LFnDTA translocation from the endosomal lumen, we reasoned that other cellular proteins were required for facilitated delivery through the PA63(7) transmembrane pore. In this report, we provide evidence to support a role for the endosomal vesicle associated chaperone GRP78 (BiP) in the delivery of LFnDTA, LF, and EF to the eukaryotic cell cytosol.
GRP78 and GRP94 are protein chaperones that are constitutively expressed in eukaryotic cells, and elevated levels of each protein has been observed in cells that are under stress (Kozutsumi et al., 1988; Zhang & Kaufman, 2004). Although each chaperones possess a KDEL endoplasmic reticulum (ER) retrieval signal accounting for the high abundance in this cellular compartment, the presence of GRP78 and GRP94 on the eukaryotic cell surface has been well documented (Takemoto et al., 1992; Davidson et al., 2005; Triantafilou & Triantafilou, 2003; Xiao et al., 1999; Gonzalez-Gronow et al., 2007). Furthermore, both chaperones have been shown to be internalized into endosomal vesicles (Kim et al., 2006; Arnold-Schild et al., 1999; Singh-Jasuja et al., 2000; Berwin et al., 2002; Wassenberg et al., 1999).
GRP78 and GRP94 are known to be part of a chaperone system that assists in the retrieval and degradation of mis-folded proteins by facilitating their retro-translocation through the Sec61 or Derlin-1 complex from the lumen of the ER back to the cytosol (Plemper et al., 1997; Nishikawa et al., 2001; Molinari et al., 2002; Hegde et al., 2006; Christianson et al., 2008; Wiertz et al., 1996; Wahlman et al., 2007). Using an in vitro protease protection assay, we show that both GRP78 and GRP94 have unfoldase activity and are able to convert the recombinant fusion protein LFnDTA from a trypsin resistant to trypsin sensitive conformation at neutral pH. In contrast, native LF appears to be unfolded to a trypsin sensitive conformation at neutral pH only by GRP78. In addition, we demonstrate by immuno-magnetic selection and immuno-electron microscopy that GRP78 and GRP94 are co-localized in the lumen of both Rab5 and Rab7 positive endosomes. Finally, we show in vivo that siRNA knock down of GRP78 in murine J774.A1 cells confers an anthrax toxin resistant phenotype, suggesting that GRP78 functions as a denaturing chaperone or unfoldase for LF and EF under mildly acidic conditions and permitting cytosolic delivery from the lumen of early endosomal vesicles. Based upon observations described in the present work and earlier findings, we discuss a working model of anthrax toxin entry into eukaryotic cell cytosol.
Results
We have previously demonstrated that the in vitro translocation of LFnDTA from the lumen of LFnDTA/PA63(7) pre-loaded endosomal vesicles to the external medium required the addition of ATP and COPI coatomer complex proteins to the reaction mixture (Tamayo et al., 2008). Since it was apparent that the process by which LFnDTA was translocated from the endosomal vesicle lumen to the cytosol was more complex than initially proposed (Young & Collier, 2007) and required the participation of eukaryotic cell factors, we initiated a detailed analysis of endosomal vesicle associated proteins by mass spectrometry sequencing. This analysis was conducted in anticipation of identifying additional cellular proteins that might be uniquely associated with LFnDTA/PA63(7) in toxin pre-loaded vesicles and play a direct role in the entry process. Both control and LFnDTA/PA63(7) pre-loaded vesicles were isolated by discontinuous sucrose gradient ultracentrifugation in the presence of the (v)ATPase inhibitor Bafilomycin A1 as previously described (Ratts et al., 2003). The partially purified anthrax toxin loaded endosomal vesicles were then further resolved by continuous sucrose velocity sedimentation ultracentrifugation. Individual fractions from these gradients were analyzed for ADP-ribosyltransferase activity, and by immunoblot using anti-Rab5 and anti-clathrin (heavy chain) antibodies as probes (Supplemental Material, Fig. S1). In addition, partially purified endosomal vesicles were characterized by immunoblot using antibodies directed toward early endosomal, Golgi, endoplasmic reticulum, and lysosomal markers as probes (Supplemental Material, Fig. S2).
The protein composition of these enriched endosomal vesicle populations were analyzed by electrophoresis in 4–20% denaturing SDS-polyacrylamide gels. As shown in Figure 1, silver stained gels revealed a complex mixture of proteins. No apparent differences were observed in the total protein banding patterns between native and LFnDTA/PA63 preloaded endosomal vesicles following silver staining (data not shown). Nonetheless, in order to further characterize proteins that were associated with these preparations of endosomal vesicles individual protein bands were excised from silver stained gels, digested in-gel with trypsin, and the resulting peptides were extracted and analyzed by MALDI-TOF/TOF mass spectrometry. A number of proteins known to be associated with endosomal vesicles (e.g., myoferlin [Bernatchez et al. 2009], transferrin receptor 1, NSF, and Rab6) were identified (Fig. 1). In addition, we identified the chaperones GRP78 and GRP94 in preparations of both native and toxin pre-loaded endosomal vesicles. The presence of the full length form of these two chaperones was then confirmed by immunoblot analysis using anti-GRP78 and anti-GRP94 antibodies (data not shown). GRP78 and GRP94 are chaperones that are well known to participate in the ERAD-ER mis-folded protein degradation response (Plemper et al., 1997; Nishikawa et al., 2001; Molinari et al., 2002; Hegde et al., 2006; Christianson et al., 2008; Wiertz et al., 1996; Wahlman et al., 2007). Since both GRP78 and GRP94 have been shown to facilitate the unfolding and retro-translocation of mis-folded proteins from the ER lumen through the Sec61 pore to the cytoplasm (Nishikawa et al., 2001; Molinari et al., 2002; Hegde et al., 2007; Christiansen et al., 2008), we reasoned that these two chaperones might function in a similar fashion and facilitate the translocation of LFnDTA and/or LF through the PA63(7) trans-endosomal membrane pore.
Figure 1.
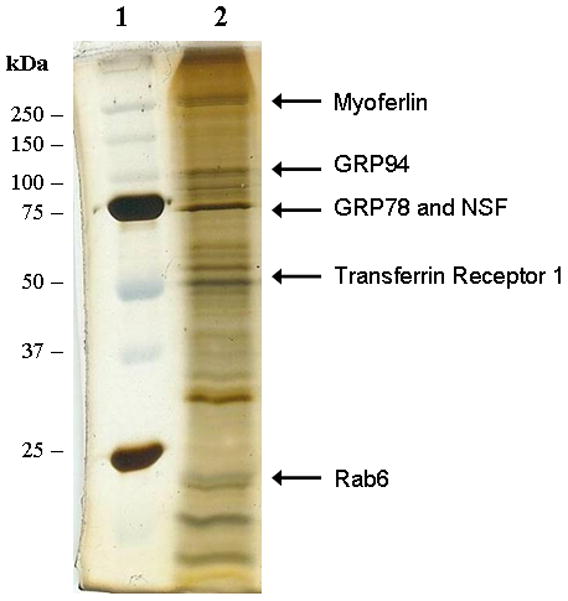
Silver stained SDS-polyacrylamide gel analysis of purified endosomal vesicles that were pre-loaded with LFnDTA/PA63(7). Following electrophoresis, individual protein bands were excised, protease digested in situ and identified by mass spectrometry. Lane 1: molecular weight markers; Lane 2: endosomal vesicle associated proteins.
We first asked whether either GRP78 or GRP94 might also serve as a protein isomerase or “unfoldase” to promote changes in tertiary structure of LFnDTA. Since it is well known that LFnDTA is highly resistant to proteolytic degradation at neutral pH, this recombinant protein was pre-incubated in the absence or presence of either GRP78 or GRP94 for 30 min at 37°C at pH 7.6. Trypsin was then added to the reaction mixture, which was then incubated for an additional 30 min at 37°C. At the end of the incubation period, the reaction was stopped by the addition of reducing SDS-polyacrylamide gel loading buffer and boiling for 5 min. Individual reaction mixtures were then analyzed by SDS-polyacrylamide gel electrophoresis and immunoblot using anti-DTA as the probe. As demonstrated in Figure 2, LFnDTA is completely resistant to trypsin digestion at pH 7.6. In marked contrast, preincubation with either GRP78 or GRP94 before the addition of trypsin to the reaction mixture converts LFnDTA to a more highly protease sensitive conformation. When protease protection assays were conducted at pH 6.5 and 5.5, conditions that mimic early and late acidified endosomal compartments, LFnDTA becomes increasingly sensitive to trypsin attack. Preincubation of LFnDTA with either GRP78 or GRP94 under these conditions further enhances its relative sensitivity to trypsin degradation (Fig. 3). Taken together, these experiments strongly suggest that both GRP78 and GRP94 are able to effect the unfolding of LFnDTA in vitro. In contrast, co-incubation of PA63 with either GRP78 or GRP94 at neutral pH, followed by the addition of trypsin to the reaction mixture, did not result in the conversion of PA63 from a trypsin resistant to sensitive conformation (Supplemental Material, Fig. S3).
Figure 2.
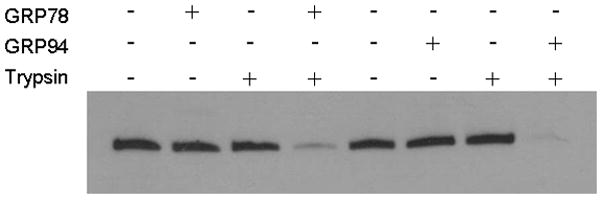
Incubation of LFnDTA with either GRP78 or GRP94 at pH 7.6 results in the conversion of LFnDTA from a trypsin resistant to sensitive conformation in vitro. Following incubation, proteins were denatured in SDS-loading buffer, electrophoresed in SDS-polyacrylamide gels, and following transfer to nitrocellulose membranes blots were probed with anti-DTA antibodies.
Figure 3.
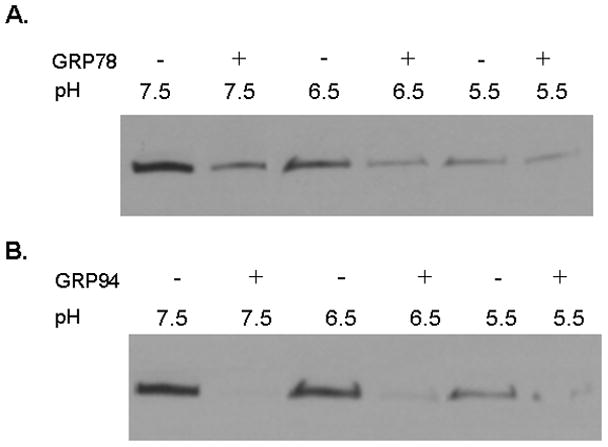
Immunoblot analysis of LFnDTA following incubation in the absence or presence of GRP78 or GRP94. (A) LFnDTA was incubated in buffer at pH 7.5, 6.5, or 5.5 in the absence or presence of GRP78 and then exposed to trypsin. (B) LFnDTA was incubated in buffer at pH 7.5, 6.5, and 5.5 in the absence or presence of GRP94 and then exposed to trypsin. Following the addition of trypsin to the reaction mixture, samples were denatured in SDS-loading buffer, electrophoresed in SDS-polyacrylamide gels, and following transfer to nitrocellulose blots were probed with anti-DTA antibodies.
In an attempt to rule out the possibility that the fusion protein LFnDTA might be recognized as a partially misfolded protein by GRP78 and GRP94, we next examined the effect of these chaperones on native LF. As shown in Figure 4, at pH 7.5, native LF is resistant to proteolytic degradation by trypsin; however, preincubation of LF with GRP78 prior to the addition of trypsin results in its conversion from a protease resistant to sensitive conformation. While native LF is not as sensitive to GRP78 mediated unfolding as LFnDTA, we consistently find that it is converted from a protease resistant to sensitive conformation (p = 0.0002). In marked contrast, however, preincubation of LF with GRP94 at neutral pH prior to the addition of trypsin did not result in its conversion to a trypsin sensitive conformation (data not shown).
Figure 4.
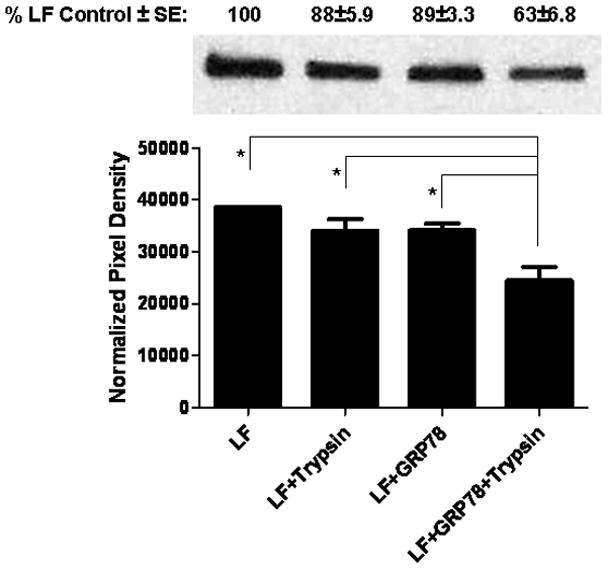
Incubation of recombinant LF with GRP78 at pH 7.5 results in its conversion from a trypsin resistant to sensitive conformation in vitro. LF (100 ng; List Biologicals) was incubated in the presence or absence of GRP78 (538 ng; Enzo Life Sciences) in unfolding buffer (25 mM Tris-HCl, 150 mM NaCl, 10 mM KCl, pH 7.5) for 10 min at 37°C, before the addition of trypsin (2.5 ng; Promega) or buffer alone and then incubated for 1 hr at 37°C. Digestion reactions were stopped by addition of 2X SDS-PAGE loading buffer with β-mercaptoethanol and boiled for 5 min prior to electrophoresis and detection on immuno-blots probed with anti-LF (Pierce Antibodies). Pixel density was assessed using Kodak 1D v.3.6.0 software. *Statistical analysis was performed using GraphPad Prism v.5.01. ANOVA followed by the Tukey Test revealed p = 0.0002 (n = 7).
The observed selectivity of GRP78 and GRP94 in the unfolding of LFnDTA and LF in in vitro protease digestion assays caused us to question whether these chaperones facilitate the process by which LFnDTA and LF entered the cytosol from the endosomal vesicle lumen. We first examined whether GRP78 and GRP94 could be detected in the lumen of partially purified endosomal vesicles. Following partial purification by sucrose density gradient ultracentrifugation, endosomal vesicles were further purified by immuno-affinity magnetic separation. In these experiments, we mixed anti-Rab5 antibodies with Protein A-conjugated 50nm magnetic nano-particles. The antibody adsorbed particles were washed and then incubated with LfnDTA/PA63 pre-loaded endosomal vesicles that were isolated by sucrose density centrifugation. Following incubation the mixture was placed in a magnetic field to immobilize the nano-particles and antibody bound target vesicles, the column was washed extensively, and the immuno-reactive Rab5 positive endosomes were then released and analyzed by SDS-polyacrylamide gel electrophoresis and immunoblot. As shown in Figure 5, the lysates from the immunoaffinity bead purified endosomes were found to contain Rab5, GRP78, and GRP94. These vesicles also were found to contain high levels of LFnDTA ADP-ribosyltransferase activity (data not shown).
Figure 5.
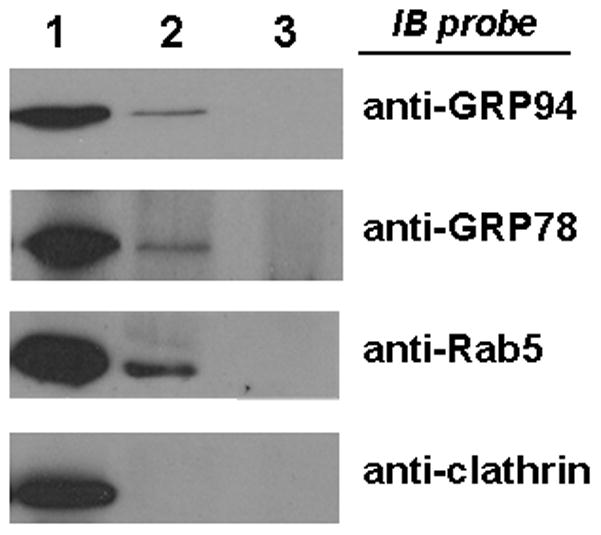
Anti-Rab5 immuno-affinity magnetic bead purified endosomal vesicles are immunoblot positive for GRP78 and GRP94, but not clathrin. Lane 1: partially purified endosomal vesicles; Lane 2: Rab5 immuno-affinity purified endosomal vesicles; Lane 3: isotype control antibody immuno-affinity purified endosomal vesicles.
We next analyzed these partially purified endosomal vesicles by cryo-electron microscopy and immuno-gold labeling. Endosomal vesicles were cut into ultra-thin frozen sections and then exposed to anti-GRP78 prior to incubation with 15nm gold-labeled Protein A. As anticipated we found GRP78 immunoreactivity to be located mostly within the vesicle lumen (Fig. 6; Supplemental Material, Fig. S4). We next probed thin sections with10nm gold-labeled anti-Rab7, and as anticipated this label was found to be associated with the cytosolic surface of the vesicle membranes. Control immuno-histochemical labeling experiments with an isotype matched IgG antibody failed to detect GRP78 (Supplemental Material, Fig. S4).
Figure 6.
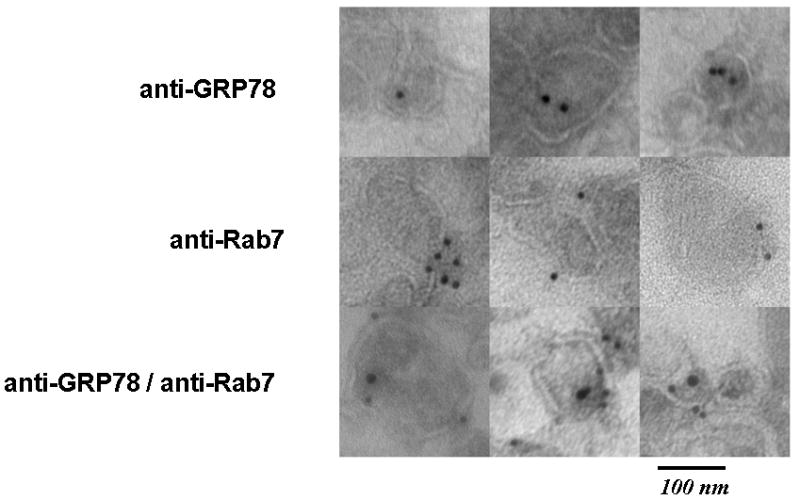
Immunohistochemical analysis of partially purified endosomal vesicles with 15nm gold-labeled anti-GRP78 and/or 10nm gold-labeled anti-Rab7 antibodies, followed by visualization by transmission electron microscopsy (TEM).
Since the observations from the in vitro experiments described above suggested that GRP78 was able to effect the unfolding of LFnDTA and LF, and that GRP78 was readily detected in the vesicle lumen by immuno-histochemical cryo-electron microscopic analysis, we next asked whether this chaperone plays a role in the in vivo process by which LF enters the eukaryotic cell cytosol. siRNA was used to knock down expression of GRP78 and GRP94 independently and together in mouse macrophage J774A.1 cells. Five days after the introduction of silencing siRNA to either chaperone, cells were challenged with lethal toxin and cell viability was measured. In these dose response experiments the concentration of PA was held constant (11 nM); whereas, the concentration of LF was varied between 0.01 – 1.0 nM. As shown in Figure 7A knock down of GRP78 resulted in an approximate 2-fold shift in dose response curve. The reduced level of GRP78 in siGRP78 treated J774A.1 cells was confirmed by immunoblot analysis using anti-GRP78 antibodies as the probe (Fig. 7B).
Figure 7.
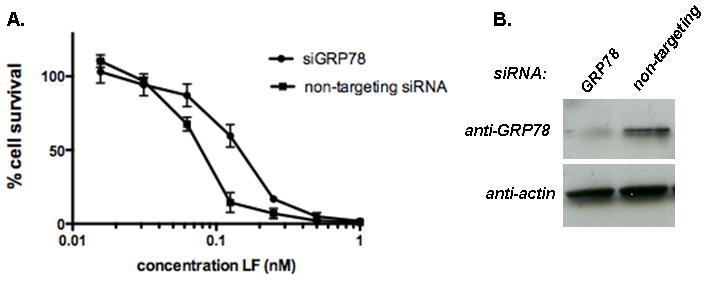
siRNA knock down of GRP78 expression in J774A.1 murine macrophage cells confers resistance against anthrax toxin challenge in vivo. (A) Dose response analysis of anthrax toxin challenge in J774A.1 cells following exposure to either non-targeting siRNA or siGRP78. In these experiments, cells were exposed to 11 nM PA and increasing concentrations of LF (as shown). (B) Immunoblot analysis of J774A.1 cell cytosol following exposure to non-targeting siRNA and siGRP78 using anti-GRP78 and anti-actin antibodies as probes. (N = 4)
To lend further support to a direct role of GRP78 in the entry of anthrax toxin into the cell, rather than a subsequent downstream process event related to the protease activity of LF, we tested the effect of GRP78 knock down in J774A.1 cells by challenge with anthrax edema toxin. After five days of GRP78 knock down, edema toxin was added to the cells and cAMP concentration in the cell lysate was measured by ELISA. We found that GRP78 knock down inhibited the increase in cAMP catalyzed by edema factor (Fig. 8), suggesting that GRP78 also facilitates entry of edema factor.
Figure 8.

siRNA knock down of GRP78 expression in J774A.1 murine macrophage cells confers resistance to anthrax edema toxin in vivo. Concentration of cAMP in cell lysates following exposure to non-targeting siRNA and siGRP78 and challenge with anthrax edema toxin (11 nM PA and 1.7 nM EF).
Since GRP78 also resides in the ER and participates in the secretory pathway, knock down of this chaperone potentially could affect the cycling of the anthrax toxin receptors to the cell surface. To determine if these receptors were still present and functional on the surface of J774A.1 cells after GRP78 knock down, we tested whether lethal toxin was able to kill cells when cytosolic delivery of LF occurred directly from the plasma membrane in a manner that bypasses the normal endocytic entry process. We tested whether lethal toxin was able to kill cells when cytosolic delivery of LF occurred directly from the plasma membrane after binding toxin to its receptors and exposing the cells to a low pH wash to trigger PA pore formation and LF entry.
Cells were incubated with lethal toxin at 4°C, washed, and then incubated at 37°C in pH 5.0, buffer for two minutes to allow PA pores to insert into the plasma membrane and cytosolic delivery of LF to occur. In addition, we blocked normal lethal toxin entry through the receptor-mediated endocytotic route by pretreating cell cultures with Bafilomycin A1, which inhibits endosomal acidification. We found that GRP78 knockdown did not affect the level of LF that was able to enter the cytosol from the plasma membrane surface and kill the cells (Fig. 9). In contrast, when cells were pulsed at pH 7.0 (which does not induce PA pore formation and thus cytosolic delivery of LF), rather than pH 5.0, or when cells were incubated with an antibody to ATXR2 (CMG2), there was minimal cell death. Finally, the presence of isotype matched control antibody in the culture medium failed to protect cells from lethal toxin challenge. These experiments demonstrate that in the presence of GRP78 knockdown, there are still sufficient receptors on the cell surface to bind anthrax lethal toxin and mediate cell death from LF entry through the plasma membrane. Taken together, these experiments demonstrate that GRP78 knockdown does not markedly affect the cycling of at least one of the dominant anthrax toxin receptors, CMG2, to the cell surface.
Figure 9.
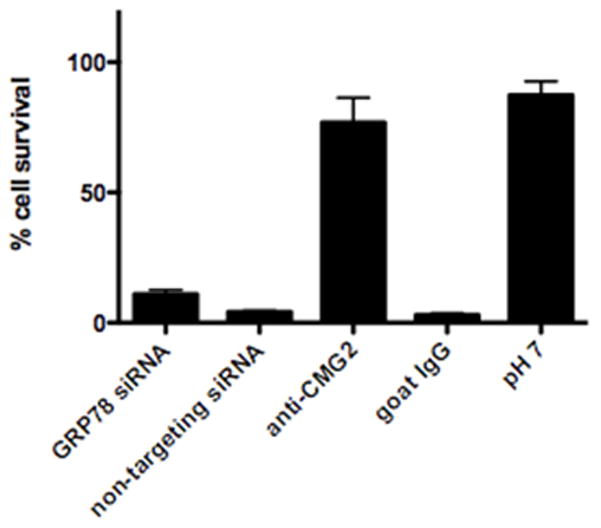
siRNA knock down of GRP78 expression in J774A.1 murine macrophage cells does not inhibit cytosolic entry of lethal toxin through PA pores in the plasma membrane. Percent J774A.1 cell survival after incubation with PA and LF at 4°C, followed by pore formation and translocation for 2 min at pH 5 and 37°C, and incubation in complete medium in the presence of Bafilomycin A1, with pre-treatments as indicated or with translocation medium adjusted to pH 7.0.
Although the results of siRNA knock down of GRP78 suggest that this chaperone plays an in vivo role in facilitating the delivery of LF via the endocytic pathway to the eukaryotic cell cytosol, previous results have shown that low pH (pH 5.5) alone is sufficient to spontaneously unfold both LFnDTA and LF (Fig. 3). We therefore hypothesized that GRP78 might enhance the unfolding of LF in a mildly acidic early endosomal compartment. In order to test this hypothesis, we used ammonium chloride, a weak base that has previously been used to investigate the pH requirements for PA pore formation in early and late endosomes (Rainey et al., 2005). Prior work has demonstrated that a weak base, ammonium chloride (NH4Cl), when present in the culture medium is able to delay the lowering of pH (< 6.0) typical in late endosomal vesicles and thereby block PA-receptor dissociation, and pore formation in this compartment, without suppressing PA pore formation and translocation from the early endosome. Using this method to block toxin translocation from the late endosome, we asked whether GRP78 exerts its predominant effect in early endosomal LF delivery. We found that the addition of 1.25 mM NH4Cl to the cell culture medium markedly enhances the protective effect of GRP78 knockdown compared to control cells treated with the non-targeting siRNA in the presence of ammonium chloride (Fig. 10). For these experiments, we used a high concentration of LF (1 nM) to obtain complete killing of cells in the presence of siRNA knockdown of GRP78 in order to elicit any synergistic benefit from NH4Cl treatment (Fig. 7A). While there is much to be done to further characterize these effects, under the experimental conditions used, the simplest explanation for these results support the argument that in the presence of a weak base, 1.25 mM ammonium chloride, the contribution made by GRP78 in facilitating the entry of LF in a mildly acidic early endosomal vesicle is enhanced.
Figure 10.
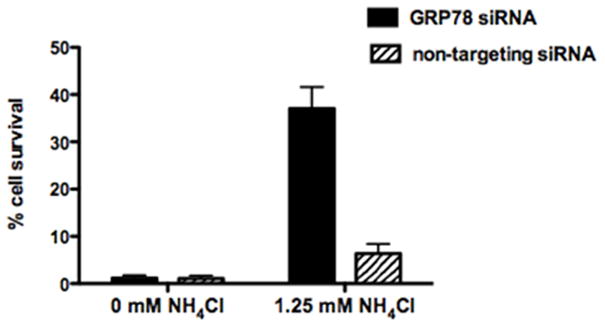
In the presence of minimally protective concentrations of ammonium chloride (1.25 mM) GRP78 knock down cells show markedly enhanced protection against challenge with anthrax lethal toxin (11 nm PA, 1 nM LF); whereas, in the absence of ammonium chloride, both GRP78 knock down and non-targeting siRNA treated J774A.1 cells are equally sensitive to toxin challenge.
Consistent with in vitro unfolding experiments which show GRP94 is unable to mediate LF unfolding, we did not observe any change in lethal toxin sensitivity with GRP94 knockdown (data not shown). These experiments were however, complicated by the long half-life of this chaperone as well as toxicity associated with its knockdown. Although reduction of GRP94 transcript levels 48 hrs post-transfection could be detected by qRT-PCR, cells began to show significant toxicity with only slightly reduced protein levels at this time. We also detected a strong apparently compensatory up-regulation of expression of GRP78 upon repeated attempts to knockdown GRP94 expression. Thus, a limitation(s) imposed by using an siRNA approach prevents us from clearly defining or excluding a role for GRP94 in the LF entry process at this time.
Discussion
GRP78 and GRP94 are well studied multifunctional chaperones that have been shown to play a role in the retro-translocation of mis-folded proteins from the ER lumen through the polypeptide conducting channel formed by Sec61 or Derlin-1 to the cytosol (Plemper et al., 1997; Christianson et al., 2008). Whilst these chaperones have been best studied in the ER, there are numerous reports which document the presence of GRP78 and GRP94 on the cell surface (Takemoto et al., 1992; Davidson et al., 2005; Triantafilou & Triantafilou, 2003; Xiao et al., 1999; Gonzalez-Gronow et al., 2007), as well as their internalization into endosomal vesicles (Kim et al., 2006; Arnold-Schild et al., 1999; Singh-Jasuja et al., 2000; Berwin et al., 2002; Wassenberg et al., 1999). Deinhardt et al. (2006) also have shown that GRP78 (Hsp70) is found in Rab7 positive endosomal vesicles which contain the Hc domain of tetanus neurotoxin. We have confirmed and extended these earlier observations by demonstrating the presence of GRP78 and GRP94 in Rab5- and Rab7- associated endosomal vesicles that have been pre-loaded with LFnDTA/PA63(7) in the presence of Balfilomycin A1. Furthermore, we have shown by immuno-electron microscopy that GRP78 and GRP94 (data not shown) are located in the lumen, rather than on the cytosolic surface of the endosomal vesicle membrane.
We have demonstrated that GRP78 and GRP94 have unfoldase activity and are able to mediate the conversion of LFnDTA from a trypsin resistant to sensitive conformation at neutral pH in vitro. In these studies, we have confirmed and extended the observation that LFnDTA is resistant to proteolytic attack by tryspin at neutral pH. However, at neutral pH, co-incubation of LFnDTA with either GRP78 or GRP94 followed by the addition of trypsin to the reaction mixture results in rapid degradation of this recombinant fusion protein in vitro. As anticipated, even a modest reduction of the pH which mimics the transition that occurs in the endosomal vesicle lumen increases the sensitivity of LFnDTA to trypsin digestion. Nonetheless, under all conditions tested co-incubation of LFnDTA at these pH values in the presence of GRP78 or GRP94 enhances its relative sensitivity to proteolytic attack. In marked contrast, however, only GRP78 appears to be able to mediate the unfolding of native LF at neutral pH in vitro. Since LF is not converted to a protease sensitive conformation by GRP94, it is possible that GRP94 is able to recognize the recombinant fusion protein LFnDTA as being mis-folded protein and, as such, is uniquely able to mediate its denaturation in vitro.
We have used siRNA methods to knock down GRP78 expression in order to demonstrate in vivo a functional role for this chaperone in the process by which LF and EF enter the cytosol. Knock down of GRP78 resulted in increased survival of J774A.1 macrophages following challenge with lethal toxin and decreased cAMP levels following challenge by edema toxin, suggesting that this chaperone plays an active role in the process by which both LF and EF is delivered from the endosomal vesicle lumen to the eukaryotic cell cytosol. We have shown that knock down of GRP78 does not affect the cycling of anthrax receptors to the cell surface by demonstrating that lethal toxin is able to bind to cell surface receptors and that LF may be delivered directly to the cytosol following a brief low pH pulse. In these experiments, receptor mediated endocytosis of lethal toxin and cytosolic delivery of LF from lumen of endosomal vesicles was blocked by Bafilomycin A1. Finally, we have shown that the addition of 1.25 mM ammonium chloride to the cell culture medium can enhance the protective effect of siRNA knock down of GRP78, even under high concentrations of LF that otherwise mask the protective effect afforded by its knockdown.
The intoxication process is initiated by the binding of PA83 to one of two different cell surface receptors: ANTXR1 (Bradley et al., 2001) and ANTXR2 (Scobie et al., 2003). Once bound to its receptor PA83 is cleaved by the endoproteinase furin and a 20 kDa fragment is removed to yield PA63 which then oligomerizes to its heptameric form. Up to three molecules of either LF or EF, or combination of both are then able to bind to the heptamer forming the holotoxin. Anthrax toxin is then internalized in clathrin coated pits and enters an early endosomal vesicle (EEV) compartment. Acidification of the vesicle lumen by the (v)ATPase is known to trigger a dynamic change in the structure of PA63 in which a single anti-parallel beta strand from each monomer contributes to the formation of a β-barrel which spontaneously inserts into the vesicular membrane forming a pore. In the absence of either ANTXR1 or ANTXR2, PA63 has been shown to spontaneously form pores in artificial membranes; however, once bound to either of these cell surface receptors a low pH environment is required for pore formation. The pH threshold for PA pore formation depends upon the class of receptor: a pH of ~ 6.5 is required when PA is bound to ANTXR1 versus a pH of ~ 5.5 when it is bound to ANTXR2 (Santelli et al., 2004; Lacy et al., 2004; Scobie et al., 2007).
In addition to the pH triggered receptor and structural changes required for pore formation, both LF and EF must also be fully denatured in order to pass through the PA pore and be delivered to the cytosol. In contrast to ANTXR1 and ANTXR2, where much is known of the amino acid residues that influence the pH threshold for the release of PA (Scobie et al., 2007), little is known of the requirements for LF and EF denaturation. Indeed, the denaturation process has been assumed to be largely due to the low pH environment of the endosomal lumen (Young & Collier, 2007).
Over the past several years our understanding of the process by which anthrax toxin delivers LF and EF to the eukaryotic cell cytosol has become increasingly complex and under physiologic conditions requires the participation of additional cellular components. For example, two previous reports have shown that a functional COPI complex was required for anthrax lethal toxin action. Using cells that carried a temperature sensitive mutation in ε-COP, Abrami et al. (2004) demonstrated that cells grown at the non-permissive temperature were resistant to anthrax toxin. We have recently extended this observation by demonstrating that COPI plays an essential role in the delivery of LFnDTA-associated ADP-ribosyltransferase activity from the endosomal vesicle lumen to the external medium (Tamayo et al., 2008). Furthermore, analogous to the GRP78 knock down data presented here, shRNA knock down of the coatomer subunits β′-COP, ζ-COP, and γ2-COP expression were also found to block the action of anthrax toxin in J774A.1 cells (L. Slater & D. Hung, unpublished observations). In the case of the diphtheria toxin-related fusion protein DAB389IL-2, Trujillo et al. (2010) have shown that COPI complex proteins interact with ε-amino moieties in the multiple KXKXX signature rich region of the transmembrane domain in order to facilitate the cytosolic entry of the catalytic domain. Since the N-terminal portions of both LF and EF also carry multiple KXKXX motifs, the essential role played by COPI complex proteins in the anthrax toxin entry process may be analogous to that previously observed in the entry of the diphtheria toxin catalytic domain. Thus, like diphtheria toxin, the cytosolic delivery of LFnDTA, LF, and EF appears to require an ordered series of interactions between a number of cellular proteins and the toxin.
Taken together, the experiments described in this communication challenge the concept that the cytosolic delivery of anthrax toxin is only the result of exposure to a low pH environment which triggers the denauration of LF and its translocation through the trans-endosomal vesicle membrane pore formed by PA. Whilst this hypothesis was the reasonable conclusion from studies with planar lipid membranes and purified anthrax toxin exposed to low pH conditions, it does not take into consideration the multiple cellular factors that have been subsequently shown to be required for optimal LF entry from either partially purified endosomal vesicles or in intact cells. Further, experiments described here suggest that the cytosolic delivery of both LF and EF is more complex than originally anticipated and, rather than being mediated by a single triggering event, the entry process is likely to occur over a range of conditions and vesicular compartments. Based upon observations reported here and the known pH requirements for PA release from its cell surface receptors (Rainey et al., 2005), we conclude that GRP78 plays a significant role in the unfolding of both LF and EF in a mildly acidified endosomal compartment, presumably where PA bound to ANTXR1 has been released and forms a trans-endosomal membrane pore; most likely early in the endocytic pathway where the pH has not dropped low enough to denature the catalytic proteins LF and/or EF. The relative role of this chaperone in the LF/EF entry process is likely reduced in late endosomal vesicles where low pH alone may be sufficient to trigger both the release of PA bound to ANTXR2 and affect the denaturation of LF/EF.
Finally, it is of interest to note that cell surface GRP78 and GRP94 have been shown to directly interact with other virulence factors and viral surface proteins. For example, GRP78 is known to be a receptor element for serotype 2 dengue virus and coxsackievirus A9 (Jindadamrongwech et al., 2004; Triantaflou et al., 2002), and GRP94 has been shown to interact with the Listeria monocytogenes cell surface protein virulence factor, Vip (Cabanes et al., 2005). More recently, Na et al. (2008) have reported that Clostridium difficle Toxin A also binds to GRP94 on HT colonic epithelial cells. In this study, toxin A/GRP94 complex was shown to be translocated into the cytosol and pretreatment of target cells with anti-GRP94 inhibited both cell surface binding and entry into the cytosol. While the role of these two chaperones in either the viral entry process or the potential delivery of Vip or Toxin A to the cytosol have not been clearly defined, in the light of observations reported here it is tempting to postulate that in each case either GRP78 and/or GRP94 on the cell surface may be directly involved in the entry processes for each of these virulence factors.
Experimental procedures
Sucrose density gradient fractionation of anthrax toxin pre-loaded endosomal vesicles
Endosomal vesicles were prepared as previously described (Tamayo et al., 2008). Briefly, HuT102/6TG cells were pretreated with 1 μM Bafilomycin A1, and then incubated with 1 nM each of PA63 and LFnDTA or LF, or with media alone as a mock intoxication. In some cases, 5 mg/ml horse radish peroxidase (HRP) was co-loaded into endosmal vesicles along with toxin as previously described (Tamayo et al., 2008). Cells were lysed and the post nuclear supernatant fluid fractions were subjected to a sucrose density step gradient ultracentrifugation. The 25 – 36% sucrose interface containing endosomal vesicles was isolated. The total protein concentration was determined by Bradford reagent was ca. 0.1 μg/ml. Five hundred microliter aliquots of endosomal vesicles were adjusted to 50% sucrose and loaded beneath 3 ml of a 20–40% continuous sucrose gradient and centrifuged for 1 hr at 4°C at 150,000 × g. One hundred microliter fractions were collected from the top of the gradient and each fraction was analyzed for total protein concentration by Bradford reagent, NAD+-dependent ADP-ribosyltransferase activity and HRP activity as previously described (Tamayo et al., 2008) in the presence of 0.1% Triton X-100 (Sigma). Fractions were also analyzed by immuno-blot with anti-Rab5 (BD), anti-Rab7 (Santa Cruz), anti-GRP78 (Abcam) and anti-clathrin heavy chain (Abcam).
Mass Spectrometry
Protein from silver stained SDS-polyacrylamide gels were excised and processed for in gel digestion as previously described (Rosenfeld et al., 1992; Wilm et al., 1996). Briefly, individual protein bands were excised and the gel was cut into small uniform pieces; the gel pieces were then dehydrated in acetonitrile and rehydrated with 100 mM ammonium bicarbonate. Samples were then reduced by the addition of DTT to10 mM and then treated with 50 mM iodoacetamide. Gel pieces were then washed twice with ammonium bicarbonate and dried by acetonitrile. After complete dehydration with acetonitrile, the gel pieces were then suspended in 12.5 ng/μl trypsin in 50 mM ammonium bicarbonate. In gel digestion was carried out at 37°C for 10–12 hrs and the peptides were extracted from the gel in 50% acetonitrile and 5% formic acid. The extract was concentrated under reduced pressure and finally desalted by passage through a C-18 containing Zip-Tip (Millipore). Trypsin digested peptides were analyzed by MALDI-TOF-TOF (ABI-4800).
MALDI-TOF-TOF analyses were performed using the following parameters: source 1 voltage at 80 kV, source 2 voltage at 15 kV, source 1 focus at 4.3 kV, source 1 lens at 3.7 kV, Y1 deflector at 0.08 kV, grid source 1 voltage ration of 0.91, mirror 2 to mirror 1 voltage ration of 1.7, and the mirror 2 to source 2 voltage ration of 1.23. MS spectra were recorded in a mass range of 800 – 4,000 daltons.
Mass Spectrometry Data Analysis
Mass spectrometry data was analyzed and interpreted using the MASCOT (Ver. 2.0.04) and Protein Prospector (ver 4.27.2 basic and Ver. 5.0, UCSF) database search algorithms. The database search parameters were as follows: for the NCBInr database; SwissProt, tryptic digest, 2 missed cleavages, and a mass tolerance of 20–50 ppm, 5 peptides were required for identification, MS/MS data was analyzed by MASCOT.
Immuno-magnetic isolation of Rab5 positive endosomal vesicles
Two μg of monoclonal mouse anti-human Rab5 primary antibody (BD) or IgG isotype control antibody were incubated with 50 μl Protein A superparamagnetic 50 nm microbeads (Miltenyl Biotec) for 1 hr at room temperature. Beads were bound to a magnetic column (Miltenyl Biotec) and washed three times with 1 ml 10 mM HEPES, pH 7.6. Antibody conjugated beads were eluted from the column with 50 μl 10 mM HEPES, pH 7.6, and incubated for 2 hrs at room temperature with 20 – 30 μg partially purified endosomal vesicles. Beads with affinity bound endosomal vesicles were bound to a magnetic column and the bound flow through material was collected for further analysis. The column was washed three times with 1 ml 10 mM HEPES, pH 7.6. The column was then removed from the magnet and the Rab5 affinity purified endosomes were eluted with 50 μl 10 mM HEPES, pH 7.6. The unbound and bound endosomal vesicles were then analyzed by immunoblot and assayed for ADP-ribosyltransferase activity as previously described (Tamayo et al., 2008). Primary antibodies used for immunoblot are as follows: anti-Rab5 (BD), anti-GRP78 (Abcam) and anti-clathrin heavy chain (Abcam).
Immuno-electron microscopy
LFnDTA/PA63 loaded endosomal vesicles were prepared as before, and then centrifuged at 100,000 × g for 45 min at 4°C. The supernatant fluid fraction was removed and the pellet was fixed with 2% formaldehyde. After 2 hr fixation at room temperature the pellet was washed with PBS containing 0.2 M glycine. Prior to freezing in liquid nitrogen the endosomal vesicle pellet was perfused with 2.3 M sucrose in phosphate buffered saline (PBS) for 15 min. Ultrathin sections were cut at 120°C with a cryo-diamond knife or at -90°C with a glass knife. Sections were picked up from the knife with a loop dipped in a 2:1 mixture of 2.3 M sucrose and 2% methylcellulose and transferred to a formvar/carbon coated copper grid. Individual specimens were immuno-gold labeled as follows: specimens were blocked in 1% bovine serum albumin (BSA) for 15 min, incubated with Protein A-gold (PAG) in 1% BSA for 20 min and washed with 4 drops of PBS for 15 min. In the case of double labeled sections, sections were fixed in 1% glutaraldehyde for 5 min followed by quenching in 4 drops of 0.15 M glycine/PBS and then probed with different primary antibodies followed by washes and PAG incubation, and finished by a wash with 6 drops of water for 20 min. Antibodies used in labeling experiments: anti-Rab7 (Abcam), anti-GRP78 (Abcam) and IgG isotype matched antibodies as controls. Sections were viewed with a Tecnai G2 Spirit BioTWIN transmission electron microscope (TEM).
Protease protection assays
LFnDTA was cloned into the p-GEX-6p vector (GE Healthcare) using standard methods. Recombinant GST-LFnDTA fusion protein was expressed and purified from the BL21(DE3)pLysE strain of Escherichia coli as previously described (Tamayo et al., 2008). LFnDTA was cleaved from GST-LFnDTA which was bound to glutathione sepharose beads with PreScission protease (GE) in cleavage buffer (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) for at least 2 hrs at 4°C according to the manufacturer’s instructions. As noted, 27 nM of purified LFnDTA was incubated in the absence or presence of recombinant GRP78 (Assay Designs) or recombinant GRP94 (Assay Designs) in a 30 μl reaction volume for the time indicated at 37°C in buffer (10 mM HEPES, pH as indicated, 25 mM KCl, 2 mM MgCl2). One ng trypsin (Sigma) or buffer alone were added to the reaction mixture and incubated for 30 min at 37°C. SDS-polyacrylamide gel loading buffer was added and the samples were boiled for 5 min and then analyzed by SDS-polyacrylamide gel electrophoresis and immunoblot. Trypsin sensitivity of LFnDTA was measured by immunoblot using anti-DTA antibodies (Abcam).
siRNA knock down of GRP78 and GRP94 expression
J774A.1 murine cells were obtained from American Type Culture Collection (ATCC) (Manassas, VA) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Carlsbad, CA) containing L-glutamine and 10% (v/v) fetal bovine serum (Hyclone, Logan, UT) and maintained at 37°C with a humidified atmosphere containing 5% CO2. Small interfering RNA siGENOME was obtained from Dharmcon (Lafayette, CO). The siRNA target sequence used for Grp78 was GGAAUGACCCUUCGGUGCA (accession number NM_022310, catalog number D-040337-02); the catalogue number for the non-targeting siRNA is D-001206-14. The day before siRNA transfections, cells were harvested and replated in 6-well format at 4 × 105 cells per well, or in 96-well format at 1.3 × 104 cells per well. On the day of transfection, medium was replaced with 0.5 ml fresh culture medium and 0.5 ml transfection mix (6-well), or 50 μL culture medium and 50 μL transfection mix (96-well). Transfection mixes contained 60 pmol siRNA duplex and 9 μl Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) in 0.5 ml OPTI-MEM-I reduced serum medium (Gibco) and were made up according to the manufacturer’s instructions. After 24 hours, transfection medium was replaced with fresh culture medium and cells incubated for a further 60 hrs (6-well), or 72 hrs (96-well). Cells transfected in 6-well plates were harvested and replated in 384-well format with 4000 cells per well in 30 μL, and incubated overnight.
LF/PA83 assay
Cells were challenged with lethal toxin (11 nM PA83; 0.01 – 1 nM LF) for 3.5 hr at 37°C. Cell viability was measured by adding 30 μL Cell Titer Glo (Promega, Madison, WI) and reading luminescence on a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA).
Western blot analysis
Cells were lysed in RIPA buffer (20 mM Tris HCl pH 8.0, 1% Triton X-100, 150 mM sodium chloride, 0.1% SDS, 0.5% sodium deoxycholate) containing “Complete Mini” protease inhibitor cocktail (Roche, Indianapolis, IN) and lysates were clarified by centrifugation at 16,000 rpm for 30 min at 4°C. Six micrograms of cell lysate was separated by SDS-PAGE and transferred to PVDF followed by immunoblot with rabbit polyclonal IgG antibody to GRP78 (Abcam, Cambridge, MA) or rabbit polyclonal IgG antibody to actin (Sigma, St. Louis, MO) and secondary HRP goat-anti rabbit antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Blots were visualized using SuperSignal West Femto chemiluminescent substrate (Pierce, Rockford, IL).
EF/PA83 assay
EF was obtained from List Biological Laboratories, Inc. (Campbell, CA). Cells were treated with edema toxin (11 nM PA83 and 1.7 nM EF) for 4 hrs at 37°C, and the concentration of cAMP was measured by cAMP enzyme immunoassay according to the manufacturer’s instructions (Enzo Life Sciences, Plymouth Meeting, PA).
LF/PA63 cell-based translocation assay
Cells treated with antibody were incubated with 40 μg/ml Goat IgG (Santa Cruz Biotechnology, Santa Cruz, CA) or goat anti-mouse CMG2 antibody (R&D Systems, Minneapolis, MN) for 1 hr at 4°C. Cells were washed twice in cold PBS, and incubated with 3.5 μg/ml PA63 diluted in OPTI-MEM-I for 2 hrs at 4°C. Cells were washed twice in cold PBS, and incubated with 3.5 μg/ml LF diluted in OPTI-MEM-1 for 2 hrs at 4°C. Cells were washed twice in cold PBS and incubated with translocation buffer (20 mM MES, 150 mM NaCl, 5 mM gluconic acid) at pH 5 for 2 min at 37°C. Cells were washed 3X in cold PBS, and then incubated for 4 hrs at 37°C in complete medium containing 100 nM Bafilomycin A1. Cell viability was measured using Cell Titer Glo, as described previously.
Supplementary Material
Acknowledgments
We thank Dr. Maria Ericsson, Harvard Medical School, for performing the transmission electron microscopy. This work was supported by Public Health Service grant AI-021628 from the National Institute of Allergy and Infectious Diseases (JRM) and by National Institute of Allergy and Infectious Disease Regional Center of Excellence grant AI-057159 (JRM, DTH).
References
- Abrami L, Lindsay M, Parton RG, Leppla SH, van der Groot FG. Membrane insertion of anthrax protective antigen and cytoplasmic delivery of lethal factor occur at different stages of the endocytic pathway. J Cell Biol. 2004;166:645–651. doi: 10.1083/jcb.200312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrami L, Liu S, Cosson P, Leppla SH, van der Goot FG. Anthrax toxin triggers endocytosis of its receptor via a lipid raft-mediated clathrin-dependent process. J Cell Biol. 2003;160:321–328. doi: 10.1083/jcb.200211018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold-Schild D, Hanau D, Spehner D, Schmid C, Rammensee HG, de la Salle H, Schild H. Cutting edge: receptor-mediated endocytosis of heat shock proteins by professional antigen-presenting cells. J Immunol. 1999;162:3757–3760. [PubMed] [Google Scholar]
- Bernatchez PN, Sharma AA, Kodaman P, Sessa WC. Myoferlin is critical for endocytosis in endothelial cells. Am J Cell Physiol. 2009;297:C484–492. doi: 10.1152/ajpcell.00498.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwin B, Rosser MF, Brinker KG, Nicchitta CV. Transfer of GRP96(GP96)-associated peptides onto MHC class I molecules. Traffic. 2002;3:357–366. doi: 10.1034/j.1600-0854.2002.30505.x. [DOI] [PubMed] [Google Scholar]
- Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- Cabanes D, Sousa S, Cebriá A, Lecuit M, Portillo F, Cossart P. Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. EMBO J. 2005;24:2827–2738. doi: 10.1038/sj.emboj.7600750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson DJ, Haskell C, Majest S, Kherzai A, Egan DA, Walter KA, Schneider A, Gubbins EF, Solomon L, Chen Z, Lesniewski R, Henkin J. Kringle 5 of human plasminogen induces apoptosis of endothelial and tumor cells through surface-expressed glucose-regulated protein 78. Cancer Res. 2005;65:4663–4672. doi: 10.1158/0008-5472.CAN-04-3426. [DOI] [PubMed] [Google Scholar]
- Finkelstein A. The channel formed in planar lipid bilayers by the protective antigen component of anthrax toxin. Toxicology. 1994;87:29–41. doi: 10.1016/0300-483x(94)90153-8. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gronow M, Kaczowka SJ, Payne S, Gawdi G, Pizzo SV. Plasminogen structural domans exhibit different functions when associated with cell surface GRP78 or the voltage-dependent anion channel. J Biol Chem. 2007;282:32811–32820. doi: 10.1074/jbc.M703342200. [DOI] [PubMed] [Google Scholar]
- Hegde NR, Chevalier MS, Wisner TW, Denton MC, Shire K, Frappier L, Johnson DC. The role of BiP in endoplasmic reticulum-associated degradation of major histocompatibility complex 1 heavey chain induced by cytomegalovirus proteins. J Biol Chem. 2006;281:20910–20919. doi: 10.1074/jbc.M602989200. [DOI] [PubMed] [Google Scholar]
- Jindadamrongwech S, Thepparit C, Smith DR. Identification of GRP78 (BiP) as a liver cell expressed receptor element fo dengue virus serotype 2. Arch Virol. 2004;149:915–927. doi: 10.1007/s00705-003-0263-x. [DOI] [PubMed] [Google Scholar]
- Kim Y, Lillo AM, Steiniger SC, Liu Y, Ballatore C, Anichini A, Mortarini R, Kaufmann GF, Zhou B, Felding-Habermann B, Janda KD. Targeting heat shock proteins on cancer cells: selection, characterization, and cell-penetrating properties of a peptidic GRP78 ligand. Biochemistry. 2006;45:9434–9444. doi: 10.1021/bi060264j. [DOI] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- Krantz BA, Trivedi AD, Cunningham K, Christensen KA, Collier RJ. Acid-induced unfolding of the amino-terminal domains of the lethal and edema factors of anthrax toxin. J Mol Biol. 2004;344:739–756. doi: 10.1016/j.jmb.2004.09.067. [DOI] [PubMed] [Google Scholar]
- Lacy DB, Wigelsworth DJ, Melnyk RA, Harrison SC, Collier RJ. Structure of heptameric protective antigen bound to an anthrax receptor: a role for receptor in pH-dependent pore formation. Proc Natl Acad Sci, USA. 2004;101:13147–113151. doi: 10.1073/pnas.0405405101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kati W, Chen CM, Tripathi R, Molla A, Kohlbrenner W. Use of a fluorescence plate reader for measuring kinetic parameters with inner filter effect correction. Anal Biochem. 1999;267:331–335. doi: 10.1006/abio.1998.3014. [DOI] [PubMed] [Google Scholar]
- Miller CJ, Elliot JL, Collier RJ. Anthrax protective antigen: prepore—to-pore conversion. Biochemistry. 1999;38:10432–10441. doi: 10.1021/bi990792d. [DOI] [PubMed] [Google Scholar]
- Molinari M, Galli C, Piccaluga V, Pieren M, Paganetti P. Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J Cell Biol. 2002;158:247–257. doi: 10.1083/jcb.200204122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na X, Kim H, Moyer MP, Pothoulakis C, LaMont JT. gp96 is a human colonocyte plasma membrane binding protein for Clostridium difficle toxin A. Infect Immun. 2008;76:2862–2871. doi: 10.1128/IAI.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimental RA, Christensen KA, Krantz BA, Collier RJ. Anthrax toxin complexes: heptameric protective antigen can bind lethal factor and edema factor simultaneously. Biochem Biophys Res Commun. 2004;322:258–262. doi: 10.1016/j.bbrc.2004.07.105. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Böhmler S, Bordalo J, Sommer T, Wold DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Ratts R, Trujillo C, Bharti A, vanderSpek J, Harrison R, Murphy JR. A motif in transmembrane helix 1 of diphtheria toxin mediates catalytic domain delivery to the cytosol. Proc Natl Acad Sci, USA. 2005;102:15635–15640. doi: 10.1073/pnas.0504937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratts R, Zeng H, Berg C, Blue C, McComb ME, Costello CE, Murphy JR. The cytosolic entry of diphtheria toxin catalytic domain requires a host cell cytosolic translocation factor complex. J Cell Biol. 2003;160:1139–1150. doi: 10.1083/jcb.200210028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey GJ, Wigelsworth DJ, Ryan PL, Scobie HM, Collier RJ, Young JA. Receptor-specific requirements for anthrax toxin delivery into cells. Proc Natl Acad Sci, USA. 2005;102:13278–13283. doi: 10.1073/pnas.0505865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- Santelli E, Bankston LA, Leppla SH, Liddington RC. Crystal structure of a complex between anthrax toxin and its host receptor. Nature. 2004;430:905–908. doi: 10.1038/nature02763. [DOI] [PubMed] [Google Scholar]
- Scobie HM, Rainey GJ, Bradley KA, Young JA. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc Natl Acad Sci, USA. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobie HM, Marlett JM, Rainey GJA, Lacy DB, Collier RJ, Young JA. Anthrax toxin receptor 2 determinants that dictate the pH threshold of toxin proe formation. PLoS ONE. 2007;2(3):e329. doi: 10.1371/journal.pone.0000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh-Jasuja H, Hilf N, Scherer HU, Arnold-Schild D, Rammensee HG, Toes RE, Schild H. The heat shock protein gp96: a receptor-targeted cross-priming carrier and activator of dendritic cells. Cell Stress Chaperones. 2000;5:462–470. doi: 10.1379/1466-1268(2000)005<0462:thspga>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto H, Yoshimori T, Yamamoto A, Miyata Y, Yahara I, Inoue K, Tashiro Y. heavy chain binding protein (BiP/GRP78) and endoplasmic are exported from the endoplasmic reticulum in rat exocrine pancreatic cells, similar to protein disulfide-isomerase. Arch Biochem Biophys. 1992;296:129–136. doi: 10.1016/0003-9861(92)90554-a. [DOI] [PubMed] [Google Scholar]
- Tamayo A, Barti A, Trujillo C, Harrison R, Murphy JR. COPI coatomer complex proteins facilitate the translocation of anthrax lethal factor across vesicular membrane in vitro. Proc Natl Acad Sci, USA. 2008;105:5254–5259. doi: 10.1073/pnas.0710100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou K, Triantafilou M. Lipid raft microdomains: key sites for Coxsackievirus A9 infectious cycle. Virology. 2003;317:128–135. doi: 10.1016/j.virol.2003.08.036. [DOI] [PubMed] [Google Scholar]
- Trujillo C, Taylor-Parker J, Harrison R, Murphy JR. Essential lysine residues within transmembrane helix 1 of diphtheria toxin facilitate COPI binding and catalytic domain entry. Mol Microbiol. 2010;76:1010–1019. doi: 10.1111/j.1365-2958.2010.07159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlman J, DeMartino GN, Skach WR, Bulleid NJ, Brodsky JL, Johnson AE. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell. 2007;129:943–955. doi: 10.1016/j.cell.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenberg JJ, Dezfulian C, Nicchitta CV. Receptor mediated and fluid phase pathways for internalization of the ER Hsp90 chaperone GRP94 in murine macrophages. J Cell Sci. 1999;112:2167–2175. doi: 10.1242/jcs.112.13.2167. [DOI] [PubMed] [Google Scholar]
- Wesche J, Elliot JL, Falnes PO, Olsnes S, Collier RJ. Characterization of membrane translocation by anthrax protective antigen. Biochemistry. 1998;37:15737–15746. doi: 10.1021/bi981436i. [DOI] [PubMed] [Google Scholar]
- Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US1 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Brell S, Schweigerer L, Fotsis T, Mann T. Femtomole sequencing of proteins from polyacrylamide gels by nano-spray mass spectroscopy. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Xiao G, Chung TF, Pyun HY, Fine R, Johnson RJ. KDEL proteins are found on the surface of NG108-15 cells. Brain Res Mol Brain Res. 1999;72:121–128. doi: 10.1016/s0169-328x(99)00188-6. [DOI] [PubMed] [Google Scholar]
- Young JAT, Collier RJ. Anthrax Toxin: receptor binding, internalization, pore formation, and translocation. Ann Rev Biochem. 2007;76:243–265. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RL. Signaling the unfolded protein response from the endoplasmic reticulum. J Biol Chem. 2004;279:25935–25938. doi: 10.1074/jbc.R400008200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


