Abstract
Selectively breeding lines of mice and rats to differ in alcohol intake has proven useful for defining which traits correlate with high alcohol drinking behavior, as well as for creating animal models of alcoholism. This study reports the derivation of two novel sets of selected lines, High Alcohol Preferring (HAP) and Low Alcohol Preferring (LAP) replicate 2 and 3 lines. Mice were mass-selected using the same procedure as in the replicate 1 lines: using HS/Ibg as a progenitor, mice were selected for differences in 2-bottle choice intake of 10% alcohol during a 4-week testing period. In addition, another high drinking line, the crossed HAP (cHAP) line was selectively bred from a progenitors that were a cross of replicate 1 (S27) X replicate 2 (S21) HAP lines. All lines were characterized for saccharin intake. Overall, the response to selection of the HAP and LAP replicate 2 and 3 lines was quite similar. As anticipated, following selection, the cHAP line drank more than either parent HAP line (consuming 26.0 g/kg per day of alcohol by S11), suggesting that this method of crossing replicate lines and selecting from that cross captures more alleles than any single selected line, as well as producing a line with exceptionally high voluntary alcohol intake. As expected, saccharin consumption was highly associated with alcohol consumption; data from all 8 lines (HAP 1, 2, and 3, LAP 1, 2, and 3, HS/Ibg, and cHAP) indicated a genetic correlation between 10% alcohol and 0.32% saccharin intake of 0.917. Overall, these findings show the practicality of developing replicate lines divergent in alcohol preference, and validate a novel procedure for generating very high-drinking mouse populations.
Keywords: alcoholism, ethanol, drinking, selected line, genetic, sweet preference
Considerable effort has been devoted to developing laboratory animal models of human alcoholism. Given that alcoholism is heritable (Goldman et al. 2005), genetic approaches have been used to begin investigating exactly how genes mediate individual differences in risk for this disorder. One genetic approach that is appealing both in terms of simplicity and face validity is to use selective breeding for differences in free-choice alcohol drinking. Beginning in the 1960’s, at least 6 pairs of high and low alcohol drinking rat lines have been developed (Eriksson 1968; Lumeng et al. 1977; Mardones and Segovia-Riquelme 1983; Li et al. 1993; Colombo 1997; Le et al. 2001), as well as at least 2 sets of mouse lines (Belknap et al. 1997; Grahame et al. 1999). Lines resulting from these selections reliably show profound differences in alcohol intake resulting from fixation of divergent alleles at loci related to alcohol drinking. Mice are an appealing species for selection due to their low housing cost and well-characterized genomes. On the other hand, their high rate of ethanol metabolism, about 0.78 mg/dl/hr (Grisel et al. 2002), means that very high intakes are required to reach pharmacologically relevant blood alcohol concentrations. This makes it all the more important to utilize populations with ethanol consumption sufficient to overwhelm their high ethanol metabolism.
Inbred strains, especially the C57Bl/6J mouse, have also been widely used to understand the genetics of alcohol intake through the use of animal models. Unlike selected lines, inbred strains are not bred for a specific phenotype, but are instead created through at least 20 successive generations of brother-sister mating, resulting in fixation of all alleles. Although some inbred mouse strains, such as C57/Bl/6J, can drink ~16 g/kg/day unsweetened 10% ethanol (e.g. (Yoneyama et al. 2008) there may be interpretational difficulties with data gathered from a single inbred strain. Because all animals within an inbred strain are genetically identical, and because members of an inbred strain are homozygous at both trait relevant and irrelevant loci, many strains are required to assess relationships among phenotypes (Crabbe 1989). In addition, inbreeding results in a unique and rather unusual genotype that may have numerous behavioral and physiological abnormalities (Crawley 2007), so results obtained with one inbred strain cannot necessarily be generalized to a population as a whole (Crabbe et al. 1990; Falconer and Mackay 1996).
While high alcohol intake is a useful phenotype for understanding the mechanisms of alcohol drinking in an animal model, one of the most compelling reasons to create pairs of selected lines differing in drinking is to assess genetic differences in other phenotypes that may be related to alcohol drinking. These related phenotypes are known as correlated responses to selection. If a correlated response is detected, it may be attributed to the pleiotropic effects of trait-relevant alleles, and can yield clues as to the mechanisms underlying differences in the target response, in this case alcohol consumption. One must be cautious, however, when interpreting correlated responses observed between only one pair of selected lines. Due to genetic drift that inevitably arises in small populations such as selected lines, trait-irrelevant alleles become differentially fixed between the lines, and these alleles can also result in phenotypic differences that are not true genetic correlations (Crabbe et al. 1990). Therefore, it is important to maintain replicate lines that are selected from the same progenitor population as the original set of lines, and under the same environmental conditions. Detection of similar correlated responses between the lines increases confidence that these phenotypes result from pleiotropic actions of genes recruited by selection (Crabbe et al. 1990).
Despite their acknowledged limitations, there has still been a great deal of excellent work done comparing single high- and low- drinking lines. One reason this is so is that as with inbred strains, over time, selected lines become more and more well-characterized, allowing predictable behavioral and physiological differences to be observed between them, and leading to an understandable reluctance to utilize other (and perhaps not so well-characterized) animal models. For example, perhaps the most widely used rat model of alcohol drinking, the alcohol Preferring (P) line (and its low-drinking counterpart the NP line), while showing the desired phenotype of very high alcohol consumption, is highly inbred and lacks a replicate line. Compared to NPs, the P line also shows desirable correlated traits, such as increased alcohol tolerance (Waller et al. 1983) and signs of dependence following free-choice access (Waller et al. 1982), and these lines continue to be widely utilized (see (Murphy et al. 2002) for a review). Nonetheless, in the absence of replicate lines derived from the same population, the extent to which correlated responses are replicable remains unknown, as these pairs of lines are essentially a unique resource.
Another viewpoint that has not been widely leveraged in the alcohol field is that pairs of selected lines derived from a common progenitor population are not a unique resource, but a replicable outcome of a scientific experiment: the product of a procedure (the selection protocol) being applied to a sample of a population (the progenitor line). From this standpoint, the question is whether repeatedly and correctly applying the selection protocol to the progenitor line will result in lines that are relatively uniform with respect to the selection phenotype, its correlated responses, and the genetic architecture responsible for both. In this manuscript, we will report on the selection of two new sets of replicate lines, each with a High- (HAP) and Low (LAP) Alcohol Preferring line, all bidirectionally selectively bred based upon intake of alcohol during four weeks of free-choice drinking, and all derived from the HS/Ibg progenitor population. In combination with the original replicate 1 HAP and LAP mice (Grahame et al. 1999), these 2 new replicates make a total of three times that HAP and LAP have been separately derived. Of interest will be the extent to which these lines show similarities in both the target phenotype as well as a single correlated response, saccharin intake.
Opposing the uniformity expected for given sets of replicate lines, discussed above, is inbreeding, or genetic drift. The effect of selective breeding is to fix trait relevant alleles. However, over time, genetic drift can also fix alleles. Because the way in which inbreeding fixes alleles is more or less random, most of these alleles can be presumed to be trait irrelevant, and can lead to divergence between lines in loci unrelated to the selection phenotype. This may lead to relatively less uniformity among the lines. Inbreeding may also lead to the “loss” of alleles that might be important to drinking, to the extent that such alleles are fixed in an undesirable configuration prior to their being captured and recruited by selective breeding (Falconer and Mackay 1996). To the extent that this is true, two high-drinking lines may collectively contain a somewhat longer list of “high-drinking” alleles than any particular line alone, depending upon the extent of inbreeding encountered during selective breeding. In other words, alleles lost prior to their capture by selection in any single line may nonetheless have been recruited in another line. As a direct test of this theoretical construct, as well as a practical means to the end of obtaining the highest drinking mouse line possible, we herein describe an additional line derived from a cross and subsequent selective breeding of replicate 1 and replicate 2 HAP lines. Our hypothesis is that some different trait-relevant alleles were captured in each of the two lines, and that therefore the crossed line (which we name crossed HAP, or cHAP) will show a response to selection over and above either parent line, eventually drinking higher amounts of ethanol than either parent line. Such behavior could make them an attractive animal model of high free-choice alcohol drinking. Continued selection of HAP1/LAP1 will be presented for illustrative purposes.
Our third hypothesis is that selection for high drinking will result in the correlated response of greater preference for sweet solutions in multiple lines. Sweet preference/consumption has been reported to be correlated with alcoholism in humans (Kampov-Polevoy et al. 1997), rats selected for high alcohol drinking (Sinclair et al. 1992; Stewart et al. 1994; Kampov-Polevoy et al. 1996), and HAP1 mice (Grahame et al. 1999). The currently reported sets of selectively bred HAP and LAP lines, available in triplicate, are an excellent way to test for whether genetic differences in alcohol intake (analogous to family history positive in human studies) predicts differences in sweet preference. Therefore, we will test a range of saccharin concentrations to observe whether shifts in intake towards the sweeter solutions are predicted genetic differences in alcohol consumption.
Methods
Measurement of Alcohol Preference
Prior to preference testing, mice were housed in groups of four in polycarbonate cages (27.9 × 9.5 × 12.7 cm) with pine chip bedding, at an ambient temperature of 21 ± 1°C. Water and lab chow were available at all times in the home cage. During preference testing, mice were housed individually in the polycarbonate cages with microisolator tops. Lights were on from 0600 to 1800. At 45 days of age, mice were singly housed and allowed to drink from two 25–50-ml graduated cylinders. One cylinder contained 10% alcohol (v/v) in distilled water, and the other cylinder contained distilled water. Volumes consumed were visually read to a resolution of ± 0.05 ml, and the cylinder containing alcohol was switched three times per week. Mice were weighed once per week. The average score in g/kg over four weeks was used as the selection phenotype.
Selective Breeding: Animals and Protocol
The same mass selection procedure previously reported for the first 10 generations of HAP1/LAP1 lines (Grahame et al. 1999) was used to create the new HAP2/LAP2 and HAP3/LAP3 lines. Breeding pairs of HS/Ibg stock were obtained from the Institute of Behavior Genetics at Boulder CO, and bred to produce a progenitor stock of over 320 mice. High- drinking males and females were mated to produce the high drinking line and the low-drinking males and females were used to produce the low drinking line. For each generation, 8–12 breeding pairs were selected, aiming at a target population for each generation of 80 mice. The selection phenotypes were mean ethanol intakes of greater than 5.0 g/kg/day for HAP and less than 1.5 g/kg/day for LAP. However, these criteria became less meaningful within 10 generations, when line means surpassed the high- and low-cutoff values, at which time the best phenotypes available were used to determine breeders. First-cousin matings were avoided, and this took precedence over the drinking score, leading to occasional exceptions to selection criteria in order to preserve outbreeding. Breeder number was maintained at >20 per line per generation when possible.
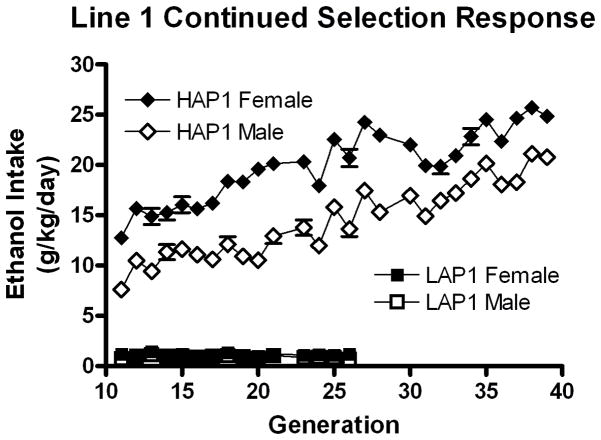
HAP1/LAP1 selection continued in this manner through generation 39, but the LAP1 line ended after generation 26 due to fecundity issues. Although the selection response for the first 10 generations of HAP1/LAP1 mice has been previously reported (Grahame et al. 1999), ethanol drinking data for generations 11–39 are illustrated in figure 1 for comparison to HAP2/LAP2 and cHAP selection response.
Figure 1.
Mean ethanol intakes in selected HAP1 (diamonds) and LAP1 (squares) lines of mice show a continued response to bidirectional selection for alcohol intake in a 24-hr access 2-bottle choice procedure for generations 11–39. Generations 0–10 have been previously reported and are not shown here. Females (filled) are shown separately from males (open).
Exp. 1: HAP2/LAP2 Selection
Eighty HS/Ibg (40 male) from the 57th generation were bred to produce 373 offspring (174 male), which comprised the progenitor population. From these, the 36 highest drinkers (12 male) and the 36 lowest drinkers (12 male) became the breeders for the first generation of HAP2 and LAP2, respectively. Breeder males were generally mated with two breeder females (so-called first- and second-choice, based upon their phenotype) to maximize offspring production. Where possible, only first-choice litters entered the breeding pool for the subsequent generation. Bidirectional selection was performed on every subsequent generation except for generation 22. Also, the LAP2 mice were not assessed at generation 15. In both these situations, mice failed to produce sufficient numbers in all litters for selection to be practical, so animals were mated following outbreeding rules, but otherwise unselected. For generations 1 through 32, the mean number of breeders was 26 (range, 16–35) for HAP2 and 23 (range, 14–34) for LAP2.
Exp. 2: HAP3/LAP3 Selection
Sixty HS/Ibg (20 male) from the 73rd generation were bred to produce 325 offspring (162 male) which comprised the progenitor population. From these, the 30 highest drinkers (10 male) and the 30 lowest drinkers (10 male) became the breeders for the first generation of HAP3 and LAP3, respectively. Bidirectional selection was performed on every subsequent generation. For generations 1 through 10, the mean number of breeders was 23 (range, 18–28) for HAP3 and 23 (range, 18–29) for LAP3.
Exp. 3: cHAP Production and Selection
A reciprocal cross of HAP1 (generation 27) and HAP2 (generation 21) was performed to produce the cHAP line. Eight HAP1 males were mated with 13 HAP2 females to produce 132 offspring. Seventeen HAP1 females were mated with 9 HAP2 males to produce 151 offspring. These offspring were pooled and formed the progenitor population. The highest drinkers were used as breeders, maintaining the existing HAP/LAP mating and phenotyping rules. The mean number of breeders for generations 1 through 9 was 25 (range, 20–30). The cHAP line was always phenotyped in the same room, by the same experimenter (C. Best) and at the same time as the replicate 1 line, meaning that comparisons between the cHAP and HAP1 mice are especially reliable. Note that drinking data were not collected for HAP1 generation 29 and HAP2 generation 22 for reasons unrelated to this experiment.
Correlated Response: Saccharin Drinking
Progenitors and selectively bred mice were assessed on saccharin intake to demonstrate the predicted correlation between sweet consumption and alcohol intake. Saccharin drinking was tested in ascending half-log concentrations of 0.032, 0.1, 0.32, and 1.0% (w/v) in a two-bottle choice procedure. Graduated cylinders containing either plain water or water plus saccharin were read to a resolution of ± 1.0 ml every other day, and positions were alternated. Means of four to six days’ drinking were used for data analysis. A two-bottle assessment of water drinking was performed for 2 days after the saccharin data were gathered. Regardless of saccharin concentration, saccharin preference was typically high, so we instead focus on intake (in ml/kg) as our principle dependent variable.
Exp. 4: HS/Ibg, HAP1, and cHAP Saccharin Intake
Forty HS/Ibg (20 male) from generation 73, 40 HAP1 (20 male) from generation 30, and 40 cHAP (20 male) from generation 3 were tested for saccharin consumption. The HS/Ibg mice were ~8 months old at testing, and the HAP1 and cHAP mice were ~3 months old. The HAP1 and cHAP mice were used as breeders, and as such had been previously assessed for ethanol preference.
Exp. 5: LAP2 and HAP2 Saccharin Intake
Forty LAP2 (20 male) and 40 HAP2 (20 male), both from generation 24, were tested for saccharin intake. Both lines were ~4 months old at the beginning of testing.
Exp 6: LAP3 and HAP3 Saccharin Preference
20 LAP3 (10 male) and 18 HAP3 (8 male), both from generation 12, were tested for saccharin intake. Both lines were 73 to 76 days old at the beginning of the experiment.
Statistics
Between-subjects ANOVA with Generation and Sex as factors was performed on ethanol intake (g/kg/day) and weight. Interactions were followed by Bonferroni-corrected t-tests to detect differences as appropriate. Statistics were not be performed on early line 1 selection, as they have previously been reported (Grahame et al. 1999). For the cHAP derivation, between-line t-tests comparing cHAPs to their respective parent lines were used to gauge the success of this selection, with Bonferroni correction performed by setting alpha to 0.05 divided by number of generations (α = 0.00454) for experiment 3. Selection response rates were compared with slopes derived from linear regressions, with non-overlapping 95% confidence intervals considered significantly different.
For assessment of changes in drinking over the four weeks of access, we subjected the last generations of the two novel selections (Line 2 S33 and Line 3 S10) to a Line X Sex X Readings mixed ANOVA. Readings were the 12 readings that occurred over the four weeks of access; the first reading was always on a Monday after 72 h access, while the remaining were either two or three days. All data are presented in g/kg per day of alcohol access. Where appropriate based on interactions with the Readings factor, we assessed changes in intake within each line over time, first with simple main effect of Reading, and then Pairwise t-tests assessing whether each reading differed from the preceding one.
For the saccharin drinking studies, mixed ANOVA with Line and Sex as between-subjects factors and concentration as a within subjects factor were performed on saccharin intake (ml/kg/day). Additionally, each selected line was compared back to the non-selected HS/Ibg progenitor line using Line X Sex X Concentration ANOVAs as orthogonal contrasts, to determine whether each line shows a correlated response to selection. To derive a genetic correlation between saccharin intake and ethanol intake, each line’s alcohol drinking score at the generation for which saccharin intake was assessed was correlated with their saccharin consumption at the highest-consumed (0.32%) concentration. Finally, all lines were compared to the HS/Ibg line to assess changes in water intake with selective breeding.
Results
HAP2/LAP2 Response to Selection
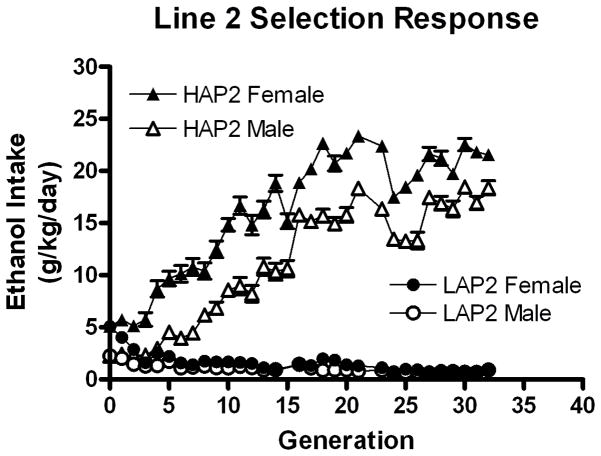
Figure 2 illustrates the response to selection observed in the HAP2 and LAP2 mouse lines for generations 0–33. Response to selection was strongest earlier, and as selection continued and exhausted allelic diversity, the response to selection tended to diminish. Heritability, or h2, was calculated as response to selection (R) divided by selection differential (S) after Falconer and Mackay (1996). To wit, R is the difference in mean ethanol intake between one generation and the next, and S equals the difference between the generation mean and the mean for the breeders selected from that generation. A heritability estimate was derived for each line and generation, and those estimates were summed by line and averaged across generations. For the first 10 generations, the mean heritability estimate was 0.141 (range −1.36 to 0.70). Generation 4 LAP2 mice showed increased drinking relative to generation 3, which was a negative response to selection. The R/S calculation is very sensitive to negative selection response, and therefore produced the large negative heritability value −1.36 for the 3rd generations; this was greater than 4 standard deviations less than the mean of the others. Disregarding that value, the mean heritability was 0.308 for the first 10 generations.
Figure 2.
Mean ethanol intakes in selected HAP2 (triangles) and LAP2 (circles) lines of mice show a robust response to bidirectional selection for alcohol intake in a 24-hr access 2-bottle choice procedure. Females (filled) are shown separately from males (open). Generations 0 (progenitor) through 32 (selected) are illustrated here.
The degree of inbreeding in a mass-selected line such as the HAP and LAP can estimated with the formula ΔF = 1/(2N) where N = number of breeders per generation (Falconer and Mackay 1996). The cumulative inbreeding coefficient for generations 0–33 is 0.658 and 0.756 for HAP2 and LAP2 lines, respectively. In truth, this formula may somewhat overestimate inbreeding, for although we did not use a rotational selection procedure, we rigorously avoided first-cousin matings.
Line (HAP2 and LAP2) X Sex X Generation ANOVA on ethanol intake in g/kg for generations 1–33 revealed robust effects of Line F(1,5512) = 20.39, p < 0.001, Sex F(1,5512) = 957, p < 0.001, Generation F(31,5512) = 129, p < 0.001 and interactions of Line X Sex F(1,5512) = 683, p < 0.001, Line X Generation F(30,5512) = 181, p < 0.001, Generation X Sex F(31,5512) = 2.02, p = 0.001, and Line X Sex X Generation F(30,5512) = 2.79, p < 0.001. These data are illustrated in figure 2. The female HS/Ibg progenitors drank more than the males t(369) = 7.77, p < 0.001, and this pattern continued throughout selection in the high-drinking lines, while decreasing in low-drinking lines. For example, at generation 33, female HAP2 mice continued to drink more than males (a mean difference of 4.47 g/kg/day), while intakes of LAP2 mice were similar (the sexes differed by only 0.08 g/kg/day. This resulted in an interaction of Line X Sex at this generation, F(1, 133) = 16.58, p < 0.001. Follow-up comparisons showed a strongly significant sex difference in HAP2 mice, p < 0.001, but only marginally significant in LAP2 mice, p = 0.032.
Alcohol preference, expressed as a ratio of total fluid consumed, is usually correlated with ethanol intake in g/kg. Alcohol preference was tightly correlated with ethanol intake in progenitors (Pearson’s; r = 0.899), as well as in S33 (r = 0.973). When aggregated, the correlation across all individuals in all generations was 0.960. Selection for alcohol intake produced a strong correlated response of alcohol preference such that progenitors showed a preference ratio of 0.19 ± 0.01, while generation 33 HAP2 mice showed 0.84 ± 0.01 and S33 LAP2 mice showed 0.04 ± 0.001.
Another correlated trait that may be relevant to ethanol intake is body weight. A Line X Sex X Generation ANOVA on body weight revealed main effects of Line F(1,5515) = 22.45, p < 0.001 and Generation F(31,5515) = 29.24, p < 0.001, but not Sex (p = 0.30). An interaction of Line X Generation F(30,5515) = 30.17, p < 0.001 was also detected, but Line X Sex, Sex X Generation, and Line X Sex X Generation were not significant (ps > 0.87). Significant main effects of Generation were observed when Lines were analyzed separately (collapsed across Sex), HAP: F(31,3027) = 35.38, p < 0.001 and LAP: F(30,2551) = 14.45, p < 0.001. Inspection of data by generation revealed that LAP2 mice weighed more than HAP 2 mice for generations S8–S23, which appeared to drive most of the observed effects (data not shown), as the difference waned later in selection. Mean weight for progenitors was 25.5 ± 0.18 g, while HAP2 and LAP2 weights at generation 33 were 25.5 ± 0.27 and 26.3 ± 0.40, respectively. Independent t-tests between progenitors and both HAP2 and LAP2 in generation 33 showed no difference (ps > 0.11).
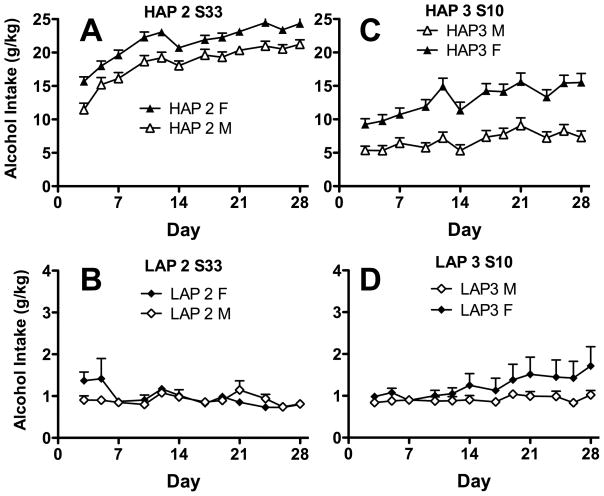
Examining alcohol intake over time during phenotyping revealed that HAP 2 mice were already drinking much more alcohol than LAP 2 mice at the first reading. Furthermore, HAP 2 mice increased alcohol drinking over time, while LAP 2 were generally abstinent throughout the four weeks of access (see Figure 3, Panels A and B). A Line X Sex X Readings ANOVA showed no 3-way interaction, but 2-way interactions of Reading X Line, F (11, 1287) = 47.13, p < 0001, Line X Sex, F (1, 117) = 8.40, p < 0.005, and main effects of Reading and Line, ps < 0.01. Follow up analysis indicated a main effect of Reading in HAP2 mice, F (11, 814) = 76.72, p < 0001, and pairwise comparisons showed that with each reading, HAP 2 mice drank more than the prior reading up through Reading 5, or Day 12 of selection, ps < 0.005. For LAPs, there was also a main effect of Reading, F (11, 495) = 2.36, p < 0.01, but little evidence in pairwise comparisons for a consistent change in level of intake over time, and intakes were generally low and stable. A t-test indicated that lines differed on the first reading, p < 0.001.
Figure 3.
Mean ethanol intake during the four weeks of free-choice access of phenotyping. Triangles depict HAP2 S33 (Panel A) and HAP3 S10 (Panel C) mice, which increased alcohol intake over time. Conversely, diamonds depict LAP2 S33 (Panel B) and LAP3 S10 (Panel D) mice, which maintained low consumption throughout phenotyping.
HAP3/LAP3 Response to Selection
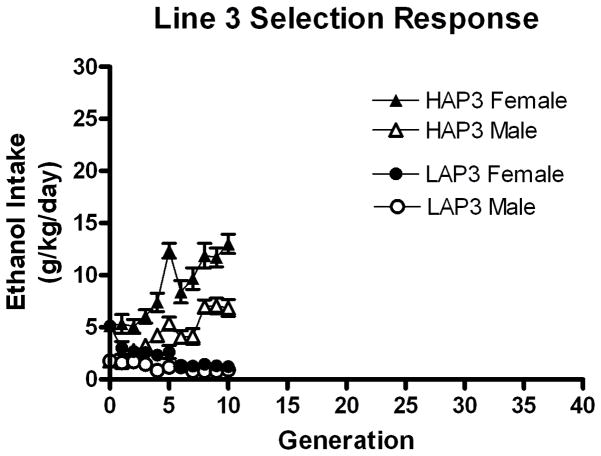
The selection response of HAP/LAP3 generations 0–10 is illustrated in figure 4. The selection response was robust and similar in magnitude to the line 2 mice. Mean heritability for HAP3/LAP3 mice over the first 10 generations was 0.310. The inbreeding coefficient for the same span was 0.224 and 0.218 for HAP3 and LAP3, respectively.
Figure 4.
Mean ethanol intakes in selected HAP3 (triangles) and LAP3 (circles) lines of mice show a robust response to bidirectional selection for high alcohol intake in a 24-hr access 2-bottle choice procedure. Females (filled) drank more than males (open), similar to experiment 1. Generations 0 (progenitor) through 10 (selected) are illustrated here.
Line (HAP3 and LAP3) X Sex X Generation ANOVA on ethanol intake in g/kg for generations 1–10 revealed robust effects of Line F(1,1539) = 761, p < 0.001, Sex F(1,1539) = 188, p < 0.001, Generation F(9,1539) = 11.48, p < 0.001 and interactions of Line X Sex F(1,1539) = 88.02, p < 0.001, Line X Generation F(9,1539) = 21.76, p < 0.001, and Line X Sex X Generation F(9,1539) = 2.06, p < 0.030. The Sex X Generation interaction was not significant (p = 0.14). The selection for line 3 resulted in a similar sex difference as observed in line 2.
Examining alcohol intake over time during phenotyping revealed that as with replicate 2 mice, HAP 3 mice were already drinking much more alcohol than LAP 3 mice at the first reading. Furthermore, HAP 2 mice increased alcohol drinking over time, while LAP 2 were generally abstinent and did not change their intake throughout the four weeks of access. A Line X Sex X Readings ANOVA showed a 3-way interaction, F(11, 1738) = 2.34, p < 0.01. To follow up on this interaction, orthogonal 2-factor ANOVAs of Sex X Reading were conducted on each line. HAP3 mice showed a Reading X Sex interaction, F(11, 847) = 3.02, p = 0.001, while LAPs showed no interaction nor any main effect of Sex or Reading, ps > 0.05. To follow up on the 2-way interaction in HAP3 mice, main effects of reading were assessed in each sex of HAPs, and both showed significance, ps < 0.001, consistent with a general increase in intake over time in both sexes of HAP 3 mice.
The selection of lines 2 and 3 was separated by about 8 years. In spite of presumed allelic fixation in the progenitor HS/Ibg line maintained at Boulder, and some environmental differences, the response to selection between replicate lines over the first 10 selected generations was very similar. A Replicate (2 or 3) X Selection (high or low) X Sex X Generation ANOVA including HAP2/LAP2 and HAP3/LAP3 generations 1–10 on ethanol intake did not detect a main effect of Replicate (p = 0.85) nor interactions with Selection, Sex, Sex X Selection, Selection X Generation, Sex X Generation, or Selection X Sex X Generation (ps > 0.16). This analysis did detect a significant interaction of Replicate X Generation F(9,3646) = 48.32, p = 0.035 which appeared to be driven by unusually high HAP3 drinking in generation 5.
The similarity in response to selection was corroborated by linear regression of Replicate Line (line 2 or 3) and Selected Line (HAP or LAP) of mean ethanol intake per day. This analysis showed no difference in the rate of response to selection between Replicate lines. The slope (± 95% confidence interval) for the HAP2, HAP3, LAP2, and LAP3 lines were 0.84 ± 0.17, 0.72 ± 0.24, −0.14 ± 0.10, and −0.16 ± 0.06 g/kg/day/generation, respectively (rs > 0.75). Overall, these results point towards a reliable and replicate outcome of selective breeding for differences in alcohol drinking from the HS/Ibg progenitor.
cHAP Response to Selection
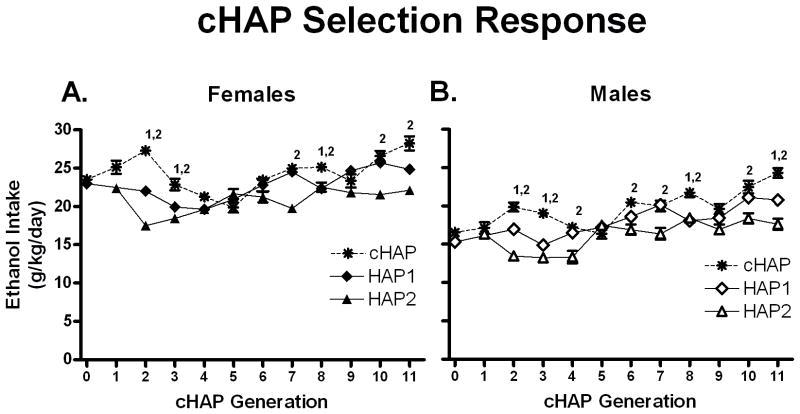
HAP1 generation 27 and HAP2 generation 21 were crossed to create cHAP progenitors. Analysis of the efficacy of this selection involves comparison between the cHAP lines and the two parental lines, all three of which continued to have selection pressure applied. Of interest was whether cHAPs showed increases in drinking relative to the two parental lines, consistent with the hypothesis that a longer list of high-drinking alleles is present in the cHAP line than in either parental line. Therefore the cHAP generations 0–11 were compared with the corresponding generations of HAP1 (28–39) and HAP2 (22–33). A Line (cHAP, HAP1, or HAP2) X Sex X Generation ANOVA revealed main effects of Line F(2,2589) = 457, p < 0.001, Sex F(1,2589) = 974, p < 0.001, Generation F(12,2589) = 46.89, p < 0.001, and interactions of Line X Generation F(20,2589) = 68.38, p < 0.001, Sex X Generation F(12,2589) = 5.87, p < 0.001, and Line X Sex X Generation F(20,2589) = 1.94, p = 0.007. The interaction of Line and Sex was not significant (p > 0.20). Data were stratified by Sex, and differences by Line were detected by t-tests within generation. Figure 5, panels A and B, show the response to selection in cHAP females and males, respectively. cHAP females drank more ethanol than HAP1 females in 3 different generations, and differed from HAP2 females in 6 different generations. These differences were first apparent at generation 2. As stated above, HAP1 and cHAP mice were always tested at the same time and in the same room. cHAP males drank more ethanol than HAP1 males in 4 different generations, and differed from HAP2 males in 8 different generations, with differences also first appearing at generation 2.
Figure 5.
Note that cHAP generations 0–11 correspond to HAP1 gen. 28–39 and HAP2 gen. 22–33. A. Mean ethanol intakes of female cHAP mice (asterisks), the selected descendents of a HAP1 X HAP2 cross, were higher than either line in some generations for gen. 0–11. B. Male cHAP mice did not drink as much female cHAP mice, but still exceeded both progenitor lines in 4 generations (1 = cHAP differs from HAP1, 2 = cHAP differs from HAP2, p < 0.00454).
To compare rate of selection response, linear regression was performed on g/kg/day ethanol intakes by generation and compared to corresponding HAP1 and HAP2 generations. Regressions were each forced to the means of the equivalent generations to cHAP progenitor (HAP1 gen. 28 and HAP2 gen. 23). Generation 23 was used in HAP2, as generation 22 data were not recorded. Slopes were calculated as mean ± 95% confidence interval, which were 0.38 ± 0.19, 0.28 ± 0.18, and −0.30 ± 0.14 g/kg/day ethanol intake for cHAP, HAP1, and HAP2 respectively. cHAP slope did not differ from HAP1, but it was steeper than the slope for HAP2. The mean ethanol intake for HAP2 generations 21 and 23 was unusually high, which created an apparent negative response to selection. Most likely, the overall pattern of selection response for line 2 over this period reflects a non-response to selection that would be expected given prior fixation of high-drinking alleles in this population.
As in the previously reported HAP1 selection and HAP2 selection reported here, females drank substantially more than males. Generally, HAP1 mice drank more ethanol than HAP2, and cHAP drank more than both of them, although differences were not apparent at every generation. Response to selection appeared to be stronger initially in female cHAPs vs. male cHAPs, but that difference lessened in later generations. Interestingly, cHAP drinking appears to be increasing in the last 3 generations, which suggests that trait-relevant allelic fixation is not complete.
Saccharin Intake
Overall, findings from saccharin drinking studies confirmed the association between alcohol consumption and sweet fluid intake. Specifically, selection for differences in alcohol drinking resulted in differences in saccharin intake, and all high-drinking lines drank more than the HS/Ibg progenitor stock. The analyses below support and expand these statements.
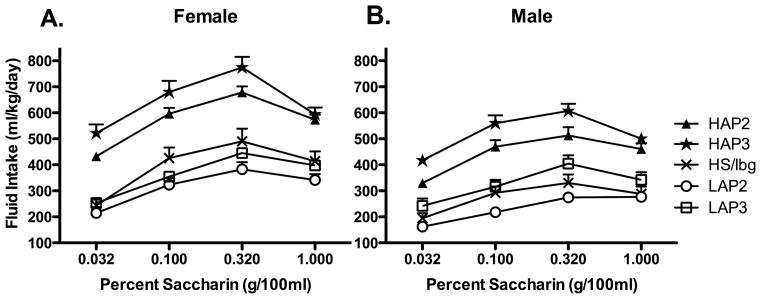
Our first question was whether selection for differences in alcohol preference yields differences in saccharin intake. This analysis focuses on replicate 2 and 3 lines. HS/Ibg lines are unselected, and therefore were not included in the overall analysis, while cHAPs are dealt with in separate analyses (see below). Saccharin intake of these lines is shown in Figure 6. A mixed Line (HAP, LAP) X Sex X Replicate (2, 3) X Concentration (0.032, 0.1, .32, and 1.0%) ANOVA on saccharin consumption revealed no 3- or 4-way interactions. Significant 2-way interactions were Line X Sex, F(1, 109) = 4.32, p < .05, Concentration X Line, Concentration X Replicate, and Concentration X Sex, Fs(3, 327) ≥ 3.96, ps ≤ 0.01. There were also main effects of Line, Sex, and Replicate, Fs(1, 109) ≥ 21.83, ps ≤ .001, as well as a main effect of Concentration, F(3, 327) = 224.32, p < 0.001. These effects indicate that overall, HAP mice drank more than LAP mice. Further examination of the data suggests that the Concentration X Sex interaction was driven by the females being more sensitive to changes in concentration than males. These data are broadly supportive of saccharin intake as a correlated response to selection for alcohol preference.
Figure 6.
Saccharin consumption in 4 selected lines and the non selected HS/Ibg mice. Alcohol preference generally predicted saccharin preference. A. Female mice: generally, selectively bred HAP lines drank more than LAP and HS/Ibg lines; LAP 2 mice drank less than HS/Ibg, while LAP 3 mice differed only from HAP3, not HS/Ibg. B. Male mice generally drank less than female mice, but broadly speaking, showed similar genetic differences in saccharin intake as female mice. Statistical tests are not illustrated here for figure clarity.
Our second question was whether selection altered saccharin intake, comparing each of the selected lines back to the unselected progenitor line, HS/Ibg. Target data for these analyses are also shown in Figure 6, panels A and B, excepting HAP 1, which is depicted in Figure 7. We excluded the cHAP from these analyses because their progenitor lines are HAP1 and HAP2, rather than HS/Ibg. To assess this divergence, we performed a series of orthogonal contrasts composed of Line (each selected vs. HS/Ibg) X Sex X Concentration ANOVAs. In the HAP lines, these ANOVAs revealed strong effects of Line for all 3 HAP lines, ps < 0.001, as well as Line X Concentration interactions for all but HAP 2 mice, ps ≤ 0.01. Main effects of Sex indicated that female mice drank more saccharin than male mice, ps < 0.001. Follow-up t-tests indicated that collapsed across Sex, all HAP lines differed from HS/Ibg at all concentrations of saccharin, ps < 0.001. For the LAP lines, the picture was not as straightforward. The ANOVA comparing LAP 2 to HS/Ibg showed a main effect of Line, F (1, 74) = 6.00, p < 0.02 and Sex, F (1, 74) = 16.24, p < 0.001, as well as a Concentration X Line interaction, F (3, 222) = 5.12, p < 0.005. Collapsing across Sex, t-tests indicated that LAP 2 mice drank less saccharin at the 0.10 and 0.32% concentrations, ps ≤ 0.025. On the other hand, LAP 3 mice did not differ from HS/Ibg, as shown by no effects of Line or Line X Concentration interactions, ps >0.14, although overall, females continued to drink more than males, F (1, 55) = 5.87, p < 0.02. These findings indicate that selection for high alcohol intake more reliably elevates saccharin consumption than selection for low intake depresses it, although it is quite possible that a correlated response will emerge in LAP 3 mice with continued selection.
Figure 7.
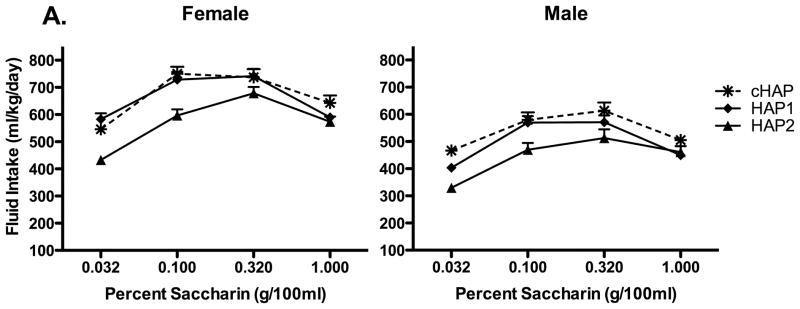
Saccharin consumption in the cHAP line and its progenitors, HAP 1 and HAP 2. Consistent with their higher intake of alcohol, cHAP mice drank more saccharin than HAP 1 mice at 0.032 and 1% saccharin, and more than HAP 2 at all concentrations tested. Again, female mice drank more overall than male mice.
Given that cHAP mice drank more alcohol than its progenitors, HAP 1 and HAP 2, a third question for these data was whether cHAP mice would also show a correlated response of higher saccharin intake than its progenitor lines. These data are broken out in Figure 7. A Line (cHAP, HAP 1, or HAP 2) X Sex X Concentration ANOVA showed a significant 3-way interaction, F (6, 342) = 2.23, p < 0.05, as well as Concentration X Line and Concentration X Sex interactions, ps < 0.001, and main effects of Concentration, Line, and Sex, ps < 0.001. To follow up on the 3-way interaction, we compared cHAP to each of its progenitor lines separately in separate Line X Sex X Concentration ANOVAs. These analyses showed that cHAP mice drank more than both progenitor lines, as shown by main effects of Line, Fs (1, 76) ≥ 21.78, ps < 0.001, as well as Line X Concentration interactions, Fs (3, 228) ≥ 7.80, ps < 0.001, and main effects of Sex, Fs (1, 76) ≥ 36.54, ps < 0.001. In neither case were 3-way interactions of Line X Sex X Concentration significant, so t-tests were used to compare cHAP to each of its progenitors at each saccharin concentration. These showed less intake of saccharin in cHAP mice than HAP1 mice at 0.032% saccharin, t(78) = 5.55, p< 0.001, but greater intake at 1% saccharin, t(78) = 2.05, p < 0.05. The other concentrations did not differ, ps > 0.50. The cHAP line drank more saccharin than the HAP2 line at all concentrations tested, ts(78) ≥ 2.13, ps < 0.05. These results show that overall, the higher alcohol intake in the cHAP line was accompanied by a correlated response of higher saccharin intake.
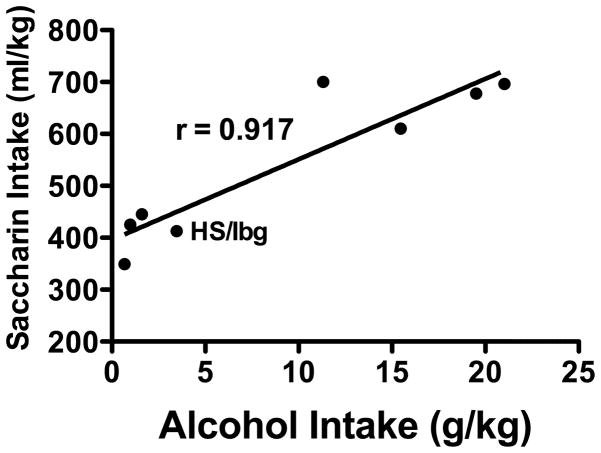
To obtain a global picture of the strength of the genetic correlation between alcohol intake and saccharin intake, we took the line mean of alcohol intake for the generation that corresponded to the one at which we assessed saccharin intake, and performed a bivariate correlation on the corresponding mean for 0.32% saccharin intake, the highest-consumed concentration in most lines. To increase power, we included all lines in the current manuscript, as well as the already-published data on LAP1 mice, resulting in 8 lines for the analysis. This showed a strong correlation between intake of 10% ethanol and 0.32% saccharin of r = 0.917, p < 0.001 (see Figure 8).
Figure 8.
The genetic correlation between free-choice 10% alcohol drinking and 0.32% saccharin intake. Each point represents a line mean for alcohol and saccharin intake assessed at the same generation. All seven lines assessed in this paper (HAP 1–3, HS/Ibg, and LAP 2–3) were included, along with the previously-published data on LAP 1 mice to create this regression model with 8 points.
Finally, we examined water intake conducted with a single water bottle, following the saccharin test. Water intake for each line during this period was HAP 1: 349 ± 11.3 (mean ml/kg ± SEM), HAP 2: 317 ± 10.6, HAP 3: 301.1 ± 14.9, cHAP: 351 ± 12.2, HS/Ibg: 245.8 ± 12.2, LAP 2: 222 ± 5.00, LAP 3: 291.4 ± 14.9. A Line (cHAP, HAP1, 2, 3, HS/Ibg, LAP 2, 3) X Sex ANOVA showed no interaction, but did show main effects of Line, F(6, 221) = 26.48, p < 0.001 and Sex, F(1, 221) = 42.24, p < 0.001. T-tests further assessed whether each line differed from the non-selected HS/Ibg line. These showed that all the high-drinking lines (cHAP, HAP 1, HAP 2, and HAP 3) mice all drank more water than HS/Ibg, ps ≤ 0.01. LAP 2 mice did not differ from HS/Ibg in water intake, while LAP 3 mice drank somewhat more than HS/Ibg, t(57) = 2.27, p < 0.05. These findings imply that selection for high alcohol consumption increases water consumption when that is the only source of fluid, but that selection for low drinking has relatively little effect. Because water intake was affected by selection for alcohol drinking differences in some of the replicates, we inquired whether saccharin intake continued to differ between HAP and LAP lines when differences in water intake were taken into account. Replicate 2 mice showed the largest difference in water intake, so we assessed the difference in saccharin intake between HAP2 and LAP2 mice when that intake was expressed as a percentage of water intake (percent = [saccharin intake/water intake] × 100). A Line X Sex X Concentration ANOVA indicated a main effect of Line, F(1, 75) = 26.84, p < 0.001, as well as Concentration, F(3, 225) = 174.03, p < 0.001. T-tests showed that HAP2 mice drank more than LAP2 mice at all concentrations tested (ps < 0.001), with saccharin intake peaking in both lines at the 0.32% concentration at 191.6% of water in HAP2 mice, and 148.5% of water intake in LAP2 mice. These data indicate that even if the difference in water intake is taken into account, the lines continue to differ in saccharin consumption.
Conclusions
The result of these selection experiments is the emergence of replicate lines that will be useful in the analysis of correlated responses, as well as in testing of behaviors where high alcohol consumption is a useful phenotype. Alcohol intakes observed here are among the highest reported in the mouse literature for 24-h, 2-bottle choice. That this high-drinking behavior is observed in outbred lines, as opposed to a single inbred strain, makes the use of these animals important when investigators wish to obtain results that are representative of wider populations (Crabbe et al. 1990; Grahame 2000). These findings show that the derivation of High- and Low- Alcohol Preferring selected lines is highly replicable, and consistent in its progression each time these lines are derived. This further indicates that as the HAP 3 line continues to respond to selection, its intake is likely to approach the other HAP lines that have, by now, reached a plateau for selection. The results of these selections are lines that diverge greatly in alcohol consumption, and include some of the highest average free-choice alcohol intakes observed in mice.
The course of selection and resulting heritability values of about 0.3 are similar to those observed earlier when using the same selection methodology and progenitor population to derive HAP and LAP mice (Grahame et al. 1999). The realized heritability observed here is similar to that seen in the other published study using selective breeding to alter voluntary alcohol intake in mice with concurrent access to 10% ethanol and water (Belknap et al. 1997), although HAP lines drink considerably more than the high line in that short-term selection study. Interestingly, heritability in the present study was over three times that observed using unidirectional selection for high blood alcohol levels in mice following the 2-h, single bottle drinking in the dark (DID) procedure (Crabbe et al. 2009). There are many possible reasons for differences in heritability, but among them are the bidirectional selection used in the present lines, which would tend to increase heritability scores, or lower measurement error in the present selections that relied on intake averaged over a month, as compared to one that relies on blood alcohol levels assessed after a single 2-h drinking period. Nonetheless, the heritability values obtained here hew closely to those observed among panels of inbred strains tested for 10% alcohol, including a panel of inbreds derived from an F2 cross of C57Bl/6 J X DBA/2J (BXD panel, heritability of 0.34; (Phillips et al. 1994; Rodriguez et al. 1995), a panel of 15 inbred strains (0.23; (Belknap et al. 1993), but lower than a more recent study of 22 inbred strains (0.49; (Yoneyama et al. 2008). Overall, these results suggest that 24-h 2-bottle choice alcohol consumption is more heritable than DID intake. The magnitude of heritability is important when considering whether to initiate a search for alleles responsible for these genetic differences, or when seeking to better understand genetic mechanisms that are responsible for heritable individual differences.
In spite of being a useful description of how these lines responded to selection, heritability should not be regarded as implying a simple, deterministic relationship between altered allele sequence and behavior, for a number of reasons. First, we cannot distinguish here between genetic and epigenetic sources of heritable variance. It remains quite possible that experience with ethanol alters parental behavior, physiology, or epigenetics in a way that induces enduring and heritable change in offspring. Second, the heritability estimate here was obtained in a particular population (HS/Ibg) under a particular set of environmental circumstances. Changes in the environment would likely alter the estimate that we obtained in unpredictable ways. In other words, a heritability score doesn’t specify the extent to which these behavioral observations are deterministic, because the importance of environmental or epigenetic alterations could be quite different in different rearing or environmental conditions. Nonetheless, heritability does provide a way to assess the efficacy of selection: that is, spaced over long periods of time, we were able to repeatedly alter behavior in these animals by applying selection pressure, with largely similar results each time we conducted the experiment. Heritability is also useful when considering whether to proceed with further exploration of the source of these heritable differences. Were heritability scores considerably lower, such explorations would be more difficult to justify.
Novel in the current study was the derivation and selection of a line, the crossed HAP (cHAP) from two already high drinking progenitor lines. Of interest here was whether their alcohol intake would diverge from the parent lines, possibly due to inclusion of high-drinking alleles in this population that were present in only one of the two parent lines, HAP 1 and HAP 2 mice. As shown in Figure 4, this line did drink more than both the parent lines. In fact, cHAP mice, which by S11 consumed 24.3 (male) to 28.2 (female) g/kg per day of 10% ethanol drink much more than the widely used C56BL/6J (B6) strain, more 10% ethanol than the hybrid B6 X NZB F1 cross discovered by Blednov and colleagues (2005), and more 10% ethanol than any of the other F1 inbred strain crosses tested by this group consuming a range of concentrations from 3–36% (v/v) (Blednov et al. 2010). Thus, the cHAP line is at least a candidate for the highest ethanol consuming mouse population known with respect to 24-h, two-bottle choice access. At some point, it seems possible that these mice will have reached a “ceiling effect” for alcohol intake – a point at which alcohol consumption is self-limiting, due to pharmacological or taste factors. Such a ceiling might explain why cHAP mice do not more consistently differ from their parent lines. At the same time, it isn’t clear where such a ceiling would be, as these lines already drink considerably more than they might be expected to metabolize easily. Furthermore, without characterization of blood alcohol levels for all lines, we can’t be sure how close these animals are to reaching ethanol concentrations that might alter the capacity for further ethanol consumption. Future studies are needed to examine behavior and blood ethanol concentrations of these animals during free-choice consumption.
While our working hypothesis for the high intake in the cHAP line is that our crossing and subsequent selection procedure allowed for the inclusion of more high-drinking alleles than would be possible in any single selected line, another possibility is heterosis, or overdominance. In overdominance, heterozygotes show superiority over either homozygote. Such superiority (in this case, higher alcohol consumption) has been shown for alcohol drinking, as demonstrated by Blednov and colleagues who showed overdominance in the F1 cross of B6 X FVB and B6 X SJL strains (2010). While we cannot discard this possibility, the pattern of our findings is not entirely consistent with overdominance. As in the Blednov study, evidence for overdominance would be expected right away in the F1 cross, but in the case of the cHAPs, the F1 cross of HAP 1 X HAP 2 drank no more than the parent lines. Instead, their superiority relative to the parent lines emerged in the S1 and S2 generations, and continued (albeit somewhat intermittently) through the 11 generations of selection reported here. The fact that selection pressure was required to see the emergence of cHAP superiority suggests accumulation of alleles, rather than overdominance, is responsible for the high intake in this line. This is because heterozygosity would be presumed most prevalent at the time of the initial cross, and would tend to decline thereafter due to inbreeding and its concomitant fixation of alleles. However, we should note that selection in cHAPs may have acted to preserve heterozygosity, as has been argued by others (Phillips et al. 2002). Regardless of cause, the present outcome supports the idea that crossing and subsequent selection from the cross of two outbred, selectively bred lines results in a superior phenotype than observed in either parent line.
Thus far, detailed QTL analysis of drinking in HAP and LAP mice has only been completed for the replicate 1 set (Bice et al. 2006; Bice et al. 2009). However, QTLs on Chromosomes 2 and 9 found in B6 X DBA/2J crosses as well as replicate 1 HAP and LAP mice were also confirmed as present in replicate 2 mice (Bice et al. 2006). Certainly, these QTLs have a large effect size, which is likely what has made them detectable thus far. While our supposition is that the cHAP line may have a longer list of QTLs than either parent line alone, it is highly likely that these additional loci would not be detectable in a conventional QTL screen that relies on substantial effect size for each individual QTL to allow detection (Flint et al. 2005). Presumably, the loci with the strongest effect sizes would be captured fairly early in selection, meaning that those alleles differing between HAP 1 and HAP 2 would tend to be the ones with smaller effect sizes. Thus, it would be very difficult to directly assess whether the cHAP line actually contains alleles unique to HAP 1 or HAP 2, or to determine the loci of those alleles.
The second key finding in the present study is the replication of the previously observed genetic association between alcohol intake and saccharin intake. This association has been observed previously in lines of rats and mice selectively bred for differences in alcohol intake (Stewart et al. 1994; Grahame et al. 1999; Kampov-Polevoy et al. 1999; Foroud et al. 2002; Tampier and Quintanilla 2005). On the other hand, additional studies suggest that genetic differences in alcohol drinking in animals (Agabio et al. 2000) or differences in family history of alcoholism in humans (Kranzler et al. 2001; Tremblay et al. 2009) do not always predict differences in sweet preference. Another recent study in 5–12-year old children showed that a family history of alcoholism in addition to self-reported symptoms of depression were required to see shifts in sucrose preference towards sweeter solutions (Mennella et al. 2010). Speaking to the complexity of these sorts of cross-species comparisons, Lu et al. (2005) showed that while variations in taste receptor sequence account for variation in saccharin avidity in mice, they appear to play little role in behavioral differences in rats.
While the current study supports the predominant findings in the literature that saccharin and alcohol consumption are genetically correlated, clearly the literature doesn’t always support that perspective, and the reasons for differences between species and laboratories in the existence of this covariation aren’t always clear. The association has also been observed across inbred strains that vary spontaneously in these phenotypes, although the strength of the genetic correlation assessed in this manner has been much more modest than that seen here (83%), with covariance ranging from 8–15% (Phillips et al. 1994; Yoneyama et al. 2008). It is not clear why observed covariation in this trait is less in inbred strains, although the present association may be caused in part by the use of divergent selected lines which enriched correlations by inclusion of extreme scores. Nonetheless, every high drinking line we tested here showed higher intake than the HS/Ibg progenitor as well as the low-drinking LAP lines. Selection for low drinking only lowered saccharin consumption in one of the two LAP lines tested (LAP 2), although this difference could still emerge in LAP 3. Nonetheless, these data are the first indicating that selection for low alcohol intake results in a correlated response of suppressed saccharin intake relative to a progenitor line. Many of the high-drinking lines showed increased intake of plain water as compared to the HS/Ibg line as well as the LAP lines, suggesting that selection for alcohol preference may have affected overall fluid intake, though the magnitude of the effect on water intake was less than changes in saccharin intake, and much less than differences in alcohol intake. Moreover, changes in saccharin intake following selection were often concentration-dependent. Interestingly, cHAP mice only drank more than HAP 1 mice at the highest saccharin concentration. We note that the shift in saccharin intake occurring only at the highest concentrations is similar to previous studies in which a family history of alcoholism (Kampov-Polevoy et al. 1997; Kampov-Polevoy et al. 1999), or a family history of alcoholism plus depression symptoms (Mennella et al. 2010) is most likely to influence preference for the sweetest sucrose solutions.
We should add that by presenting a genetic correlation between saccharin and alcohol intake, we do not seek to deny other inputs. As with any genetic correlation, these findings should be thought of as particular to the environment and genetic background in which these observations were conducted. We did not manipulate environment other than by altering saccharin concentration, and were therefore unable to examine more complex interactions than the simple bivariate correlation reported here. However, as with any correlation, the current findings are useful in allowing predictions. Given a constant environment, heritable sources of variation in alcohol intake are useful in predicting differences in sweet intake, implying an overlap at the physiological level between heritable factors that alter alcohol drinking and those that alter saccharin consumption.
While differences in saccharin intake between the lines may be due to differences in taste receptor function, other behaviors not so easily chalked up to taste sensitivity, such as impulsivity and cocaine self-administration, also covary with differences in saccharin consumption (Carroll et al. 2008; Oberlin and Grahame 2009), in that lines with higher saccharin intake are more likely to show a pro-addictive phenotype. Such behavioral results indicate that the genetic association between alcohol drinking and saccharin intake likely involves more than just the tongue, and indicate common neurobiological responses to ethanol and saccharin.
Evident with both saccharin and alcohol was higher intake in female mice. Greater saccharin and alcohol intake was observed in female progenitor HS/Ibg line mice, as well as in females of the HAP and LAP lines throughout selection, although the magnitude of the effect tended to diminish as selection progressed. The magnitude of the sex difference was strikingly similar between replicate 2 and 3 lines. The appearance of this sex difference in lines that presumably do not drink alcohol to pharmacologically relevant levels (i.e., LAP and HS/Ibg) as well as the high-drinking lines, means its presence is not related specifically to genes that contribute to high alcohol consumption. Additionally, the fact that the trait is present prior to selection indicates that if anything, selection tended to minimize sex differences as both sexes were brought further and further towards a floor for alcohol consumption (in the LAP lines) or towards the highest intake we could obtain (in the cHAP line), although the sex difference, relatively speaking, persisted more in the HAP lines than the LAP lines. The most reasonable explanation for this asymmetry is that LAP lines are essentially drinking the lowest detectable amount of alcohol, such that a floor effect would minimize any observable differences between the sexes. Sex differences were somewhat larger for saccharin drinking than alcohol drinking. Presumably at some point, alcohol consumption begins to be limited by its pharmacological effects, but this is not true for saccharin drinking. This may allow the full magnitude of sex differences in intake phenotypes to be revealed in saccharin, but not ethanol intake.
While saccharin drinking was the only correlated response we examined in this manuscript, a recent meta-analysis suggests a strong likelihood that the differences in alcohol intake between HAP and LAP lines would also be reflected in line differences in (in descending order of tightness of genetic correlation) operant oral self-administration, conditioned taste aversion, and conditioned place preference (Green and Grahame 2008). Indeed, we have previously shown the expected line difference in conditioned taste aversion, with HAP replicate 1 and 2 mice being less sensitive to this measure of the aversive effects of ethanol than LAP 1 and 2 mice, respectively (Chester et al. 2003). The availability of this array of lines, including two complete sets of HAP and LAP lines, will allow investigators to reliably assess which traits are genetically correlated with free-choice alcohol consumption. Following the suggestions of Crabbe and colleagues that replicate lines be used to increase confidence in genetic correlations (1990), investigators will be able to use replicate 2 and 3 HAP and LAP lines to uncover these associations. Given the high volitional intake of the HAP lines, and especially the cHAP line, we hope that investigators will seek to utilize them in studies where high alcohol intake is useful. These include investigation of potential pharmacotherapies for alcoholism, as well as studies where continuous, high-dose exposure to ethanol is required, such as ethanol toxicology and teratology. We note that the volitional alcohol intakes reported here are in the same range as those reported to lead to dependence and withdrawal in liquid diet studies using C57Bl/6J mice (e.g., Jacquot et al., 2008), but without the potential interpretational issues that arise from liquid diet experiments.
In conclusion, the current findings add to the list of selected lines divergent in free-choice alcohol intake that are available to researchers interested in using animal models of voluntary alcohol consumption. Alcohol intakes reported here are among the highest known in murine models of alcoholism, and these lines will likely prove useful in studies of causes and effects of high alcohol intake. These results also strongly buttress previous findings of a genetic correlation between alcohol intake and intake of sweet solutions. Future studies will focus on further characterization of these novel lines.
Acknowledgments
This work was supported by the Indiana Alcohol Research Center (P60 07611) to David Crabb, U01 AA13483 to NJG, and F31 AA016430 to BGO. Thanks are also extended to Lawrence Lumeng, who initiated selection of replicate 1 HAP and LAP mice.
Bibliography & References Cited
- Agabio R, Carai MA, et al. Dissociation of ethanol and saccharin preference in sP and sNP rats. Alcohol Clin Exp Res. 2000;24(1):24–29. [PubMed] [Google Scholar]
- Belknap JK, Crabbe JC, et al. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology (Berl) 1993;112(4):503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Richards SP, et al. Short-term selective breeding as a tool for QTL mapping: ethanol preference drinking in mice. Behav Genet. 1997;27(1):55–66. doi: 10.1023/a:1025615409383. [DOI] [PubMed] [Google Scholar]
- Bice P, Foroud T, et al. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm Genome. 1998;9(12):949–955. doi: 10.1007/s003359900905. [DOI] [PubMed] [Google Scholar]
- Bice P, Valdar W, et al. Genomewide SNP screen to detect quantitative trait Loci for alcohol preference in the high alcohol preferring and low alcohol preferring mice. Alcohol Clin Exp Res. 2009;33(3):531–537. doi: 10.1111/j.1530-0277.2008.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bice PJ, Foroud T, et al. Identification of QTLs influencing alcohol preference in the High Alcohol Preferring (HAP) and Low Alcohol Preferring (LAP) mouse lines. Behav Genet. 2006;36(2):248–260. doi: 10.1007/s10519-005-9019-6. [DOI] [PubMed] [Google Scholar]
- Blednov YA, Metten P, et al. Hybrid C57BL/6J x FVB/NJ mice drink more alcohol than do C57BL/6J mice. Alcohol Clin Exp Res. 2005;29(11):1949–1958. doi: 10.1097/01.alc.0000187605.91468.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Ozburn AR, et al. Hybrid mice as genetic models of high alcohol consumption. Behav Genet. 2010;40(1):93–110. doi: 10.1007/s10519-009-9298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Walker D, et al. Perception of sweet taste is important for voluntary alcohol consumption in mice. Genes Brain Behav. 2008;7(1):1–13. doi: 10.1111/j.1601-183X.2007.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr LG, Habegger K, et al. Analyses of quantitative trait loci contributing to alcohol preference in HAD1/LAD1 and HAD2/LAD2 rats. Alcohol Clin Exp Res. 2003;27(11):1710–1717. doi: 10.1097/01.ALC.0000097161.51093.71. [DOI] [PubMed] [Google Scholar]
- Carroll ME, Morgan AD, et al. Selective breeding for differential saccharin intake as an animal model of drug abuse. Behav Pharmacol. 2008;19(5–6):435–460. doi: 10.1097/FBP.0b013e32830c3632. [DOI] [PubMed] [Google Scholar]
- Chester JA, Lumeng L, et al. High- and low-alcohol-preferring mice show differences in conditioned taste aversion to alcohol. Alcohol Clin Exp Res. 2003;27(1):12–18. doi: 10.1097/01.ALC.0000046340.06154.9F. [DOI] [PubMed] [Google Scholar]
- Colombo G. ESBRA-Nordmann 1996 Award Lecture: ethanol drinking behaviour in Sardinian alcohol-preferring rats. Alcohol Alcohol. 1997;32(4):443–453. doi: 10.1093/oxfordjournals.alcalc.a008279. [DOI] [PubMed] [Google Scholar]
- Crabbe JC. Genetic animal models in the study of alcoholism. Alcohol Clin Exp Res. 1989;13(1):120–127. doi: 10.1111/j.1530-0277.1989.tb00296.x. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Metten P, et al. A line of mice selected for high blood ethanol concentrations shows drinking in the dark to intoxication. Biol Psychiatry. 2009;65(8):662–670. doi: 10.1016/j.biopsych.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, et al. Estimation of genetic correlation: interpretation of experiments using selectively bred and inbred animals. Alcohol Clin Exp Res. 1990;14(2):141–151. doi: 10.1111/j.1530-0277.1990.tb00461.x. [DOI] [PubMed] [Google Scholar]
- Crawley JN. What’s Wrong with my Mouse? Behavioral Phenotyping of Transgenic and Knockout Mice. Hoboken, NJ: Wiley-Interscience; 2007. [Google Scholar]
- Dess NK, Badia-Elder NE, et al. Ethanol consumption in rats selectively bred for differential saccharin intake. Alcohol. 1998;16(4):275–278. doi: 10.1016/s0741-8329(98)00010-x. [DOI] [PubMed] [Google Scholar]
- Eriksson K. Ethyl alcohol consumption: valid measurement in albino rats. Science. 1968;161(836):76–77. [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Essex, Addison Wesley Longman 1996 [Google Scholar]
- Flint J, Valdar W, et al. Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet. 2005;6(4):271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- Foroud T, Bice P, et al. Mapping of QTL influencing saccharin consumption in the selectively bred alcohol-preferring and -nonpreferring rat lines. Behav Genet. 2002;32(1):57–67. doi: 10.1023/a:1014459912935. [DOI] [PubMed] [Google Scholar]
- Foroud T, Ritchotte A, et al. Confirmation of alcohol preference quantitative trait loci in the replicate high alcohol drinking and low alcohol drinking rat lines. Psychiatr Genet. 2003;13(3):155–161. doi: 10.1097/00041444-200309000-00004. [DOI] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, et al. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6(7):521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Grahame NJ. Selected lines and inbred strains. Tools in the hunt for the genes involved in alcoholism. Alcohol Res Health. 2000;24(3):159–163. [PMC free article] [PubMed] [Google Scholar]
- Grahame NJ, Li TK, et al. Selective breeding for high and low alcohol preference in mice. Behav Genet. 1999;29(1):47–57. doi: 10.1023/a:1021489922751. [DOI] [PubMed] [Google Scholar]
- Grahame NJ, Li TK, et al. Selective breeding for high and low alcohol preference in mice. Behav Genet. 1999;29(1):47–57. doi: 10.1023/a:1021489922751. [DOI] [PubMed] [Google Scholar]
- Green AS, Grahame NJ. Ethanol drinking in rodents: is free-choice drinking related to the reinforcing effects of ethanol? Alcohol. 2008;42(1):1–11. doi: 10.1016/j.alcohol.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisel JE, Metten P, et al. Mapping of quantitative trait loci underlying ethanol metabolism in BXD recombinant inbred mouse strains. Alcohol Clin Exp Res. 2002;26(5):610–616. [PubMed] [Google Scholar]
- Jacquot C, Croft AP, et al. Effects of the glucocorticoid antagonist, mifepristone, on the consquences of withdrawal from long term alcohol consumption. Alc Clin Exp Res. 2008;32(12):2107–2116. doi: 10.1111/j.1530-0277.2008.00799.x. [DOI] [PubMed] [Google Scholar]
- Kampov-Polevoy A, Garbutt JC, et al. Evidence of preference for a high-concentration sucrose solution in alcoholic men. Am J Psychiatry. 1997;154(2):269–270. doi: 10.1176/ajp.154.2.269. [DOI] [PubMed] [Google Scholar]
- Kampov-Polevoy AB, Garbutt JC, et al. Association between preference for sweets and excessive alcohol intake: a review of animal and human studies. Alcohol Alcohol. 1999;34(3):386–395. doi: 10.1093/alcalc/34.3.386. [DOI] [PubMed] [Google Scholar]
- Kampov-Polevoy AB, Kasheffskaya OP, et al. Pain sensitivity and saccharin intake in alcohol-preferring and -nonpreferring rat strains. Physiol Behav. 1996;59(4–5):683–688. doi: 10.1016/0031-9384(95)02110-8. [DOI] [PubMed] [Google Scholar]
- Kranzler HR, Sandstrom KA, et al. Sweet taste preference as a risk factor for alcohol dependence. Am J Psychiatry. 2001;158(5):813–815. doi: 10.1176/appi.ajp.158.5.813. [DOI] [PubMed] [Google Scholar]
- Le AD, Israel Y, et al. Genetic selection for high and low alcohol consumption in a limited-access paradigm. Alcohol Clin Exp Res. 2001;25(11):1613–1620. doi: 10.1111/j.1530-0277.2001.tb02168.x. [DOI] [PubMed] [Google Scholar]
- Li TK, Lumeng L, et al. Selective breeding for alcohol preference and associated responses. Behav Genet. 1993;23(2):163–170. doi: 10.1007/BF01067421. [DOI] [PubMed] [Google Scholar]
- Lu K, McDaniel AH, Tordoff MG, et al. No relationship between sequence variation in protein coding regions of the Tas1r3 gene and saccharin preference in rats. Chem Senses. 2005;30(3):231–240. doi: 10.1093/chemse/bji019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng L, Hawkins TD, et al. Alcohol and Aldehyde Metabolizing Systems. New York: Academic Press; 1977. [Google Scholar]
- Mardones J, Segovia-Riquelme N. Thirty-two years of selection of rats by ethanol preference: UChA and UChB strains. Neurobehav Toxicol Teratol. 1983;5(2):171–178. [PubMed] [Google Scholar]
- Mennella JA, Pepino MY, et al. Sweet preferences and analgesia during childhood: effects of family history of alcoholism and depression. Addiction. 2010;105(4):666–675. doi: 10.1111/j.1360-0443.2009.02865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JM, Stewart RB, et al. Phenotypic and genotypic characterization of the Indiana University rat lines selectively bred for high and low alcohol preference. Behav Genet. 2002;32(5):363–388. doi: 10.1023/a:1020266306135. [DOI] [PubMed] [Google Scholar]
- Oberlin BG, Grahame NJ. High alcohol preferring mice are more impulsive than low alcohol preferring mice as measured in the delay discounting task. Alcoholism: Clinical and Experimental Research. 2009;33(7):1–10. doi: 10.1111/j.1530-0277.2009.00955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Crabbe JC, et al. Localization of genes affecting alcohol drinking in mice. Alcohol Clin Exp Res. 1994;18(4):931–941. doi: 10.1111/j.1530-0277.1994.tb00062.x. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Shen EH, et al. Forward, Relaxed, and Reverse Selection for Reduced and Enhanced Sensitivity to Ethanol’s Locomotor Stimulant Effects in Mice. Alcohol Clin Exp Res. 2002;26(5):593–602. [PubMed] [Google Scholar]
- Rodriguez LA, Plomin R, et al. Alcohol acceptance, preference, and sensitivity in mice. II. Quantitative trait loci mapping analysis using BXD recombinant inbred strains. Alcohol Clin Exp Res. 1995;19(2):367–373. doi: 10.1111/j.1530-0277.1995.tb01517.x. [DOI] [PubMed] [Google Scholar]
- Sinclair JD, Kampov-Polevoy A, et al. Taste preferences in rat lines selected for low and high alcohol consumption. Alcohol. 1992;9(2):155–160. doi: 10.1016/0741-8329(92)90027-8. [DOI] [PubMed] [Google Scholar]
- Stewart RB, Russell RN, et al. Consumption of sweet, salty, sour, and bitter solutions by selectively bred alcohol-preferring and alcohol-nonpreferring lines of rats. Alcohol Clin Exp Res. 1994;18(2):375–381. doi: 10.1111/j.1530-0277.1994.tb00028.x. [DOI] [PubMed] [Google Scholar]
- Tampier L, Quintanilla ME. Saccharin consumption and the effect of a long-term exposure to a sweetened alcoholic solution in high- (UChB) and low- (UChA) alcohol-drinking rats. Alcohol. 2005;37(1):47–52. doi: 10.1016/j.alcohol.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Tremblay KA, Bona JM, et al. Effects of a diagnosis or family history of alcoholism on the taste intensity and hedonic value of sucrose. Am J Addict. 2009;18(6):494–499. doi: 10.3109/10550490903206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller MB, McBride WJ, et al. Induction of dependence on ethanol by free-choice drinking in alcohol-preferring rats. Pharmacol Biochem Behav. 1982;16(3):501–507. doi: 10.1016/0091-3057(82)90459-2. [DOI] [PubMed] [Google Scholar]
- Waller MB, McBride WJ, et al. Initial sensitivity and acute tolerance to ethanol in the P and NP lines of rats. Pharmacol Biochem Behav. 1983;19(4):683–686. doi: 10.1016/0091-3057(83)90345-3. [DOI] [PubMed] [Google Scholar]
- Yoneyama N, Crabbe JC, et al. Voluntary ethanol consumption in 22 inbred mouse strains. Alcohol. 2008;42(3):149–160. doi: 10.1016/j.alcohol.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]