Abstract
Background and objective
Surfaces and fluids can affect oral bacterial colonization. The aim of this study was to compare re-developing biofilms on natural teeth and dentures.
Methods
Supragingival plaque samples were taken from 55 dentate subjects and the denture teeth of 62 edentulous subjects before and after professional cleaning. Also, samples from 7 “teeth” in randomly selected quadrants were collected after 1, 2, 4 and 7 days of no oral hygiene. Samples were analyzed using checkerboard DNA-DNA hybridization. Counts and proportions of 41 bacterial taxa were determined at each time point and significant differences were sought using the Mann-Whitney test. Ecological succession was determined using a modified moving window analysis.
Results
Mean total DNA probe counts were similar pre-cleaning but were higher in dentate subjects at all post-cleaning visits (p<0.01). Pre-cleaning edentate biofilms had higher counts and proportions of Streptococcus mitis, Streptococcus oralis and Streptococcus mutans, whereas dentate subjects had higher proportions of Tannerella forsythia, Selenomonas noxia and Neisseria mucosa. By 2 days, mean counts of all taxa were higher in natural teeth and most remained higher at 7 days (p<0.01). Succession was more rapid and complex in dentate subjects. Both groups demonstrated increased proportions of S. mitis and S. oralis by 1 day. N. mucosa, Veillonella parvula and Eikenella corrodens increased in both groups but later in edentate samples.
Conclusions
“Mature” natural and denture teeth biofilms have similar total numbers of bacteria but different species proportions. Post-cleaning biofilm re-development is more rapid and more complex on natural than denture teeth.
Keywords: microbiota, biofilms, supragingival, dental plaque, tooth, dentures
INTRODUCTION
The formation of multispecies biofilms is influenced by three major factors; the nature of the surface to which the biofilm adheres, the composition of the potential colonizing species that will make up the biofilm and the bulk fluid(s) that bathe and sustain the biofilm community1. In this communication, we examined the effect of two of those factors, surface (tooth surface × denture surface) and bulk fluid (presence × absence of gingival crevicular fluid), on the sequence of species colonization in “dental” biofilms formed on natural teeth or on the “teeth” of full dentures. This analysis was made possible by comparing the results of two studies that were conducted in parallel in the same laboratory using the same clinical and laboratory methods to examine biofilm re-growth after professional cleaning in the absence of homecare procedures. In one study, Sachdeo et al 2described the changes in numbers and proportions of 41 bacterial taxa in biofilms that formed on the denture teeth of 62 fully edentulous individuals. After an initial professional cleaning, subjects refrained from oral hygiene for 7 days. Total DNA probe counts returned to pre-cleaning levels in about 4 days. Prominent species detected during the early re-colonization period included Streptococcus mitis and Streptococcus oralis. In a parallel study, Uzel et al 3 reported the changes in mean microbial counts of the same 41 taxa during 7 days of supra and subgingival biofilm re-development after professional tooth cleaning in 38 periodontally healthy and 17 periodontitis subjects. It was found that supragingival biofilm re-development was similar in the periodontally healthy and periodontitis subjects and that the mean total DNA probe counts reached pre-cleaning levels by 2 days. Prominent species that increased in total numbers during supragingival biofilm re-development included S. mitis, S. oralis, Capnocytophaga gingivalis, Eikenella corrodens, Veillonella parvula and Neisseria mucosa. Mean species counts were significantly higher after 7 days of no oral hygiene than they were in the mature biofilm harvested prior to professional dental cleaning. In a follow-up paper, Teles et al4 described analytical methods to determine species succession during biofilm re-development. They demonstrated more “significant” differences in species succession during supragingival biofilm development than subgingival biofilm re-development and found supragingival biofilm re-development to be similar in subjects who were periodontally healthy or had periodontitis. Species succession differed more profoundly between health and periodontitis when examining subgingival samples.
The parallel conduct and identical methods employed in these two studies provided an unusual opportunity to examine the effect of surface and bulk fluid on early bacterial re-colonization of “dental” surfaces. The purpose of the present investigation was to compare the changes in bacterial species numbers and proportions on natural teeth and denture teeth over a 7 day test period in the absence of oral hygiene or denture cleaning. In addition, the nature of bacterial succession was determined. Since the subgingival biofilm samples from the dentate subjects were clearly not analogous to the denture samples, data from subgingival samples were not included in this comparison.
MATERIAL AND METHODS
Subject population
The dentate population was described previously3, 4. Periodontally healthy (N=38) and chronic periodontitis (N=17) subjects were recruited at The Forsyth Institute, between February/2003 and July/2006. Included subjects were at least 20 years old, presented more than 20 teeth and were in good general health. Individuals that had periodontal or antibiotic therapy in the previous 3 months, systemic conditions which could affect periodontal disease or treatment, systemic conditions which required antibiotic coverage for periodontal procedures, soft tissue lesions as well as smokers were excluded from the study. The study was approved by The Forsyth Institute Institutional Review Board and all subjects signed informed consent prior to entering the study.
The edentate population was described earlier2. Fully edentulous subjects were recruited in the Department of Oral Medicine at Tufts School of Dental Medicine. The subject population consisted of edentulous subjects who used complete maxillary and mandibular dentures on a daily basis. To be included in the study, subjects had to be over 20 years of age, have been edentulous for at least 1 year and worn complete maxillary and mandibular dentures on a daily basis. Subjects who had received antibiotic therapy in the 3 months prior to the start of the study or who had any oral lesions or a systemic condition that required antibiotic coverage for routine dental procedures were excluded from the study.
Clinical monitoring, professional cleaning, microbial sample-taking and enumeration
Dentate subjects
The clinical monitoring, professional tooth-cleaning, microbial sampletaking and enumeration of bacterial species were described previously3, 4. In brief, after the initial clinical monitoring, samples of mature supragingival biofilms were collected from the mesiobuccal aspect of each tooth present using sterile 11–12 Gracey curettes. All periodontitis subjects received full mouth scaling and root planing (SRP) at a single visit, while periodontally healthy subjects received a dental prophylaxis. Then, immediately after cleaning, supragingival biofilm samples were taken from the mesial aspect of all teeth (excluding third molars) from 55 dentate individuals (38 periodontally healthy and 17 chronic periodontitis subjects). After professional tooth cleaning, subjects refrained from oral hygiene procedures for 7 days. Quadrants in each subject were randomly assigned to be sampled at 1, 2, 4 and 7 days. Up to 7 supragingival samples were taken from all teeth present in the assigned quadrant at those time points. Samples were analyzed individually for their content of 41 bacterial taxa using checkerboard DNA-DNA hybridization5, 6.
Denture-wearing subjects
The sampling and bacterial enumeration were essentially identical for the denture-wearing subjects2. Samples of biofilm were removed from the mesiobuccal aspect of each denture tooth prior to and immediately after denture cleaning. Subjects wore the dentures during sampling procedures. Samples were individually analyzed for their content of 41 bacterial taxa. Subjects were asked to keep their dentures in their mouths overnight and refrain from denture cleaning for 7 days. Quadrants in each subject were randomly assigned to be sampled at 1, 2, 4 and 7 days and 7 samples were taken at each time point to be analyzed individually for their content of 41 bacterial species.
Data Evaluation
Since supragingival biofilm samples did not differ markedly in counts or proportions during biofilm re-development in subjects who were periodontally healthy or exhibited periodontitis 3, 4, the 38 periodontally healthy and 17 periodontitis subjects were combined to form the “dentate” subject group for the following analyses. For all sampling time points, microbial counts of the 41 test species in individual supragingival biofilm samples were assessed separately. The counts for each species were averaged within each subject at each time point and then averaged across subjects in the 2 clinical groups separately. Up to 28 samples were averaged per subject pre tooth-cleaning and post tooth-cleaning, and 7 samples at days 1, 2, 4 and 7. Significance of differences over time (pre-cleaning, post-cleaning, and days 1, 2, 4 and 7) for each species was determined using the Friedman test. Differences between clinical groups for each species at each time point were sought using the Mann-Whitney test. The mean values for each species at each time point in the 2 clinical groups were depicted graphically as “microbial profiles” ordered according to the microbial complexes described previously7.
The % of the total DNA probe count comprised by each of the 41 taxa in each biofilm sample was computed. The % for each species was averaged within each subject at each time point and then averaged across subjects at each time point in the 2 clinical groups separately. Differences between dentate and edentate clinical groups for each species at each time point were determined using the Mann-Whitney test.
Ecological succession was sought in the 2 groups of subjects separately using the proportions comprised by each taxon at the immediate post-cleaning, 1, 2, 4 and 7 day time points. Major increases or decreases in proportion of species were sought over time using a modification4 of a “moving window” approach8. The mean proportions of each species at each time point in each group were compared from the time point immediately after tooth or denture cleaning with the mean proportions of each species in samples from the same subjects at each of the later time points and the “significance” of differences between individual species proportions at different time points tested using a t statistic.
RESULTS
Microbial species proportions in “mature” biofilms on natural and denture teeth
Samples were taken from the mesiobuccal surface of natural teeth and denture teeth prior to professional cleaning. The samples of “mature” biofilm actually represented samples of indeterminate biofilm age since the oral hygiene history of the various tooth surfaces was not known. Although the mean total DNA probe counts prior to cleaning did not differ significantly between clinical groups (see below), there were notable significant differences in mean proportions between the dentate and edentate groups for many test taxa (Fig. 1). S. mitis, S. oralis, Streptococcus sanguinis, Streptococcus mutans, Aggregatibacter actinomycetemcomitans, Fusobacterium periodonticum and Gemella morbillorum were in significantly higher mean proportions on denture teeth than on natural teeth. Four Actinomyces species, Capnocytophaga sputigena, Campylobacter rectus, Campylobacter showae, Neisseria mucosa and Selenomonas noxia were in significantly higher mean proportions in samples from natural teeth.
Fig. 1.
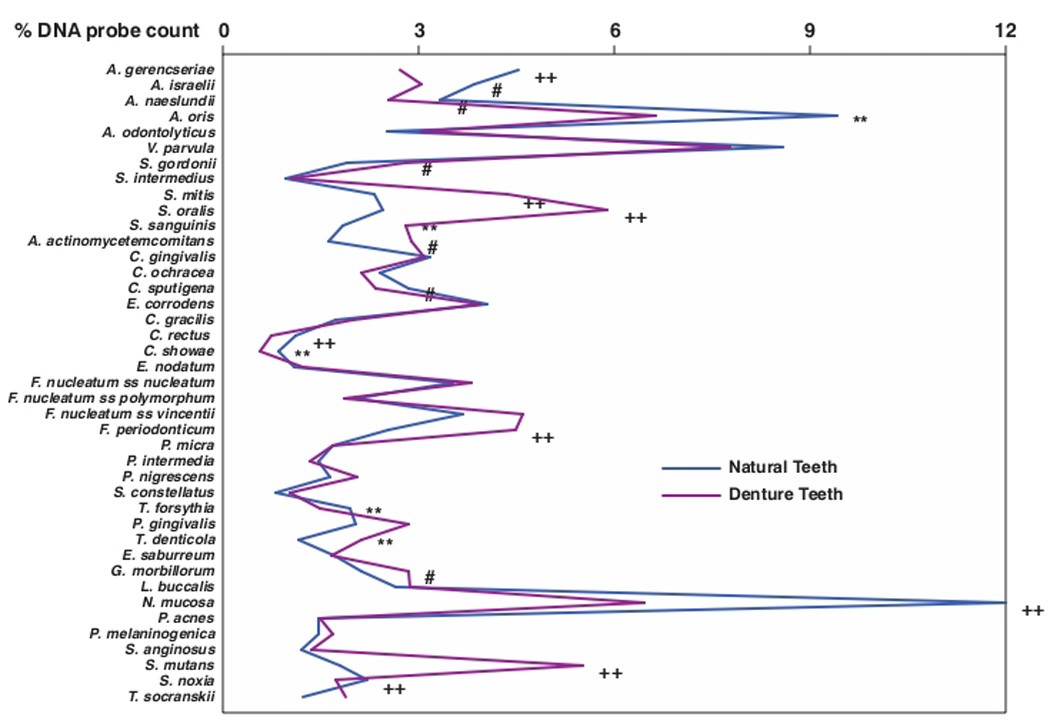
Mean % of the total DNA probe counts of 41 bacterial taxa in samples taken from 55 subjects with natural teeth and 62 subjects with full mouth dentures. Significance of differences in mean species proportions between groups was determined using the Mann Whitney test; # p < 0.05, ** p < 0.01, †† p < 0.001. The species were ordered according to previously described microbial complexes (Socransky et al. 1998)7.
Changes in microbial species counts on natural and denture teeth during biofilm redevelopment
The changes in mean total DNA probe counts of the supragingival biofilm samples obtained from the dentate subjects and the biofilm samples from the denture teeth are presented in Fig. 2. Mean total DNA probe counts of samples obtained prior to professional cleaning did not differ significantly between natural or denture teeth. Mean (× 105, ± SEM) total DNA probe counts were 60.7± 6.3, and 60.0± 6.7 in samples from dentate and edentate subjects respectively. Mean total DNA probe counts exceeded baseline levels at day 2 in the dentate subjects but required greater than 4 days for this to occur on denture teeth. A steady increase in mean total DNA probe counts was observed until day 7 in both groups. Differences between clinical groups in mean total microbial counts were statistically significant (p < 0.01) immediately after prophylaxis and at 1, 2, 4 and 7 days.
Fig. 2.
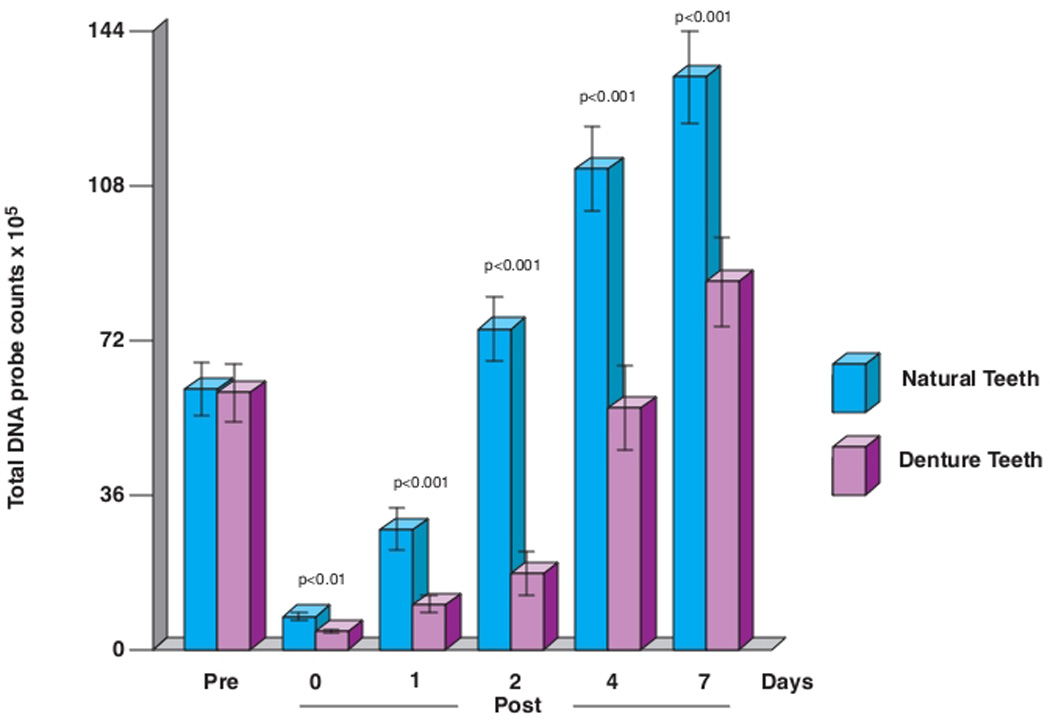
Mean total DNA probe counts (× 105, ± SEM) of supragingival biofilm samples taken at entry (pre-cleaning), immediately post-cleaning and after 1, 2, 4 and 7 days of biofilm development. Significance of differences between clinical groups at each time point was determined using the Mann Whitney test. The bars represent mean total DNA probe counts and the whiskers the SEM at each time point.
There were clear differences in the cumulative plots of mean counts of the individual species on natural and denture teeth (Fig. 3). There was a much more rapid increase in mean counts of individual species in samples from natural than denture teeth in the period starting immediately after cleaning and extending to the 2 day sampling time point. The relative increase in species counts in samples from dentate subjects slowed from 4 to 7 days but the rate increased from 2 to 7 days in the samples from dentures. While the yellow and red complex species showed as great or greater increases in mean counts on denture teeth, mean counts of the remaining complexes were lower.
Fig. 3.
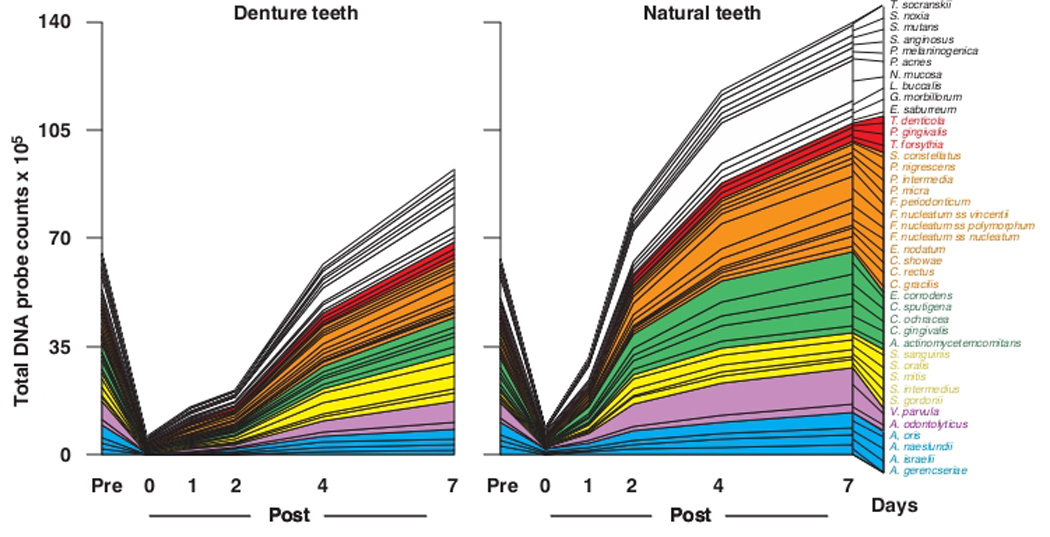
Cumulative mean counts (× 105) of 41 bacterial species in samples taken from 55 subjects with natural teeth and 62 subjects with full mouth dentures. The plots present the cumulative mean values at each time point in each clinical group. The species were ordered and color-coded according to previously described microbial complexes (Socransky et al. 1998) 7.
The mean counts (× 105) of the 41 bacterial taxa in biofilm samples taken at different time points from the natural and denture teeth were compared directly (Fig. 4). Significant differences in mean counts over time were observed for all of the test taxa in both clinical groups (Friedman test, p<0.001). At entry (prior to cleaning), counts of 8/41 taxa were significantly elevated in the dentate samples including Capnocytophaga ochracea, Capnocytophaga sputigena, Campylobacter rectus, Campylobacter showae, Fusobacterium nucleatum ss polymorphum, Tannerella forsythia, Neisseria mucosa and Selenomonas noxia. Before cleaning, 3 species, S. mitis, S. oralis and Streptococcus mutans were significantly higher in mean counts on denture teeth than on natural teeth. Multiple statistically significant higher counts of individual species were observed immediately post-cleaning and at 1 day in samples from natural teeth when compared with samples from denture teeth. They were mostly among Actinomyces and Streptococcus species. By day 2, all mean species counts were significantly higher in the samples from natural teeth than the samples from dentures. At day 7, 28 taxa remained at significantly higher mean counts in the samples from natural teeth than in the samples from dentures. However, the species that were significantly higher in mean counts on denture teeth before tooth cleaning had either reached (S. oralis, S. mutans) or significantly exceeded (S. mitis) mean counts on natural teeth.
Fig. 4.
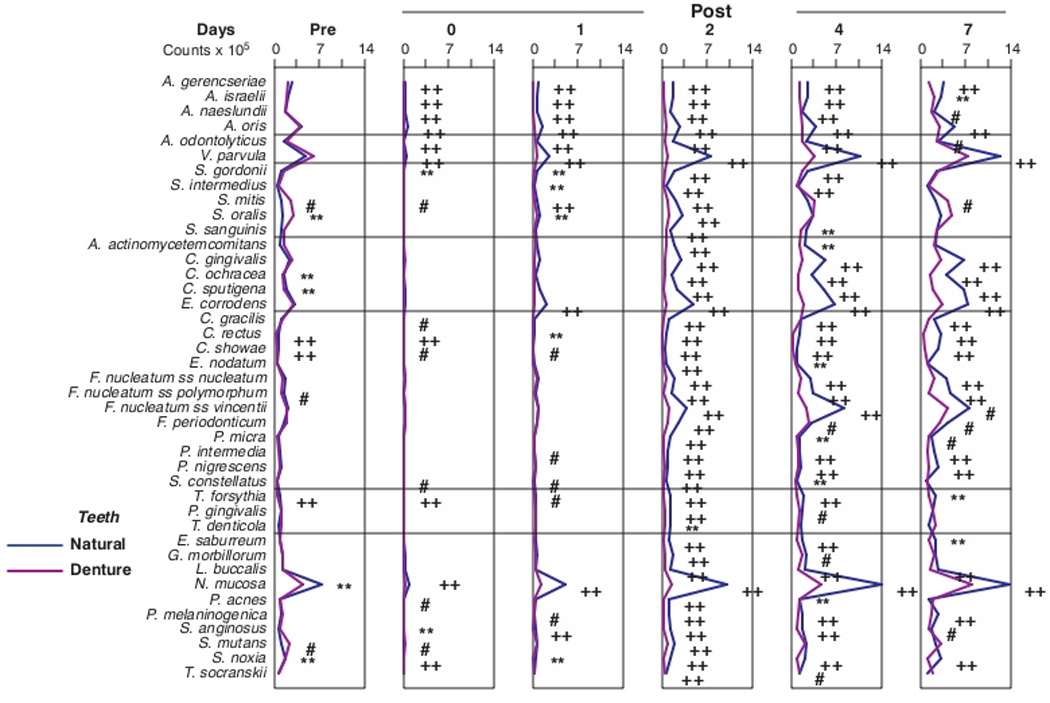
Mean counts (× 105) of 41 bacterial species in samples taken from 55 subjects with natural teeth and 62 subjects with full mouth dentures. Significance of differences in mean species counts between groups at each time point was determined using the Mann Whitney test; # p < 0.05, ** p < 0.01, †† p < 0.001. The species were ordered according to previously described microbial complexes (Socransky et al. 1998) 7.
Bacterial succession on natural teeth or denture teeth during biofilm re-development
Fig. 5 presents mean proportions of 41 bacterial taxa in biofilms obtained from natural or denture teeth immediately after professional cleaning and after 1, 2, 4 and 7 days of biofilm accumulation in the absence of self-performed oral hygiene. On natural teeth, the first species to significantly increase in mean proportions at one day, were V. parvula, S. mitis, S. oralis, E. corrodens and N. mucosa. S. mitis and S. oralis maintained their high proportions at day 2 but began to decrease in proportions thereafter. V. parvula and E. corrodens increased significantly from immediate post-cleaning mean values to higher levels at 1, 2, 4 and 7 days, while N. mucosa increased at 1 – 4 days and declined slowly thereafter. C. gingivalis began to increase significantly at 2 days and continued to increase in mean proportions at 4 and 7 days. C. ochracea and C. sputigena increased significantly in mean proportions at 4 days. At 7 days, significant increases were observed for C. rectus, C. showae, P. melaninogenica and S. noxia. Major and statistically significant decreases in mean proportion were observed for 4 Actinomyces species, E. nodatum, F. nucleatum ss nucleatum, F. nucleatum ss vincentii, P. micra, P. intermedia, S. constellatus, T. forsythia, P. gingivalis, E. saburreum and P. acnes after cleaning.
Fig. 5.
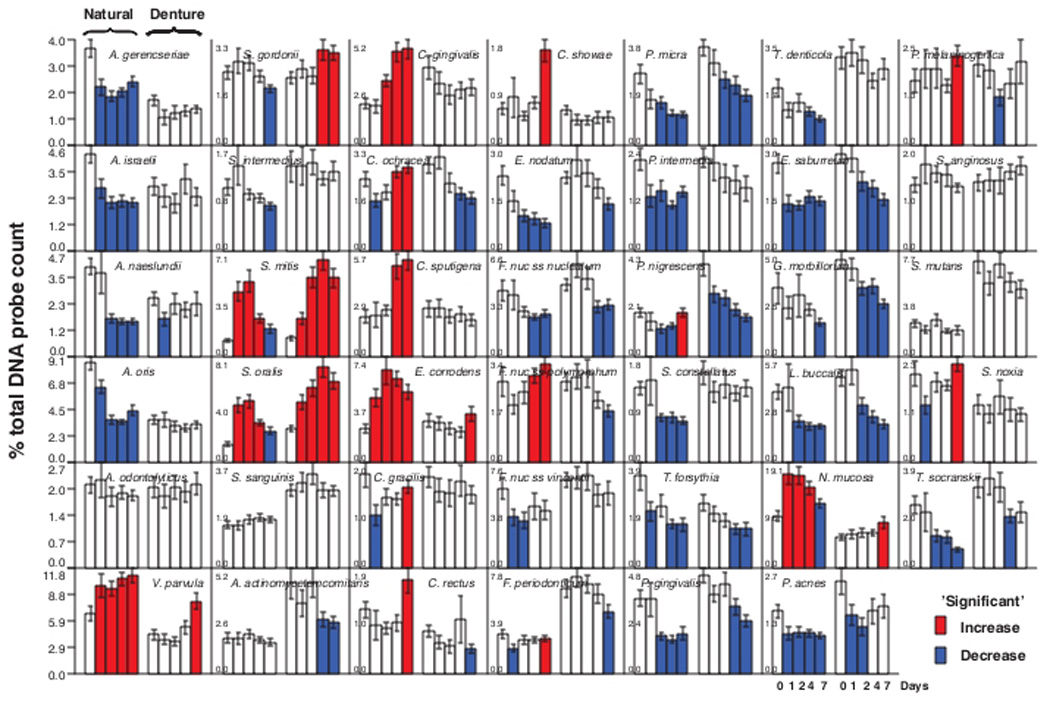
Bar charts of the mean % of the total DNA probe count of 41 bacterial species in samples of biofilm taken immediately after tooth cleaning and after 1, 2, 4 and 7 days of biofilm accumulation in the absence of oral hygiene procedures. Samples were provided by 55 dentate subjects (left set of bars in each panel) and 62 subjects with upper and lower full dentures (right set of bars in each panel). The red bars indicate a significant increase in the mean proportion from a mean value for an earlier time point for that species in that clinical group. In a similar fashion, the blue bars indicate a significant reduction in mean proportion of a species from an earlier time point.
On denture teeth, S. mitis and S. oralis increased markedly and significantly in mean proportions at 1 day and continued to increase but leveled off between 4 and 7 days. S. gordonii showed a significant increase in mean proportions at 4 and 7 days, while V. parvula, E. corrodens and N. mucosa showed significant increases in mean proportions at 7 days. Prominent declines in mean proportions in samples from denture teeth were observed for P. micra, P. nigrescens, Eubacterium saburreum, Gemella morbillorum and Leptotrichia buccalis.
The significant increases in mean proportions of taxa at 1, 2, 4 and 7 days on natural and denture teeth are summarized in Fig. 6. Many more species showed significant increases in mean proportions in samples from natural teeth than denture teeth. S. mitis and S. oralis increased significantly at 1 – 2 days in both clinical groups; but, taxa such as N. mucosa, V. parvula and E. corrodens appeared to be delayed in their proliferation on denture teeth. Other taxa that increased significantly on natural teeth did not show significant increases in mean proportions on denture teeth during the 7 day study. S. gordonii was unique in that it increased significantly on denture but not natural teeth.
Fig. 6.
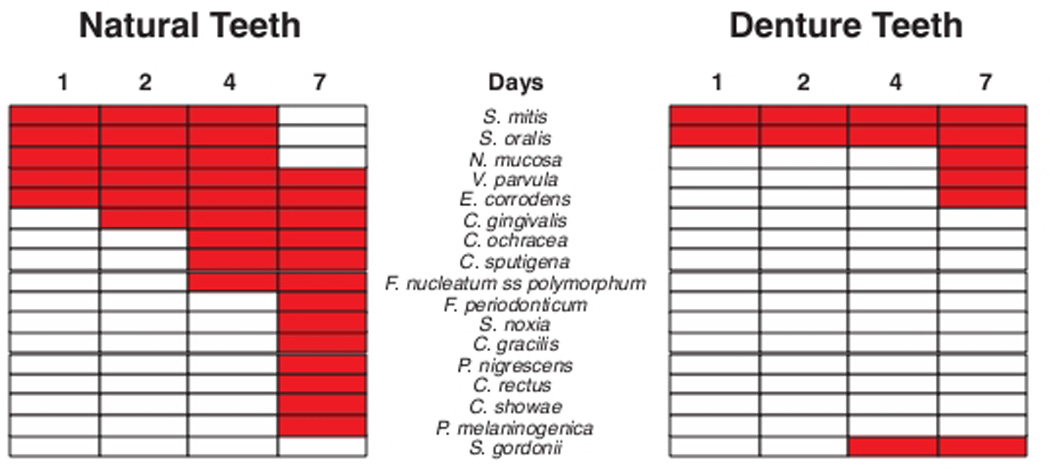
Grid plot summarizing significant increases (red rectangles) in mean proportions of bacterial taxa in biofilm samples obtained from 55 dentate subjects and 62 edentulous subjects who wore full dentures. The data were derived from Fig. 5. The red rectangles indicate a significant increase in mean proportion of that species from mean proportions of that species at an earlier time point. The species were ordered according to the order of significant increases in mean proportions of species in the dentate subjects.
Discussion
The purpose of the present investigation was to compare the changes in bacterial species numbers and proportions, as well as the patterns of microbial succession on natural teeth and denture teeth. In order to accomplish this goal, “supragingival” plaque samples were collected over a 7 day test period in the absence of oral hygiene or denture cleaning and analyzed for their microbial composition using checkerboard DNA-DNA hybridization.
Biofilm formation and composition is intrinsically related to the type of the surface to which it adheres, to the type of fluid bathing this surface (“bulk fluid”) and the microbial species available in the area1. The parallel undertaking of studies examining biofilm re-development on natural and denture teeth facilitated the examination of the effect of two of these factors-surface (tooth surface × denture surface) and “bulk fluid”(presence × absence of gingival crevicular fluid)- on the microbial composition in “mature” biofilms and the nature of bacterial succession during oral biofilm re-development in vivo.
The most obvious difference between biofilm re-development on natural or denture teeth was the rate of re-development which appeared to be far more rapid on natural than denture teeth (Figs. 2 and 3). The greater rate of biofilm re-development on natural teeth may have been due in part to differences in the size of the area of the anatomical surface that was sampled on natural teeth and dentures. This area may have been somewhat larger on the natural teeth than the denture teeth because the curette could extend into the interproximal space in the natural dentition. However, the effect may not have been excessively large because there were no significant differences in mean total counts in the pre-cleaning samples between the 2 clinical groups (Fig. 2).
The different rate of plaque formation in the dentate and edentulous individuals may also be explained, partially, by the nature of the surface to which unbound species adhered. The hydroxyapatite surface presented by natural teeth and the acrylic surface presented by denture teeth differ markedly in their chemical and physical properties as well as surface roughness. In vitro studies suggested that biofilm formation may be influenced by forces determined by the underlying substratum that can be transferred through the pellicle layer 9. Salivary glycoproteins, proline-rich proteins, phosphoproteins, histidine-rich proteins, enzymes and other molecules are some of the components of that film that can function as adhesion sites for bacteria10 and those are the first to adsorb to the tooth surface. Hence, although a surface-specific protein adsorption to restorative surfaces is conceivable9, its effects seem to be limited to pellicle formation and may not extend to biofilm development and maturation. In fact, in vivo studies have shown that protein adsorption to surfaces and bacterial adherence are mostly determined by the surface roughness rather than by other material specific physicochemical properties10,11,12, 13. In addition, it appears that rougher surfaces, including crowns, implant abutments and denture bases accumulate and retain more plaque and, after several days of undisturbed plaque accumulation, they seem to harbor a more mature plaque characterized by an increased proportion of rods, motile organisms and spirochetes10. This is in contrast with the results of the present study, where plaque development on dentures was slower in comparison with that on natural teeth and plaque composition appeared to be less mature.
Other factors that are peculiar to natural teeth might have been responsible for differences in plaque re-development in the two groups. Gingival crevicular fluid (GCF) bathed the crevice of natural teeth but not denture teeth and may have influenced the rate of biofilm redevelopment and bacterial composition. Gingival crevice fluid flow is present in subjects who are periodontally healthy as well as those who exhibit periodontitis. Indeed, the selection criteria for “periodontal health” in this study were based primarily on minimal periodontal pocketing and attachment loss, rather than level of gingival inflammation. A number of studies have shown that more plaque accumulates in the presence of local inflammation in comparison with gingival health 14–16. Hence, the increased GCF flow associated with inflamed tissues may be a major factor in promoting plaque formation17. Despite the professional cleaning and scaling and root planing performed in the beginning of the current study, it is unlikely that the inflammation exhibited by periodontal tissues at entry would be fully resolved within 7 days. In addition, the procedures associated with professional cleaning represent a mechanical insult that could trigger an increase in levels of GCF. Along with the increased volume of GCF in inflamed areas, the concomitant edema present in the gingival margin might constitute an anatomic shelter for growing plaque 18, 19.
It is also possible that more denture-wearing subjects violated protocol than subjects with natural teeth. In that scenario, although instructed to not remove their dentures overnight, some subjects might have ignored or forgotten this instruction leading to lower levels of biofilm on their denture. This notion is supported by the observed greater variation in mean counts of species in samples collected at time points after professional cleaning in the denture wearing than in dentate subjects.
As observed in Figs. 1, 3 and 4, the differences between biofilms formed around natural teeth and denture “teeth” are not only quantitative, but qualitative. Besides consistently higher plaque biomass measured at all post-cleaning time points in the dentate group, their microbial composition was a clear departure from that of edentulous individuals.
For instance, S. mitis, S. oralis and S. mutans were strikingly prominent in the samples from the denture teeth in the pre-cleaning samples. These were the only taxa that were significantly higher in mean counts and/or proportions on denture teeth. S. mitis and S. oralis also increased significantly in mean proportions during biofilm re-development on both natural and denture tooth surfaces affirming their role as talented initial colonizers of dental biofilms apparently irrespective of nature of the hard surface for attachment. The abundance of Streptococcus species in denture plaque has been reported previously. Theilade et al20, using predominant cultivable techniques, showed that streptococci comprised up to 81% of the microbiota (median of 41%) and demonstrated varying proportions of S. mutans and Streptococcus salivarius. Hoshino & Sato21 identified Streptococcus, along with Veillonella species, as the predominant taxa in denture plaque, accounting for 31% and 23% of the total cultivable microbiota, respectively. S. salivarius was the most common Streptococcus followed by S. mutans. Souza et al22 employed the checkerboard DNA-DNA hybridization technique to assess the levels of selected bacterial species colonizing denture surfaces. S. sanguinis was the most abundant species, along with Parvimonas micra, followed by S. oralis and S. mutans. The results of the present study are in line with those studies, particularly when data from “mature” plaque is taken into account (Fig.1). Unfortunately, S. salivarius was not evaluated in the present study obviating comparisons for this taxon. Our observations were also in accord with results reported by Campos et al23. Using cloning and sequencing, the authors assessed the microbial diversity in biofilms associated with soft tissues and the inner surface of dentures. They observed that Streptococcus species were prominent members of the microbiota and that S. mutans and S. mitis represented their most common representatives. In addition, members of the genera Veillonella and Selenomonas were also abundant. Further, Streptococcus and Neisseria species were detected by cultural techniques in more than 95% and 60% of the samples, respectively from the fitting surface of upper dentures24.
In the present study, members of the green complex, including E. corrodens, C.sputigena, C. gingivalis and C. ochracea as well as orange complex species, including P. intermedia and three subspecies of F. nucleatum, were prominent in dentate subjects. Those taxa could also be detected among edentulous individuals, which is in overall accord with previous reports 25, 26. However, they were all present in significantly higher levels in dentate individuals starting at 2 days post-cleaning. Interestingly, this was a key time point in studies of plaque development in dentate individuals. Between 2–4 days is typically when plaque biomass surpasses entry level 3, 27, 28. The increased levels of E. corrodens and C. gingivalis have been associated with this surge in plaque levels 29. In addition, the increased levels of F. nucleatum subspecies at that time point support their proposed role as a bridging species 30. It seems possible that those taxa may foster the faster development of a more complex biofilm in dentate individuals, in comparison with edentulous subjects.
Multiplication of bacteria initially attached to the cleaned tooth surface is likely to account for the increase in plaque mass over time 31. Availability of nutrients is essential to this multiplication. GCF appears to be a driving force in biofilm development, in large part due to its role as a nutrient source. Thus, the greater levels of the taxa listed above on natural teeth might be associated with the presence of GCF. GCF also seems to favor the growth of specific bacterial species, mostly facultative/anaerobic and non-saccharolytic taxa, such as Capnocytophaga species, Fusobacterium species and P. intermedia. For instance, the presence of orange and red complex species has been shown to be associated with higher GCF volume 32–34. As bacterial plaque increases in amount and complexity, the local microbial insult grows and further stimulates GCF production. Hence, a positive feedback is established that sustains the microbiota and the inflammation1. Since GCF was only present in dentate subjects, it is a possible factor underlying the quantitative and qualitative differences between the two clinical groups.
Besides the presence of GCF, the existence of a subgingival area adjacent to the supragingival compartment was a major difference between the two groups. Hence, it is possible that the proliferation of bacterial cells located in the superficial areas of the subgingival area, where inflammation was likely to be present, as discussed above, might have contributed to the differences in rate and composition of the biofilms on denture and natural teeth.
An additional potential explanation for the differences in plaque development and composition between dentate subjects and edentulous individuals might have been the source of bacterial cells. They could have originated from saliva, which has been shown to differ in composition depending on the periodontal status of the subject 1, 16. Rowshani et al16 showed that untreated periodontitis patients had higher numbers of bacterial cells in rinsing samples, in comparison with healthy subjects. In addition, the authors showed that, in general, individuals presenting an inflamed reduced periodontium and untreated periodontitis subjects had higher numbers of rods, motile organisms and spirochetes. Similarly, the tongue dorsum could also have represented a source of bacterial cells for colonization. It exhibits a similar microbial composition to saliva35 and may be a significant reservoir of oral microorganisms in edentulous patients36. In fact, a number of periodontal bacteria species have been detected in tongue biofilm samples from edentulous individuals, including C. rectus, P. micra, E. corrodens and Aggregatibacter actinomycetemcomitans 26, as well as a number of red and orange complex species2. Even though, in the present study, periodontitis patients had been treated in the beginning of the study, it is possible that their salivary counts remained high and contributed to biofilm development. Conversely, in edentulous patients, even though a complex tongue microbiota was present2, the environment was not conducive for their colonization of the dentures in the time frame of the study.
The present investigation demonstrated that the microbial composition of biofilms on natural and denture teeth differed substantially. Differences were observed both in the proportions of species present in “mature” biofilm present on the teeth as the individual presented as well as in the rate and nature of species colonization during seven days of no oral hygiene. These differences may be due to in part to the nature of the surface for initial bacterial attachment but probably colonization pattern may be affected even more by the availability of GCF to the organisms colonizing this supragingival biofilm on natural teeth. GCF may have not only affected the rate of biofilm accumulation, but influenced the composition of the microbiota present on the tooth surface. Certain species such as S. mitis and S. oralis appear to be pioneer species on both natural and denture teeth, but the community becomes more complex on the natural teeth during the early stages of biofilm redevelopment. The present results can assist in the understanding of the ecology of the oral cavity, highlighting the relevance of surfaces and fluids in oral biofilm formation. Further, they can foster better treatment planning and supportive care for edentulous individuals.
Summary.
Microbial biofilm composition and development in natural and denture teeth differ substantially and those differences seem to be due to in part to the nature of the surface for bacterial attachment and the availability of gingival crevicular fluid.
ACKNOWLEDGEMENTS
This work was supported in part by NIH/NIDCR grants R01-DE-14368 (S.S.), R03-DE-021742 (F.T.) and U01-DE-021127 (R.T.) and the Eleanor and Miles Shore Fellowship Program for Scholars in Medicine (The Forsyth Institute/Harvard Medical School) (F.T.).
Footnotes
The authors have no conflicts of interest.
References
- 1.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 2.Sachdeo A, Haffajee AD, Socransky SS. Biofilms in the edentulous oral cavity. J Prosthodont. 2008;17:348–356. doi: 10.1111/j.1532-849X.2008.00301.x. [DOI] [PubMed] [Google Scholar]
- 3.Uzel NG, Teles FR, Teles RP, et al. Microbial shifts during dental biofilm redevelopment in the absence of oral hygiene in periodontal health and disease. J Clin Periodontol. 2011;38:612–620. doi: 10.1111/j.1600-051X.2011.01730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teles FR, Teles RP, Uzel NG, et al. Early microbial succession in redeveloping dental biofilms in periodontal health and disease. J Periodontal Res. 2011 doi: 10.1111/j.1600-0765.2011.01409.x. In Press (Sep 5. doi:10.1111/j.1600-0765.2011.01409.x.Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin AE. "Checkerboard" DNA-DNA hybridization. Biotechniques. 1994;17:788–792. [PubMed] [Google Scholar]
- 6.Socransky SS, Haffajee AD, Smith C, et al. Use of checkerboard DNA-DNA hybridization to study complex microbial ecosystems. Oral Microbiol Immunol. 2004;19:352–362. doi: 10.1111/j.1399-302x.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 7.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 8.Legendre P, Legendre L. Vol. 20. Amsterdam: Elsevier; 2003. Numerical Ecology; pp. 637–705. [Google Scholar]
- 9.Hannig C, Hannig M. The oral cavity-a key system to understand substratum-dependent bioadhesion on solid surfaces in man. Clin Oral Investig. 2009;13:123–139. doi: 10.1007/s00784-008-0243-3. [DOI] [PubMed] [Google Scholar]
- 10.Teughels W, Van Assche N, Sliepen I, Quirynen M. Effect of material characteristics and/or surface topography on biofilm development. Clin Oral Implants Res. 2006;17:68–81. doi: 10.1111/j.1600-0501.2006.01353.x. [DOI] [PubMed] [Google Scholar]
- 11.Quirynen M, Bollen CML. The influence of surface roughness and surface free energy on supragingival and subgingival plaque formation in man - a review of the literature. J Clin Periodontol. 1995;22:1–14. doi: 10.1111/j.1600-051x.1995.tb01765.x. [DOI] [PubMed] [Google Scholar]
- 12.Quirynen M, Marechal M, Busscher HJ, Weerkamp AH, Darius PL, van Steenberghe D. The influence of surface free energy and surface roughness on early plaque formation. An in vivo study in man. J Clin Periodontol. 1990;17:138–144. doi: 10.1111/j.1600-051x.1990.tb01077.x. [DOI] [PubMed] [Google Scholar]
- 13.Siegrist BE, Brecx MC, Gusberti FA, Joss A, Lang NP. In vivo early human dental plaque formation on different supporting substances. A scanning electron microscopic and bacteriological study. Clin Oral Implants Res. 1991;2:38–46. doi: 10.1034/j.1600-0501.1991.020105.x. [DOI] [PubMed] [Google Scholar]
- 14.Brecx M, Theilade J, Attstrom R. Influence of optimal and excluded oral hygiene on early formation of dental plaque on plastic films. A quantitative and descriptive light and electron microscopic study. J Clin Periodontol. 1980;7:361–373. doi: 10.1111/j.1600-051x.1980.tb02009.x. [DOI] [PubMed] [Google Scholar]
- 15.Ramberg P, Lindhe J, Dahlen G, Volpe AR. The influence of gingival inflammation on de novo plaque formation. J Clin Periodontol. 1994;21:51–56. doi: 10.1111/j.1600-051x.1994.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 16.Rowshani B, Timmerman MF, Van der Velden U. Plaque development in relation to the periodontal condition and bacterial load of the saliva. J Clin Periodontol. 2004;31:214–218. doi: 10.1111/j.0303-6979.2004.00468.x. [DOI] [PubMed] [Google Scholar]
- 17.Daly CG, Highfield JE. Effect of localized experimental gingivitis on early supragingival plaque accumulation. J Clin Periodontol. 1996;23:160–164. doi: 10.1111/j.1600-051x.1996.tb02071.x. [DOI] [PubMed] [Google Scholar]
- 18.Walsh TF, Walmsley AD, Carrotte PV. Scanning electron microscopic investigation of changes in the dentogingival area during experimental gingivitis. J Clin Periodontol. 1991;18:20–25. doi: 10.1111/j.1600-051x.1991.tb01114.x. [DOI] [PubMed] [Google Scholar]
- 19.Quirynen M, Dekeyser C, van Steenberghe D. The influence of gingival inflammation, tooth type, and timing on the rate of plaque formation. J Periodontol. 1991;62:219–222. doi: 10.1902/jop.1991.62.3.219. [DOI] [PubMed] [Google Scholar]
- 20.Theilade E, Budtz-Jorgensen E, Theilade J. Predominant cultivable microflora of plaque on removable dentures in patients with healthy oral mucosa. Arch Oral Biol. 1983;28:675–680. doi: 10.1016/0003-9969(83)90101-2. [DOI] [PubMed] [Google Scholar]
- 21.Hoshino E, Sato M. [Predominant microorganisms of plaque on complete dentures] Nihon Hotetsu Shika Gakkai Zasshi. 1988;32:763–766. doi: 10.2186/jjps.32.763. [DOI] [PubMed] [Google Scholar]
- 22.Souza RF, Regis RR, Nascimento C, Paranhos HF, Silva CH. Domestic use of a disclosing solution for denture hygiene: a randomised trial. Gerodontology. 2010;27:193–198. doi: 10.1111/j.1741-2358.2009.00309.x. [DOI] [PubMed] [Google Scholar]
- 23.Campos MS, Marchini L, Bernardes LA, Paulino LC, Nobrega FG. Biofilm microbial communities of denture stomatitis. Oral Microbiol Immunol. 2008;23:419–424. doi: 10.1111/j.1399-302X.2008.00445.x. [DOI] [PubMed] [Google Scholar]
- 24.Sumi Y, Kagami H, Ohtsuka Y, Kakinoki Y, Haruguchi Y, Miyamoto H. High correlation between the bacterial species in denture plaque and pharyngeal microflora. Gerodontology. 2003;20:84–87. doi: 10.1111/j.1741-2358.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- 25.Kononen E, Asikainen S, Alaluusua S, et al. Are certain oral pathogens part of normal oral flora in denture-wearing edentulous subjects? Oral Microbiol Immunol. 1991;6:119–122. doi: 10.1111/j.1399-302x.1991.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 26.Fernandes CB, Aquino DR, Franco GC, Cortelli SC, Costa FO, Cortelli JR. Do elderly edentulous patients with a history of periodontitis harbor periodontal pathogens? Clin Oral Implants Res. 2010;21:618–623. doi: 10.1111/j.1600-0501.2009.01892.x. [DOI] [PubMed] [Google Scholar]
- 27.Zee KY, Samaranayake LP, Attstrom R. Predominant cultivable supragingival plaque in Chinese "rapid" and "slow" plaque formers. J Clin Periodontol. 1996;23:1025–1031. doi: 10.1111/j.1600-051x.1996.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 28.Ramberg P, Sekino S, Uzel NG, Socransky S, Lindhe J. Bacterial colonization during de novo plaque formation. J Clin Periodontol. 2003;30:990–995. doi: 10.1034/j.1600-051x.2003.00419.x. [DOI] [PubMed] [Google Scholar]
- 29.Haffajee AD, Teles RP, Patel MR, Song X, Veiga N, Socransky SS. Factors affecting human supragingival biofilm composition. I. Plaque mass. J Periodontal Res. 2009;44:511–519. doi: 10.1111/j.1600-0765.2008.01154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 31.Socransky SS, Manganiello AD, Propas D, Oram V, van Houte J. Bacteriological studies of developing supragingival dental plaque. J Periodontal Res. 1977;12:90–106. doi: 10.1111/j.1600-0765.1977.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 32.Dalwai F, Spratt DA, Pratten J. Modeling shifts in microbial populations associated with health or disease. Appl Environ Microbiol. 2006;72:3678–3684. doi: 10.1128/AEM.72.5.3678-3684.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teles R, Sakellari D, Teles F, et al. Relationships among gingival crevicular fluid biomarkers, clinical parameters of periodontal disease, and the subgingival microbiota. J Periodontol. 2010;81:89–98. doi: 10.1902/jop.2009.090397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salvi GE, Franco LM, Braun TM, et al. Pro-inflammatory biomarkers during experimental gingivitis in patients with type 1 diabetes mellitus: a proof-of-concept study. J Clin Periodontol. 2010;37:9–16. doi: 10.1111/j.1600-051X.2009.01500.x. [DOI] [PubMed] [Google Scholar]
- 35.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30:644–654. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 36.Hori R, Sato M, Kohno S, Hoshino E. Tongue microflora in edentulous geriatric denture-wearers. Microb Ecol Health Dis. 1999;11:89–95. [Google Scholar]


