Abstract
In the present study, we investigated the role of the transcription factor RUNX2 in melanomagenesis. We demonstrated that the expression of transcriptionally active RUNX2 was increased in melanoma cell lines as compared with human melanocytes. Using a melanoma tissue microarray, we showed that RUNX2 levels were higher in melanoma cells as compared with nevic melanocytes. RUNX2 knockdown in melanoma cell lines significantly decreased Focal Adhesion Kinase expression, and inhibited their cell growth, migration and invasion ability. Finally, the pro-hormone cholecalciferol reduced RUNX2 transcriptional activity and decreased migration of melanoma cells, further suggesting a role of RUNX2 in melanoma cell migration.
Keywords: Melanoma, Transcription factor, RUNX2, FAK, Migration, Cholecalciferol (Vitamin D3)
1. INTRODUCTION
It is estimated by the NCI SEER that 44,250 American men and 32,000 American women were diagnosed with invasive melanoma of the skin in 2012 and 9,180 men and women died of this disease that year. Even with the development of more effective treatments [1; 2; 3], metastatic melanoma is still associated with a poor prognosis [4; 5]. Transcription factors, traditionally considered undruggable, have become the focus of new targeting strategies in melanoma and other cancer types. [6; 7]. There is increasing evidence that transcription factors play oncogenic roles in melanoma and this has driven efforts to develop new approaches to target this class of proteins [8].
The RUNX (Runt-related transcription factor) genes comprise a family of three closely related transcription factors, RUNX1, RUNX2 and RUNX3. These genes are defined by a highly conserved 128 amino acid DNA binding/protein-protein interaction domain known as the Runt box [9]. Knock out models have implicated RUNX2 in cartilage and bone development [10; 11]. Some evidence also points to a role for RUNX2 specifically in bone metastasis in advanced breast and prostate cancer. However, the role of RUNX2 in promoting tumor development in hematopoietic lineages and in breast and prostate cancer extends beyond its pro-bone metastatic effects. RUNX2 regulates the expression of genes intimately associated with tumor progression, invasion and metastasis. These genes include osteopontin, bone sialoprotein, collagenases, and FAK/PTK2 [9; 12; 13; 14]. In addition, the pro-angiogenic effects of RUNX2 suggest a major role for this transcription factor in tumor promotion. These effects include endothelial cell proliferation, migration and invasion [15; 16], induction of VEGF expression and physical and functional interactions between RUNX2 and another major pro-angiogenic factor, hypoxia-inducible factor 1-a (HIF1-a) [17; 18].
In one study, two melanoma cell lines showed coexpression of RUNX2 and Bone Sialoprotein, whose expression in vivo correlated with local and regional melanoma spread [19]. Another study indicated that TGFβ, driving metastasis at advanced melanoma stages [20], up regulated RUNX2 expression in the 1205LU melanoma cell line [21]. In addition, the tumor suppressor p14ARF was reported to repress RUNX2 expression in melanoma cell lines. In this study, the authors speculated that increased RUNX2 resulting from p14ARF mutation might contribute to melanoma development [22]. As a first approach to studying the role of RUNX2 in melanoma development, we determined that RUNX2 was overexpressed in melanoma cell lines as compared with primary cultures of melanocytes or an immortalized melanocyte cell line. ShRNA-mediated knock down of RUNX2 in melanoma cell lines negatively affected cell growth and inhibited their migration and invasion in conjunction with a reduction in the levels of the kinase FAK/PTK2 involved in motility and adhesion. The RUNX2 DNA binding inhibitor Cholecalciferol [23] inhibited the activity of the RUNX2-responsive MMP13 promoter, and also decreased melanoma cell growth and their ability to migrate. Furthermore, we addressed the relevance of RUNX2 expression to human melanomagenesis using a melanoma tissue microarray and confirmed overexpression of RUNX2 in melanoma specimens as compared with benign nevi.
2. MATERIAL AND METHODS
2.1. Cell lines
WM1552C, WM9, WM1617, WM793, WM278, and 1205LU were kindly provided by Dr. M. Herlyn (Wistar Institute, Philadelphia, PA, USA [24]). These lines were cultured in MCDB153/L-15 (4/1 ratio) medium containing 2% FBS, 5 μg/ml Insulin and 1.7 mM Calcium Chloride. C8161 melanoma cell line was provided by Dr. Mary Hendrix (Children’s Memorial Research Center, Chicago, IL, USA [25] and was grown in D-MEM (Mediatech, 10-013-CV) containing 10% FBS. UACC903 cells were provided by Dr. Jeffrey M. Trent (Translational Genomics Research Center, Phoenix, AZ, USA [26]) and were grown in RPMI1640 (Invitrogen, 11875) containing 10% FBS. The SKMEL2 and WM35 melanoma cell lines were purchased from ATCC (American Type Culture Collection, Manassas, VA 20110, U.S.A). WM35 [24] was grown in MCDB153/L-15 (4/1 ratio) medium containing 2% FBS, 5 μg/ml Insulin and 1.7 mM Calcium Chloride. SKMEL2 was grown in MEM containing 10% FBS [27]. Primary cultures of melanocytes kindly provided by Dr M. Herlyn (Wistar Institute, Philadelphia, PA) were maintained in MCDB153 medium containing 2% FBS, 10% chelated FBS, 2 mM L-Glutamine, 20 pM cholera toxin, 60 pM basic FGF (Fibroblast Growth Factor), 100 nM Endothelin 3 and 10 ng/ml SCF (Stem Cell Factor). Two other primary cultures of melanocytes, AG22173 and AG22151, were purchased from Coriell Institute for Medical Research (Camden, NJ) and cultured in medium 254 from Invitrogen (#M254500), supplemented with human melanocyte growth supplement (#S0025). Immortalized melanocytes (PMEL/hTERT/CDK4(R24C)/p53DD) have been kindly provided by Dr. Hans Widlund and grown as previously described [28].
2.2. Cell counting
2–4 × 104 cells were seeded in 24-well plates. The following day, DMSO, 2.5 or 5 μM of cholecalciferol (Sigma, Saint Louis, MO) were added to the plates for 24 or 48 hours. Cells were then washed in phosphate-buffered saline, trypsinized and counted, using a Beckman Coulter Vi-CELL 1.00. For the analysis of cleaved caspase 3, 5 × 105 cells were seeded in 6-well-plates. The following day, melanoma cells were treated with DMSO, 2.5 or 5 μM of cholecalciferol for 24 hours before harvesting the cells and preparing lysates for immunoblotting.
2.3. Immunoblotting
Cells were harvested, washed with PBS, and lysed with cell lysis buffer in the presence of protease and phosphatase inhibitors (Roche) as previously described [29]. Equal amounts of protein were separated on polyacrylamide gel electrophoresis and transferred onto nitrocellulose membrane, and immunoblots were analyzed using antibodies against RUNX2 (Rabbit mAb, Epitomics, an Abcam Company, Burlingame, CA), GAPDH (Rabbit mAb), FAK (Rabbit Ab), cleaved caspase 3 (Rabbit Ab) all three from Cell Signaling (Danvers, MA) and Actin (Sigma Aldrich, St. Louis, MO)
2.4. RNA isolation and real time qPCR
Total RNA was isolated using Trizol (Invitrogen, Carlsbad, CA) and Direct-zol RNA miniprep kit (Zymo Research, Irvine, CA) following manufacturer’s instruction. One microgram of total RNA was used for cDNA synthesis using SuperScript II cDNA synthesis kit (Invitrogen, Carlsbad, CA) The qPCR was performed in One Step Plus qPCR instrument (Applied Biosystems Inc, Carlsbad, CA). All primers for SYBR qRT-PCR were purchased from Qiagen (Valencia, CA). Changes in gene expression were calculated using the delta delta Ct method. All experiments were independently replicated 3 times.
2.5. Detection of RUNX2 by immunofluorescence
50,000 cells were plated on Poly-L-Lysine (ScienCell Research laboratories, Carlsbad, CA) coated glass cover slip 24 hours before the experiment. After 24 hours, cells were washed twice in phosphate-buffered saline (PBS) at room temperature, fixed in 4% paraformaldehyde for 10 min, and permeabilized with 0.5% Triton X-100 in PBS for 25 min. Cells were then incubated with 1 μg/ml mouse monoclonal anti-RUNX2 antibody (Abcam, Cambridge, MA) diluted in PBS containing 5% goat serum for 1 hour in room temperature. The cells were then washed twice with PBS and incubated with the 1:200 diluted fluorescein isothiocyanate conjugated goat anti-mouse antibody (Jackson Immunoresearch Laboratories, West Grove, PA) for 1 hour at room temperature in the dark. The cells were then washed with PBS twice and 4′,6-diamidino-2-phenylindole (DAPI) was added, and digital images were obtained with a Nikon eclipse 80i fluorescence microscope.
2.6. Detection of RUNX2 by immunohistochemistry
The human melanoma tissue microarray (TMA) was purchased from US Biomax, Inc. (Rockville, MD, USA). The melanoma TMA was deparaffinized and antigen retrieval was performed using extended CC1 treatment (Cell Conditioning Solution, Ventana Medical Systems, Oro Valley, AZ). The rabbit polyclonal antibody for RUNX2 (Abcam, Cambridge, MA) was applied and incubated at 37°C for 1 or 2 hours. Donkey anti-Rabbit secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) was then applied and incubated at 37°C for 60 min, followed by chromogenic detection using the RedMap kit (Ventana Medical Systems, Oro Valley, AZ). Slides were counterstained with Hematoxylin and dehydrated and cleared before coverslipping from Xylene.
2.7. Imaging and sub-sampling (cropping)
Glass slides were digitized under 20x objective on a Trestle MedScan Whole-Slide Scanner. The resulting image archives were viewed and sub-sampled via the Trestle web service hosted on a Windows 2003 Server R2 server.
2.8. Quantification and statistics
A mixture of three colors is present on each specimen prepared for this experiment: red corresponds to the immunohistochemical stain; brown reveals the presence of melanin; and blue serves as the hematoxylin counter stain. In order to isolate the immunohistochemical signal for quantification, three representative color vectors, referred to as the principal color vectors (PCVs) [30], were systematically sampled from different areas throughout the specimen.
Using algorithms and software previously reported by our team [30], digitized TMA specimens were first registered to correctly cross-reference spatial coordinates of each of TMA core (histospot). The PCVs were subsequently used to split all imaged TMA cores into three constituent staining maps, thereby isolating the immunohistochemical signal (red map); melanin (brown map); as well as the hematoxylin signal (blue map). The hematoxylin maps were used to generate masks for the nuclear and cytoplasmic regions in each imaged histospot. For quality control, a certified pathologist reviewed all images to eliminate any artifacts that may have been introduced during this stage of processing. During the review TMA cores exhibiting over-staining for melanin were also eliminated from the study. Finally, the nuclear region masks and cytoplasmic region masks were superposed on the red map of the tissue cores to quantify the immunohistochemical signal. The ratio [average nuclei staining intensity]/[average cytoplasmic staining intensity] was used as an indicator of subcellular localization of the immunohistochemical signal.
2.9. RUNX2 knock down
We used two different human RUNX2 ShRNA targeting either the coding sequence (ShRUNX2-3) or the 3′UTR (ShRUNX2-2) in the pGIPZ lentiviral vector. The mature senses were CCAGCTGCATCCTATTTAA for ShRUNX2-2 and ACAAGGACAGAGTCAGATT for ShRUNX2-3. The mature sense was ATCTCGCTTGGGCGAGAGTAAG for the non-silencing control. This sequence does not match any known mammalian gene (has at least 3 or more mismatches against any gene as determined via nucleotide alignment/BLAST of 22mer sense sequence). 80–90 % confluent 293 amphotropic cells were transfected with 10 μg of non-silencing control or shRUNX2 plasmid and 4 μg of PREV (pcmv-dR8.2 dvpr) plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). 48 hours following transfection, the supernatant media was filtered through a 0.4uM filter for infection. Melanoma cell lines WM278, WM1617, WM35 and 1205LU were seeded at a density of 0.2 × 106 cells/ 6 well plate 24 hours prior to the infection. The cells were then infected with 1 ml viral particles and 8ug/ml polybrene and after 6 hours, 1ml fresh media was added and incubated for overnight. The following day, cells were incubated with 2 ml fresh media followed by stable selection with 3 μg/ml Puromycin for 1 to 2 weeks.
2.10. Trans-well Migration
Melanoma cells were seeded at a density of 75,000 cells per well in triplicates in serum free media into a 0.8uM pore insert chamber in 24-well plates containing medium with serum. The cells were left to migrate for 24 hours after which the inserts were washed with PBS and cells inside the inserts were cleaned with cotton tips. Cells which migrated were fixed and stained using the diff-quick stain according to the manufacturer’s protocol. Stained cells on the membrane were imaged and 5 random fields counted to obtain the average number of cells per mm2.
2.11. Invasion through Matrigel
Melanoma cells were seeded at a density of 75,000 to 100,000 cells per well in triplicates in serum free media into a 0.8μM pore BD Matrigel™ Invasion chamber (BD Biosciences) in 24-well plates containing media with serum. The cells were left to invade through Matrigel for 24 hours after which the inserts were washed with PBS and cells inside the inserts were cleaned with cotton tips. Cells which invaded through Matrigel were fixed and stained using the diff-quick stain according to the manufacturer’s protocol. Stained cells on the membrane were counted using 5 random fields to obtain the average number of cells per mm2.
2.12. Cell viability assay
Cell viability was analyzed using CellTiter 96 AQueous non-radioactive cell proliferation assay (MTS-MethaneThioSulfonate) reagent from Promega following the instructions of the manufacturers. Briefly, melanoma cells were seeded at a density of 9,000 to 10,000 cells per well in a flat bottom 96-well plate containing 100μl culture medium. After 48 hours, 15μl of MTS reagent was added and the plate was incubated for 1 to 3 hours in the tissue culture incubator before measuring the absorbance at 490 nm using Nanoquant 96 well plate reader (Tecan).
2.13. Luciferase assay
1205LU, WM793, WM1617 and WM35 cells were seeded at a density of 0.2 × 106 cells/ 6 well plate 24 hours prior to the transient transfection in duplicates. The cells were then transfected with PGL3 vector alone or MMP13-luciferase (MMP13-Luc, kindly provided by Dr. Nicola C. Partridge, New York University College of Dentistry, New York, NY [31]) plasmid in PGL3 vector backbone, and Renilla luciferase (phRL-CMV) plasmid as an internal control using lipofectamine LTX reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. For shRNA transfection, non-silencing control plasmid or shRUNX2-3 plasmid were co-transfected along with MMP13-luciferase and renilla luciferase plasmids. Following incubation at 37°C for 24 hours, cells were washed with phosphate-buffered saline twice, lysed with 1 × Passive Lysis Buffer (Promega, Madison, WI) and equal quantities of protein extracts were used to measure luciferase activity using Dual-Luciferase reporter assay system (Promega, Madison, WI) following the manufacturer’s protocol. Luciferase and renilla activities were measured using the Perkin Elmer Wallac instrument (Waltham, MA). Total firefly luciferase activity was normalized against total renilla activity or total protein from each well.
2.14. Statistical Analysis for MTS, migration, invasion and luciferase assays
Data are presented as the average ± standard error of the mean. For migration and invasion assays, each independent experiment was performed in triplicates. For MTS assays, each independent experiment was performed in quadruplicates. For luciferase assays, each independent experiment was performed in duplicates. Statistical analysis were performed by Student’s t test and P values < 0.05 were considered statistically significant.
3. RESULTS
3.1. Overexpression and high activity of RUNX2 in melanoma cell lines
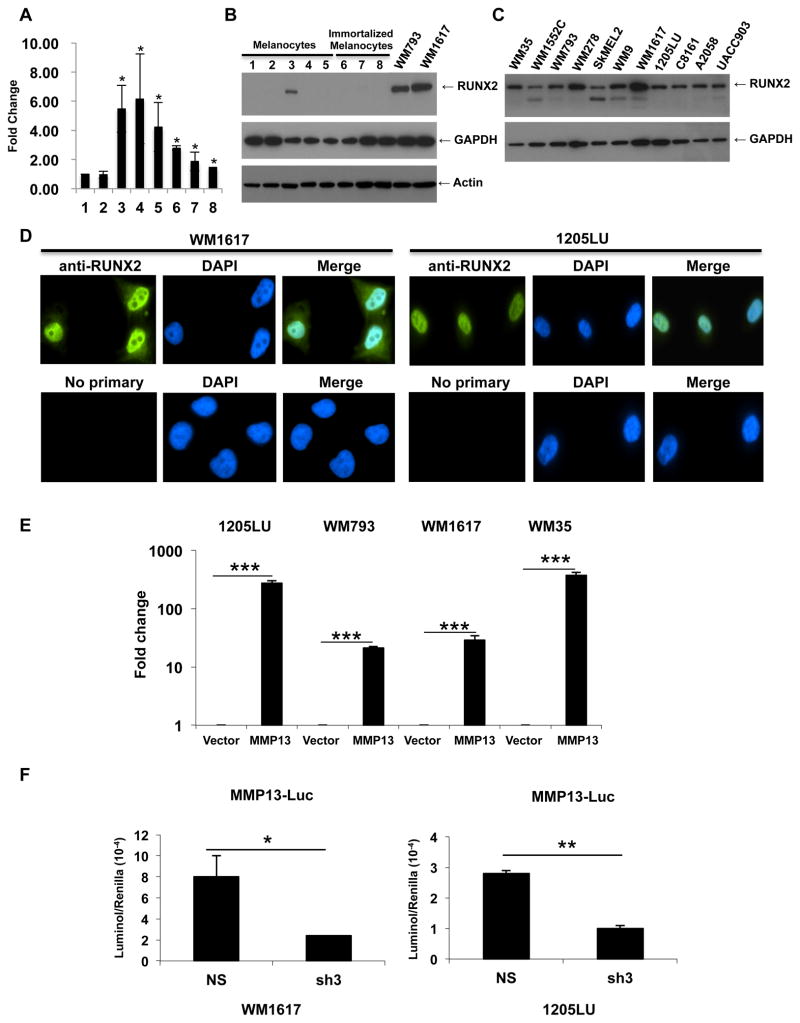
As a first step to address the possible role of RUNX2 in melanoma pathogenesis, RUNX2 mRNA expression was first assessed in two independent primary cultures of human melanocytes and six different human melanoma cell lines using real time PCR. The results showed overexpression of RUNX2 mRNA in the melanoma cell lines as compared with melanocytes (Figure 1A). By immunoblot, we tested RUNX2 expression in three different primary cultures of melanocytes and in the immortalized melanocyte cell line PMEL/hTERT/CDK4(R24C)/p53DD [28]. Except for one lysate showing some RUNX2 expression at a lower level than in the human melanoma cell lines WM793 and WM1617, RUNX2 was not detectable by immunoblot in the primary cultures of melanocytes. In addition, we could not detect significant levels of RUNX2 expression in the immortalized melanocytes. WM793 and WM1617 lysates exhibited significant levels of RUNX2 (Figure 1B). We extended our analysis of RUNX2 expression to a larger panel of melanoma cell lines. As shown in figure 1C, all the melanoma cell lines tested demonstrated varying levels of RUNX2 expression. As expected for a transcription factor, immunofluorescence analysis of RUNX2 expression in the human melanoma cell lines WM1617 and 1205LU showed mainly nuclear localization. Interestingly, in the WM1617 cell line, demonstrating the highest expression level, we could observe some RUNX2 cytoplasmic staining (Figure 1D). These results demonstrated that RUNX2 expression is overexpressed in human melanoma cells as compared to normal melanocytes or immortalized melanocytes, suggesting a role for RUNX2 in melanomagenesis. In order to determine whether RUNX2 was transcriptionally active in melanoma cell lines, we performed luciferase assays using a vector carrying the MMP13 promoter (−148/+14) fused to the luciferase gene [31] as a reporter gene. Previous work by others demonstrated that RUNX2 positively regulates the MMP13 promoter in osteoblastic cells [32]. As shown in Figure 1E, transfection of the MMP13-luciferase construct in four melanoma cell lines significantly increased luciferase activity as compared with the vector-transfected cell lines, suggesting that RUNX2 is transcriptionally active in melanoma cells. To confirm that MMP13 promoter activation is indeed due to RUNX2, we co-transfected melanoma cells with MMP13-luciferase along with either a ShRUNX2-3 expression vector, targeting the RUNX2 coding sequence (see Material and Methods for the specific sequence) or a non-silencing ShRNA expression vector. When 1205LU and WM1617 melanoma cells were transfected with ShRUNX2-3, MMP13 promoter activation, as reflected by luciferase activity, was significantly reduced as compared with cells transfected with the non-silencing control ShRNA (Figure 1F).
Figure 1. Transcriptionally active RUNX2 is overexpressed in melanoma cell lines as compared with melanocytes.
A: Quantitative PCR analysis of RUNX2 expression in independent primary cultures of melanocytes (1,2) and six melanoma cell lines 3: WM793; 4: C8161; 5: 1205LU; 6: WM9; 7: UACC903; 8: WM1617. *: P < 0.05 as compared with melanocytes (1). B: Immunoblot analysis of RUNX2 expression in the primary cultures of melanocytes AG22151: 1 and 2 (two different lysate preparations), AG22173: 3 and 4 (two different lysate preparations) and FOM-136-1: 5; in immortalized melanocytes: 6–8 (three different lysate preparations); and in the human melanoma cell lines WM793 and WM1617. GAPDH and Actin were used for normalization. C. Immunoblot analysis of RUNX2 in human melanoma cell lines. GAPDH was used for normalization. D. Immunofluorescence analysis of RUNX2 in the two human melanoma cell lines WM1617 and 1205LU. E. 1205LU, WM793, WM1617 and WM35 melanoma cells were transfected with the empty PGL3 vector (Vector) or the vector carrying the MMP13 promoter fused to the luciferase gene (MMP13), along with the Renilla plasmid. 24 hours post-transfection, firefly luciferase and renilla activities were measured and relative light units (RLU) were normalized to the renilla units per sample. The values for luciferase activity in the presence of MMP13-luciferase vector are reported as relative fold change compared to the transfection with vector control (set to 1). This figure is representative of three independent experiments. *** indicate p < 0.001 compared with the vector control. F. WM1617 (Left) and 1205LU (Right) melanoma cells were co-transfected with non-silencing ShRNA (NS) or shRUNX2-3 (Sh3) lentiviral plasmids along with the PGL3-MMP13-luc and renilla (phRL-CMV) plasmids. Firefly luciferase and renilla activities were measured 24 hours post-transfection, and relative light units (RLU) were normalized to the renilla units per sample. This figure is representative of three independent experiments with p values indicated. * indicates p < 0.05 and ** indicate p < 0.01 compared with the non-silencing (NS) vector.
3.2. Overexpression of RUNX2 in melanoma cells as compared with melanocytes in human samples
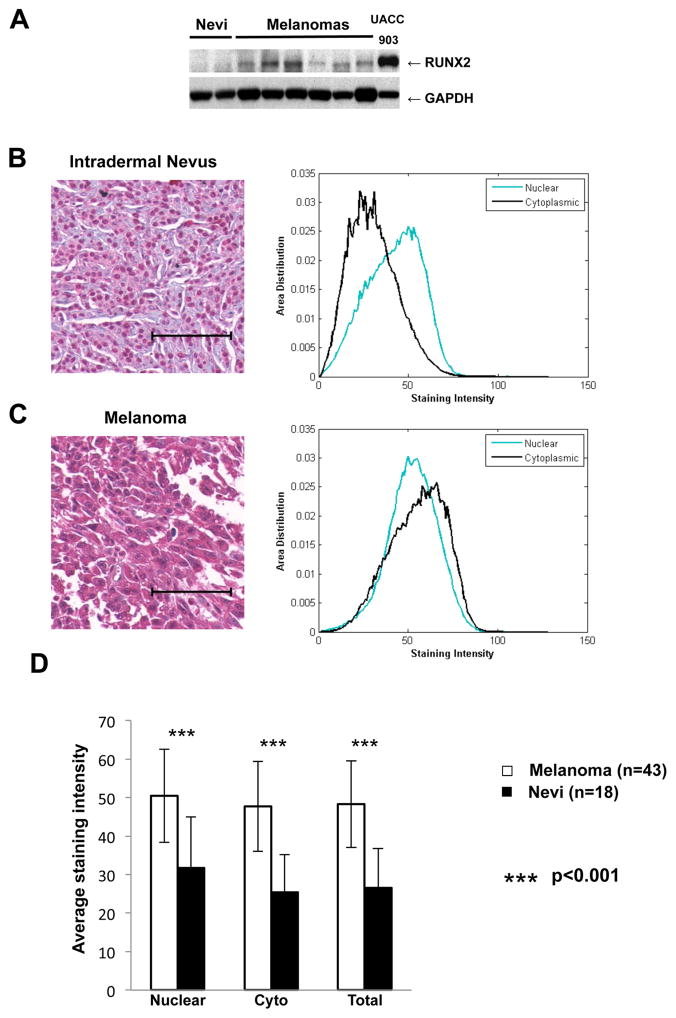
To examine the clinical relevance of RUNX2 expression in melanoma, we analyzed melanoma samples and benign nevus cells by immunoblot. As shown in figure 2A, we could not detect RUNX2 expression in the two nevi, while all melanoma samples were positive for RUNX2. We are aware that immunoblot might not be sensitive enough to detect RUNX2 in nevi.
Figure 2. RUNX2 overexexpression in melanomas as compared with benign nevi.
A. RUNX2 expression analysis in nevi and melanoma samples from one in transit metastasis and lymph nodes metastases. B, Left Panel: Representative immunohistochemical staining of a benign nevus showing mainly nuclear staining of RUNX2. Right Panel: Histogram showing area distribution of staining intensity over each measuring mask for this nevus core. The histograms were normalized to summation of 1. Green: Nuclear Staining Intensity; Black: Cytoplasmic Staining Intensity. C, Left Panel: Representative immunohistochemical staining of a malignant melanoma showing nuclear and cytoplasmic staining of RUNX2. Right panel: Histogram showing area distribution of staining intensity over each measuring mask for this melanoma core. Green: Nuclear Staining Intensity; Black: Cytoplasmic Staining Intensity. D: Average Staining Intensity in the nuclear and cytoplasmic compartments for melanomas (N=43) and nevi (N=18). Average Staining Intensity for the combined nuclear and cytoplasmic staining is also represented. Student t tests shows significantly higher staining intensities in melanoma specimens as compared with nevi with p values of 1.8e-7 (Nuclear), 2.4e-10 (Cytoplasmic) and 3.0e-10 (Total). Cyto: Cytoplasmic. The scale bar represents 100 microns.
We further performed a quantitative immunohistochemical analysis of RUNX2 expression in a melanoma tissue microarray (TMA). The analysis included 43 melanoma samples and 18 benign nevi. Figure 2C shows strong nuclear and diffuse cytoplasmic staining for the melanoma sample, while melanocytes in nevi (Figure 2B) express RUNX2 predominantly in the nucleus, with some weaker cytoplasmic staining. There is an apparent discrepancy between the expression of RUNX2 in nevi observed by immunohistochemistry and the lack of expression of RUNX2 in primary cultures of melanocytes and immortalized melanocytes in vitro. This apparent discrepancy between in vivo and in vitro data will be addressed in the Discussion.
The quantitative analysis was based on our previously reported principal color decomposition method [30] which is able to decompose the mixture of three colors present on the specimen prepared for this experiment: red corresponds to the immunohistochemical stain; brown reveals the presence of melanin; and blue serves as the hematoxylin counter stain. We computed a ratio R = Nuclear Staining Intensity/Cytoplasmic Staining Intensity for each tissue core as a measure of the subcellular distribution. A two-sample t-test demonstrates that melanoma samples had significantly lower R values than normal nevi, indicating more cytoplasmic distribution of RUNX2 in the melanoma samples as compared with nevi (p= 2.03e-5). In addition, there existed statistical differences in the average staining intensity between melanoma and nevi, in the nucleus, the cytoplasm and in the sum of both (Figure 2D). Therefore, the TMA analysis confirmed the overexpression of RUNX2 in melanomas as compared with nevi. The same analysis revealed similar staining pattern and intensity in primary cancer and metastasis (data not shown).
3.3. RUNX2 knockdown inhibits cell growth, migration and invasion of melanoma cells
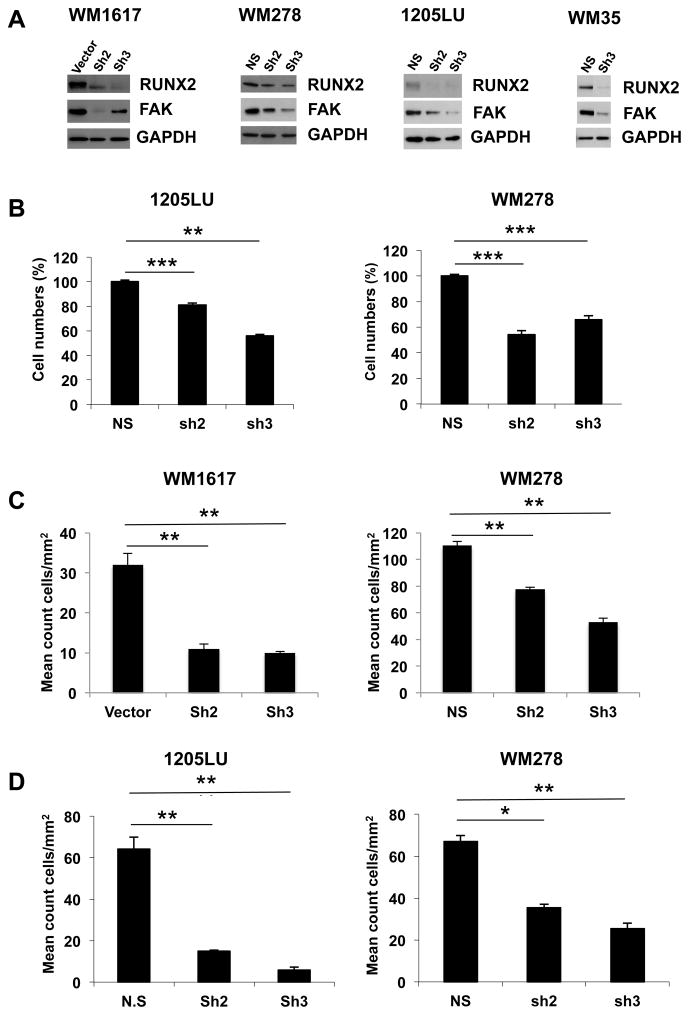
The link between RUNX2 and migration [14; 15; 33] and RUNX2 involvement in the metastatic process [9; 34] prompted us to investigate whether RUNX2 played a role in melanoma cell migration. Using RUNX2 shRNA lentiviral expression vectors, we generated stable melanoma cell lines expressing two types of RUNX2 ShRNA, called ShRUNX2-2, targeting RUNX2 3′UTR, and shRUNX2-3 targeting the RUNX2 coding sequence. As shown in Figure 3A, we obtained a reduction in RUNX2 expression levels in WM1617, WM278 and 1205LU melanoma cells expressing these two types of RUNX2 ShRNA, and in WM35 melanoma cells expressing ShRUNX2-3. The FAK/PTK2 gene has been shown to be a RUNX2 target gene, and to contribute to RUNX2-induced motility in osteosarcoma cells [14]. We found that FAK was down regulated in parallel with the RUNX2 decrease in WM1617, WM278, WM35 and 1205LU melanoma cells expressing ShRUNX2-2 and ShRUNX2-3 (Figure 3A). In WM1617 melanoma cells, it seems surprising that while RUNX2 knockdown is stronger with ShRUNX2-3 than with ShRUNX2-2, the expression of FAK remains higher in ShRUNX2-3-expressing cells than in ShRUNX2-2-expressing cells. This result could be explained by a feedback mechanism (occurring in WM1617 melanoma cells expressing ShRUNX2-3) by which FAK expression could be reactivated by other transcription factors such as c-myc or NF-kB [35]. The decrease in FAK expression observed when RUNX2 is reduced in the four melanoma cell lines suggested that RUNX2 knockdown could affect their in vitro migration and invasion abilities. Indeed, WM1617 and WM278 melanoma cells expressing ShRUNX2-2 and ShRUNX2-3 had significantly reduced migration ability as compared with WM1617 and WM278 cells expressing the empty vector or the non-silencing ShRNA (Figure 3C). In addition, 1205LU and WN278 melanoma cells expressing ShRUNX2-2 and ShRUNX2-3 had significantly reduced invasion ability as compared with 1205LU and WM278 cells expressing the non-silencing shRNA (Figure 3D). These results suggest that RUNX2 plays an active role in melanoma cells migration and invasion. In order to determine whether RUNX2 knock down also affected cell growth, we performed MTS assays. 48 hours after seeding 1205LU and WM278 melanoma cells expressing non-silencing ShRNA, ShRUNX2-2 or ShRUNX2-3, the number of viable cells was counted. As shown in Figure 3B, 1205LU and WM278 melanoma cells expressing ShRUNX2-2 or ShRUNX2-3 had reduced numbers of viable cells as compared with 1205LU and WM278 melanoma cells expressing non-silencing ShRNA, suggesting that RUNX2 is involved in melanoma cell growth.
Figure 3. Decreased RUNX2 expression results in reduced cell growth, migration and invasion of melanoma cells and lower FAK expression level.
A. RUNX2 and FAK levels in vector/non-silencing ShRNA (NS), ShRUNX2-2 (Sh2) and ShRUNX2-3 (Sh3) stable melanoma cell lines. B. Non-silencing ShRNA (NS), ShRUNX2-2 (Sh2) and ShRUNX2-3 (Sh3) stable melanoma cells were used for MTS assays in quadruplicates. 48 hours after seeding, the number of viable cells was counted. C. Vector, non-silencing ShRNA (NS), ShRUNX2-2 (Sh2) and ShRUNX2-3 (Sh3) stable melanoma cells were used for transwell migration assays in triplicates. After 24 hours, cells, which migrated, were fixed and stained. The number of cells, which migrated through the insert, was then quantified as described in Material and Methods. Results are expressed as mean count cells/mm2 +/− SEM. ** indicate p < 0.01 compared with the Vector (WM1617) or the non-silencing ShRNA (WM278). D. Non-silencing ShRNA (NS), ShRUNX2-2 (Sh2) and ShRUNX2-3 (Sh3) stable melanoma cells were used for invasion through Matrigel assays in triplicates. After 24 hours, cells were fixed and stained. The number of cells, which invaded through Matrigel, was then quantified as described in Material and Methods. Results are expressed as mean count cells/mm2 +/− SEM. * indicates p < 0.05 and ** indicate p < 0.01 compared with the non-silencing ShRNA based on Student’s t-test.
3.4. Inhibition of RUNX2 transcriptional activity reduces cell growth and migration of melanoma cells
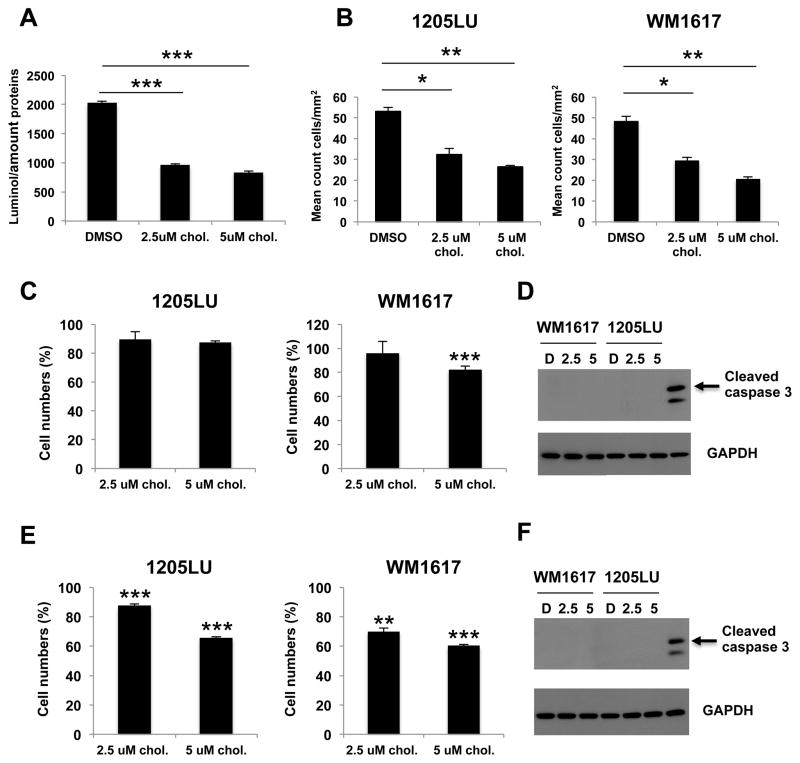
Complementary to the genetic approach, we were interested in analyzing the effect of a RUNX2 inhibitor on melanoma cells migration. The inactive vitamin D3 precursor cholecalciferol has been shown to enhance RUNX2 DNA binding in vitro at a dose of 1 nM, while doses greater than 10 nM inhibited RUNX2 DNA binding [23]. In order to determine whether cholecalciferol was able to inhibit RUNX2 transcriptional activity in melanoma cells, we performed luciferase assays in the presence or absence of micromolar concentrations of cholecalciferol. As illustrated in Figure 4A, 1205LU melanoma cells treated with 2.5 or 5 μM cholecalciferol had a significant decrease in MMP13 luciferase activity, indicating that this molecule inhibited RUNX2 transcriptional activity on the MMP13 promoter. In parallel, we showed that treatment with 2.5 or 5 μM cholecalciferol for 24 hours inhibited the migration ability of 1205LU and WM1617 cells in transwell assays (Figure 4B). In order to verify that at these concentrations, the effect of cholecalciferol on migration was not related to an effect on cell proliferation or apoptotic death, we performed cell growth experiments in the presence of 2.5 or 5 μM cholecalciferol for 24 hours and showed no major effect of the treatment on 1205LU and WM1617 cell numbers at this time point (Figure 4C). In addition, 1205LU and WM1617 cells did not show any increase in cleaved caspase 3 levels after treatment with 2.5 or 5 μM cholecalciferol for 24 hours as compared with vehicle-treated cells (Figure 4D). These results further support the hypothesis that RUNX2 activity is required for melanoma cell migration. In order to determine whether longer treatment with cholecalciferol will have an effect on melanoma cell growth, we performed cell growth experiments in the presence of 2.5 or 5 μM cholecalciferol for 48 hours. As shown in Figure 4E, 5 μM cholecalciferol significantly reduced 1205LU and WM1617 cell numbers to around 60% of DMSO-treated cells. Since 1205LU and WM1617 cells did not exhibit any increase in cleaved caspase 3 in the same conditions (48 hours in the presence of 2.5 or 5 μM cholecalciferol; Figure 4F), our results suggest that inhibiting RUNX2 transcriptional activity by cholecaciferol negatively affected cell proliferation.
Figure 4. Inhibition of RUNX2 transcriptional activity leads to reduced cell growth and migration of melanoma cells.
A. Luciferase assays using the MMP13-Luc vector, in the presence of DMSO, 2.5 or 5 μM cholecalciferol (Chol.) in 1205LU cells. *** indicate p < 0.001. B. 1205LU and WM1617 melanoma cell lines were analyzed in transwell migration assays, in the presence of DMSO, 2.5 or 5 μM cholecalciferol for 24 hours. The experiments were done in triplicates. * indicates p < 0.05, ** indicate p < 0.01 compared with DMSO. C. Cell growth assays showing 1205LU and WM1617 melanoma cell lines cultured in the presence of DMSO, 2.5 or 5 μM cholecalciferol for 24 hours. The experiments were done in triplicates. Results are represented as percents: (cells with cholecalciferol/cells with DMSO) X 100. D. Analysis of cleaved caspase 3 expression in the presence of DMSO or cholecalciferol (2.5 or 5 μM) for 24 hours in WM1617 and 1205LU cells. E. Cell growth assays showing 1205LU and WM1617 melanoma cell lines cultured in the presence of DMSO, 2.5 or 5 μM cholecalciferol for 48 hours. The experiments were done in triplicates. Results are represented as percents: (cells with cholecalciferol/cells with DMSO) X 100. F. Analysis of cleaved caspase 3 expression in the presence of DMSO or cholecalciferol (2.5 or 5 μM) for 48 hours in WM1617 and 1205LU cells.
4. DISCUSSION
In the present study, we examined for the first time the role of RUNX2 in melanoma pathogenesis. We demonstrated that RUNX2 mRNA and protein are overexpressed in melanoma cell lines as compared with normal melanocytes. Interestingly, radial growth phase (such as WM35 and WM1552c), vertical growth phase (such as WM793, WM278) and metastases (such as 1205LU, C8161)-derived cell lines had comparable levels of RUNX2, suggesting that RUNX2 is not only involved in the metastatic process, but possibly in melanomagenesis as well. Luciferase assays using the RUNX2-responsive MMP13-luciferase vector [32], showed that RUNX2 was functionally active in the radial growth phase melanoma (WM35), vertical growth phase melanoma (WM793) and metastases (1205LU)-derived cell lines. Immunofluorescence analysis for RUNX2 demonstrated mainly nuclear staining, with some cytoplasmic staining visible in the highest RUNX2-expressing melanoma cell line, WM1617. Dynamic shuttling of RUNX2 between the nucleus and the cytoplasm was reported to be dependent on microtubules in osteosarcomas cells. This nuclear-cytoplasmic shuttling likely contributes to the modulation of RUNX2 transcriptional activity [36]. Analysis of a melanoma tissue microarray confirmed that in addition to the nuclear localization of RUNX2, we observed some expression of RUNX2 in the cytoplasmic compartment of melanoma cells and to a lesser extent in melanocytes from nevi (Figure 2B, C and D). These results may appear to contradict a role for RUNX2 as an oncogenic transcription factor since the presence of such a factor in the cytoplasm usually equals inactivity. However, the possibility that cytoplasmic RUNX2 might be fulfilling another task is not excluded. In favor of this hypothesis, localization of RUNX proteins at the centrosome, through interaction with rootletin and γ-tubulin, and at the mitotic spindle and midbody was reported. These results suggest a role for RUNX proteins in the functioning of the mitotic apparatus and cytokinesis [37; 38]. These results are in accordance with the effects of RUNX2 knock down on melanoma cell growth (Figure 3B). In addition, the detection of RUNX proteins at non-transcription sites indicates that RUNX factors can be involved in processes other than transcriptional regulation. Interestingly, unphosphorylated latent STAT1 was shown to sequester RUNX2 in the cytoplasm, thereby attenuating RUNX2 activity. This study provides an example of a transcription factor (STAT1) playing an active role while in the cytoplasm by preventing another transcription factor, RUNX2, from entering the nucleus [39]. Similarly, overexpression of the transcription factor Tbx3 was reported to mislocalize RUNX2 in the cytoplasm, although the study did not analyze whether RUNX2 directly associates with Tbx3 [40]. These studies altogether expand the functions of RUNX proteins outside the nucleus.
The quantitative analysis of the melanoma tissue microarray further confirmed the overexpression of RUNX2 in melanoma cells as compared with nevic melanocytes. Although this result further suggests that RUNX2 might play a role in melanoma pathogenesis, it is surprising that we detected RUNX2 expression in melanocytes in nevi, while RUNX2 was undetectable in all but one melanocyte lysates by immunoblot. Nevi contain senescent melanocytes. Melanocytic nevi are a well-studied system for oncogene-induced senescence (OIS), a well-established tumor suppressive mechanism that impairs proliferative signals initiated by deregulated oncogenes [41]. The activating mutation V600E in BRAF observed in a large majority of nevi [42] likely plays a role in the induction of the OIS program in melanocytes by yet undefined mechanisms [43; 44; 45]. The detectable expression of RUNX2 in melanocytes in nevi suggests that in normal cells, RUNX2 may contribute to OIS, as a major role has been attributed to RUNX transcription factors in OIS [46]. In vitro, the primary cultures of melanocytes and the immortalized melanocytes may develop a growth advantage by down regulating RUNX2, which could explain the observation that RUNX2 is almost undetectable in these cells. Similarly, primary RUNX2-null osteoblasts have a growth advantage, exhibit loss of p21WAF1/CIP1 expression and fail to undergo cellular senescence [47]. Although our conclusion that RUNX2 may play a similar role in melanocyte is speculative, it seems to reconcile the results found in situ in melanocytic nevi (where RUNX2 is expressed) with the loss of RUNX2 expression in melanocytes established in culture. The concept that RUNX factors may play a pro or anti-proliferative role depending on the context is well established [46]. It is conceivable that RUNX2 plays an anti-proliferative role in melanocytes and promotes proliferation in melanoma cells. The negative effects of both the RUNX2 knock down and the 48 hour-treatment with cholecalciferol on the number of melanoma cells suggest that RUNX2 plays a pro-proliferative role on these cells. In favor of this hypothesis is the demonstration that RUNX2 is associated with the mitotic spindle [37] and the centrosomal protein γ-tubulin [38].
Our results demonstrated that RUNX2 knock down inhibited migration and invasion of melanoma cells. These results are in accordance with the role of RUNX2 in migration and invasion in other cellular systems. RUNX2 siRNA treatment of the prostate cancer cell line PC3 decreased migration and invasion through Matrigel in vitro [48]. Overexpression of Runx2 in the prostate cancer cell line C4-2B enhanced their invasiveness. In addition, Runx2 overexpression up regulated transcription factors (Sox9, SNAI2 and SMAD3) implicated in the process of epithelial to mesenchymal transition (EMT), whose features include increased motility and invasion potential. Runx2 overexpression also up regulates genes involved in cellular movement and cytoskeleton remodeling [49]. Forced expression of Runx2 or dominant negative Runx2 in the mouse osteoblastic cells MC3T3-E1 enhanced or inhibited cell migration respectively [33]. Interestingly, Runx2 stimulates the PI3K/AKT axis whose role in EMT has been documented [50]. Indeed, Runx2 up regulates the expression of the PI3K subunits p85 and p110Beta, as well as AKT [33]. The U2OS osteosarcoma cells also demonstrated reduced motility following SiRNA-mediated depletion of RUNX2. In addition, genomic promoter occupancy of RUNX2 in osteosarcoma cells identified genes involved in motility, such as FAK/PTK2 or talin (TNL1) [14]. An earlier study also demonstrated that RUNX2 regulated endothelial cells migration and invasion [15]. These studies altogether support the role of RUNX2 in cell migration and invasion.
Finally, several lines of evidence suggest that RUNX2 plays a role in melanoma development: 1) RUNX2 is overexpressed in melanoma cells as compared with melanocytes from human tissues; 2) We found that 117 melanoma samples out of 316 (37%) had complete RUNX2 gene amplification using The Cancer Genome Atlas data (TCGA, https://tcga-data.nci.nih.gov/tcga/); Interestingly, RUNX2 was also found amplified in 16 out of 21 primary conjunctival melanomas [51]; 3) RUNX2 deficiency inhibits cell growth, migration and invasion of human melanoma cells; 4) Inhibiting RUNX2 activity by treatment with cholecalciferol results in reduced cell growth and migration ability of melanoma cells; 5) Decreased RUNX2 level is associated with a reduction in FAK level, whose involvement in migration [14; 52] and melanoma metastasis is well documented [53; 54; 55]. Future experiments are underway to determine whether knocking down RUNX2 in melanoma cell lines will reduce their tumorigenicity and ability to form lung metastases in mouse xenograft models.
Acknowledgments
We thank Dr. A. Passaniti and Dr D. Medina for helpful discussions. We thank Lei Cong and the Tissue Analytical Services at the Rutgers Cancer institute of New Jersey for the immunohistochemical staining of the melanoma tissue microarray slide. We thank Seung-Shick Shin for lysates from nevi and melanoma metastases obtained in an IRB-approved and HIPAA-compliant fashion. We are grateful to Dr. Vasudeva Ginjala for his help on immunofluorescence, to Saurabh Laddha for his help on TCGA analysis of RUNX2 copy number variations and to Maung Naing Lin Shan for his help on the MTS assays. The present work was supported by a Research Development Award from the New Jersey Commission on Cancer Research 09-1143-CCR-EO, a Research Development Award from the Cancer Center Support Grant CCSG P30CA072720, a Research Scholar Grant from the American Cancer Society 116683-RSG-09-087-01-TBE (K.C.S.) and a grant from the National Cancer Institute 1RO1CA149627-01 (J. G.). Dr. Olabisi was supported by a Biomedical Science Education Postdoctoral Training Program from the National Institute of Health K12 GM093854-01. This research was funded, in part, by grants from the National Institute of Health through contract 5R01CA156386-08 and 5R01CA161375-02 from the National Cancer Institute; and contract 5R01LM011119-03 from the National Library of Medicine (D.F.).
Footnotes
Conflict of Interest Statement
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hersey P, Gallagher S. A Focus on PD-L1 in Human Melanoma. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-12-3312. [DOI] [PubMed] [Google Scholar]
- 2.Lyseng-Williamson KA, Sanford M. Ipilimumab: a guide to its use in advanced melanoma. Am J Clin Dermatol. 2012;13:349–354. doi: 10.2165/11209100-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 3.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O’Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle GM. Therapy for metastatic melanoma: an overview and update. Expert Rev Anticancer Ther. 2011;11:725–737. doi: 10.1586/era.11.25. [DOI] [PubMed] [Google Scholar]
- 5.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, Lin C, Leach C, Cannady RS, Cho H, Scoppa S, Hachey M, Kirch R, Jemal A, Ward E. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh D, Papavassiliou AG. Transcription factor therapeutics: long-shot or lodestone. Curr Med Chem. 2005;12:691–701. doi: 10.2174/0929867053202197. [DOI] [PubMed] [Google Scholar]
- 7.Yan C, Higgins PJ. Drugging the undruggable: Transcription therapy for cancer. Biochim Biophys Acta. 2013;1835:76–85. doi: 10.1016/j.bbcan.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes Dev. 2012;26:1131–1155. doi: 10.1101/gad.191999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blyth K, Vaillant F, Jenkins A, McDonald L, Pringle MA, Huser C, Stein T, Neil J, Cameron ER. Runx2 in normal tissues and cancer cells: A developing story. Blood Cells Mol Dis. 2010;45:117–123. doi: 10.1016/j.bcmd.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 11.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 12.Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 13.Chua CW, Chiu YT, Yuen HF, Chan KW, Man K, Wang X, Ling MT, Wong YC. Suppression of androgen-independent prostate cancer cell aggressiveness by FTY720: validating Runx2 as a potential antimetastatic drug screening platform. Clin Cancer Res. 2009;15:4322–4335. doi: 10.1158/1078-0432.CCR-08-3157. [DOI] [PubMed] [Google Scholar]
- 14.van der Deen M, Akech J, Lapointe D, Gupta S, Young DW, Montecino MA, Galindo M, Lian JB, Stein JL, Stein GS, van Wijnen AJ. Genomic promoter occupancy of runt-related transcription factor RUNX2 in Osteosarcoma cells identifies genes involved in cell adhesion and motility. J Biol Chem. 2012;287:4503–4517. doi: 10.1074/jbc.M111.287771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Vitolo M, Passaniti A. Runt-related gene 2 in endothelial cells: inducible expression and specific regulation of cell migration and invasion. Cancer Res. 2001;61:4994–5001. [PubMed] [Google Scholar]
- 16.Pierce AD, Anglin IE, Vitolo MI, Mochin MT, Underwood KF, Goldblum SE, Kommineni S, Passaniti A. Glucose-activated RUNX2 phosphorylation promotes endothelial cell proliferation and an angiogenic phenotype. J Cell Biochem. 2012;113:282–292. doi: 10.1002/jcb.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon TG, Zhao X, Yang Q, Li Y, Ge C, Zhao G, Franceschi RT. Physical and functional interactions between Runx2 and HIF-1alpha induce vascular endothelial growth factor gene expression. J Cell Biochem. 2011;112:3582–3593. doi: 10.1002/jcb.23289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SH, Che X, Jeong JH, Choi JY, Lee YJ, Lee YH, Bae SC, Lee YM. Runx2 Protein Stabilizes Hypoxia-inducible Factor-1alpha through Competition with von Hippel-Lindau Protein (pVHL) and Stimulates Angiogenesis in Growth Plate Hypertrophic Chondrocytes. J Biol Chem. 2012;287:14760–14771. doi: 10.1074/jbc.M112.340232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riminucci M, Corsi A, Peris K, Fisher LW, Chimenti S, Bianco P. Coexpression of bone sialoprotein (BSP) and the pivotal transcriptional regulator of osteogenesis, Cbfa1/Runx2, in malignant melanoma. Calcif Tissue Int. 2003;73:281–289. doi: 10.1007/s00223-002-2134-y. [DOI] [PubMed] [Google Scholar]
- 20.Lasfar A, Cohen-Solal KA. Resistance to transforming growth factor beta-mediated tumor suppression in melanoma: are multiple mechanisms in place? Carcinogenesis. 2010;31:1710–1717. doi: 10.1093/carcin/bgq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohammad KS, Javelaud D, Fournier PG, Niewolna M, McKenna CR, Peng XH, Duong V, Dunn LK, Mauviel A, Guise TA. TGF-beta-RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res. 2011;71:175–184. doi: 10.1158/0008-5472.CAN-10-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Packer LM, Pavey SJ, Boyle GM, Stark MS, Ayub AL, Rizos H, Hayward NK. Gene expression profiling in melanoma identifies novel downstream effectors of p14ARF. Int J Cancer. 2007;121:784–790. doi: 10.1002/ijc.22725. [DOI] [PubMed] [Google Scholar]
- 23.Underwood KF, D’Souza DR, Mochin-Peters M, Pierce AD, Kommineni S, Choe M, Bennett J, Gnatt A, Habtemariam B, MacKerell AD, Jr, Passaniti A. Regulation of RUNX2 transcription factor-DNA interactions and cell proliferation by vitamin D3 (cholecalciferol) prohormone activity. J Bone Miner Res. 2012;27:913–925. doi: 10.1002/jbmr.1504. [DOI] [PubMed] [Google Scholar]
- 24.Satyamoorthy K, Li G, Gerrero MR, Brose MS, Volpe P, Weber BL, Van Belle P, Elder DE, Herlyn M. Constitutive mitogen-activated protein kinase activation in melanoma is mediated by both BRAF mutations and autocrine growth factor stimulation. Cancer Res. 2003;63:756–759. [PubMed] [Google Scholar]
- 25.Welch DR, Bisi JE, Miller BE, Conaway D, Seftor EA, Yohem KH, Gilmore LB, Seftor RE, Nakajima M, Hendrix MJ. Characterization of a highly invasive and spontaneously metastatic human malignant melanoma cell line. Int J Cancer. 1991;47:227–237. doi: 10.1002/ijc.2910470211. [DOI] [PubMed] [Google Scholar]
- 26.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Sampas N, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Carpten J, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, Sondak V. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 27.Le MN, Chan JL, Rosenberg SA, Nabatian AS, Merrigan KT, Cohen-Solal KA, Goydos JS. The glutamate release inhibitor Riluzole decreases migration, invasion, and proliferation of melanoma cells. J Invest Dermatol. 2010;130:2240–2249. doi: 10.1038/jid.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, Lee C, Wagner SN, Li C, Golub TR, Rimm DL, Meyerson ML, Fisher DE, Sellers WR. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 29.Abushahba W, Olabisi OO, Jeong BS, Boregowda RK, Wen Y, Liu F, Goydos JS, Lasfar A, Cohen-Solal KA. Non-Canonical Smads Phosphorylation Induced by the Glutamate Release Inhibitor, Riluzole, through GSK3 Activation in Melanoma. PLoS One. 2012;7:e47312. doi: 10.1371/journal.pone.0047312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W, Reiss M, Foran DJ. A prototype for unsupervised analysis of tissue microarrays for cancer research and diagnostics. IEEE Trans Inf Technol Biomed. 2004;8:89–96. doi: 10.1109/titb.2004.828891. [DOI] [PubMed] [Google Scholar]
- 31.Lee M, Partridge NC. Parathyroid hormone activation of matrix metalloproteinase-13 transcription requires the histone acetyltransferase activity of p300 and PCAF and p300-dependent acetylation of PCAF. J Biol Chem. 2010;285:38014–38022. doi: 10.1074/jbc.M110.142141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selvamurugan N, Jefcoat SC, Kwok S, Kowalewski R, Tamasi JA, Partridge NC. Overexpression of Runx2 directed by the matrix metalloproteinase-13 promoter containing the AP-1 and Runx/RD/Cbfa sites alters bone remodeling in vivo. J Cell Biochem. 2006;99:545–557. doi: 10.1002/jcb.20878. [DOI] [PubMed] [Google Scholar]
- 33.Fujita T, Azuma Y, Fukuyama R, Hattori Y, Yoshida C, Koida M, Ogita K, Komori T. Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J Cell Biol. 2004;166:85–95. doi: 10.1083/jcb.200401138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chimge NO, Frenkel B. The RUNX family in breast cancer: relationships with estrogen signaling. Oncogene. 2013;32:2121–2130. doi: 10.1038/onc.2012.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S, Hua ZC. FAK expression regulation and therapeutic potential. Adv Cancer Res. 2008;101:45–61. doi: 10.1016/S0065-230X(08)00403-X. [DOI] [PubMed] [Google Scholar]
- 36.Pockwinse SM, Rajgopal A, Young DW, Mujeeb KA, Nickerson J, Javed A, Redick S, Lian JB, van Wijnen AJ, Stein JL, Stein GS, Doxsey SJ. Microtubule-dependent nuclear-cytoplasmic shuttling of Runx2. J Cell Physiol. 2006;206:354–362. doi: 10.1002/jcp.20469. [DOI] [PubMed] [Google Scholar]
- 37.Bonner MK, Poole DS, Xu T, Sarkeshik A, Yates JR, 3rd, Skop AR. Mitotic spindle proteomics in Chinese hamster ovary cells. PLoS One. 2011;6:e20489. doi: 10.1371/journal.pone.0020489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuang LS, Lai SK, Murata-Hori M, Yamada A, Li HY, Gunaratne J, Ito Y. RUNX3 interactome reveals novel centrosomal targeting of RUNX family of transcription factors. Cell Cycle. 2012;11:1938–1947. doi: 10.4161/cc.20278. [DOI] [PubMed] [Google Scholar]
- 39.Kim S, Koga T, Isobe M, Kern BE, Yokochi T, Chin YE, Karsenty G, Taniguchi T, Takayanagi H. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev. 2003;17:1979–1991. doi: 10.1101/gad.1119303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deepak V, Zhang Z, Meng L, Zeng X, Liu W. Reduced activity and cytoplasmic localization of Runx2 is observed in C3h10t1/2 cells overexpressing Tbx3. Cell Biochem Funct. 2011;29:348–350. doi: 10.1002/cbf.1753. [DOI] [PubMed] [Google Scholar]
- 41.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J, Salem G, Pohida T, Heenan P, Duray P, Kallioniemi O, Hayward NK, Trent JM, Meltzer PS. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 43.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, You MJ, DePinho RA, McMahon M, Bosenberg M. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, Larue L, Pritchard C, Marais R. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 45.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 46.Chuang LS, Ito K, Ito Y. RUNX family: Regulation and diversification of roles through interacting proteins. Int J Cancer. 2013;132:1260–1271. doi: 10.1002/ijc.27964. [DOI] [PubMed] [Google Scholar]
- 47.Zaidi SK, Pande S, Pratap J, Gaur T, Grigoriu S, Ali SA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proc Natl Acad Sci U S A. 2007;104:19861–19866. doi: 10.1073/pnas.0709650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akech J, Wixted JJ, Bedard K, van der Deen M, Hussain S, Guise TA, van Wijnen AJ, Stein JL, Languino LR, Altieri DC, Pratap J, Keller E, Stein GS, Lian JB. Runx2 association with progression of prostate cancer in patients: mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene. 2010;29:811–821. doi: 10.1038/onc.2009.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baniwal SK, Khalid O, Gabet Y, Shah RR, Purcell DJ, Mav D, Kohn-Gabet AE, Shi Y, Coetzee GA, Frenkel B. Runx2 transcriptome of prostate cancer cells: insights into invasiveness and bone metastasis. Mol Cancer. 2010;9:258. doi: 10.1186/1476-4598-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheng S, Qiao M, Pardee AB. Metastasis and AKT activation. J Cell Physiol. 2009;218:451–454. doi: 10.1002/jcp.21616. [DOI] [PubMed] [Google Scholar]
- 51.Lake SL, Jmor F, Dopierala J, Taktak AF, Coupland SE, Damato BE. Multiplex ligation-dependent probe amplification of conjunctival melanoma reveals common BRAF V600E gene mutation and gene copy number changes. Invest Ophthalmol Vis Sci. 2011;52:5598–5604. doi: 10.1167/iovs.10-6934. [DOI] [PubMed] [Google Scholar]
- 52.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 53.Boukerche H, Su ZZ, Emdad L, Baril P, Balme B, Thomas L, Randolph A, Valerie K, Sarkar D, Fisher PB. mda-9/Syntenin: a positive regulator of melanoma metastasis. Cancer Res. 2005;65:10901–10911. doi: 10.1158/0008-5472.CAN-05-1614. [DOI] [PubMed] [Google Scholar]
- 54.Das SK, Bhutia SK, Kegelman TP, Peachy L, Oyesanya RA, Dasgupta S, Sokhi UK, Azab B, Dash R, Quinn BA, Kim K, Barral PM, Su ZZ, Boukerche H, Sarkar D, Fisher PB. MDA-9/syntenin: a positive gatekeeper of melanoma metastasis. Front Biosci. 2012;17:1–15. doi: 10.2741/3911. [DOI] [PubMed] [Google Scholar]
- 55.Kaneda T, Sonoda Y, Ando K, Suzuki T, Sasaki Y, Oshio T, Tago M, Kasahara T. Mutation of Y925F in focal adhesion kinase (FAK) suppresses melanoma cell proliferation and metastasis. Cancer Lett. 2008;270:354–361. doi: 10.1016/j.canlet.2008.05.042. [DOI] [PubMed] [Google Scholar]