Key Points
Transcriptome analyses of human and murine reveal significant stage and species-specific differences across stages of terminal erythroid differentiation.
These transcriptomes provide a significant resource for understanding mechanisms of normal and perturbed erythropoiesis.
Abstract
We recently developed fluorescence-activated cell sorting (FACS)-based methods to purify morphologically and functionally discrete populations of cells, each representing specific stages of terminal erythroid differentiation. We used these techniques to obtain pure populations of both human and murine erythroblasts at distinct developmental stages. RNA was prepared from these cells and subjected to RNA sequencing analyses, creating unbiased, stage-specific transcriptomes. Tight clustering of transcriptomes from differing stages, even between biologically different replicates, validated the utility of the FACS-based assays. Bioinformatic analyses revealed that there were marked differences between differentiation stages, with both shared and dissimilar gene expression profiles defining each stage within transcriptional space. There were vast temporal changes in gene expression across the differentiation stages, with each stage exhibiting unique transcriptomes. Clustering and network analyses revealed that varying stage-specific patterns of expression observed across differentiation were enriched for genes of differing function. Numerous differences were present between human and murine transcriptomes, with significant variation in the global patterns of gene expression. These data provide a significant resource for studies of normal and perturbed erythropoiesis, allowing a deeper understanding of mechanisms of erythroid development in various inherited and acquired erythroid disorders.
Introduction
Mammalian erythropoiesis is an excellent example of the complex changes in temporal, developmental, and differentiation stage-specific gene expression exhibited by a single cell type.1,2 In the mammalian embryo and fetus, erythroid cells have differing developmental origins, with the primitive erythroid cell lineage developing from yolk sac–derived erythroid progenitors, and the definitive cell lineage maturing from 2 different developmentally regulated stem and progenitor cell populations.3-6 These cells have different programs of regulation, with variation in spatial, temporal, and site-specific differentiation.
In the adult, mature erythrocytes are the terminally differentiated final cellular product derived from hematopoietic stem and progenitor cells (HSPC). HSPCs undergo a series of lineage choice fate decisions, with increasingly restricted potential, ultimately committing to the erythroid lineage and beginning erythropoiesis. Traditionally, erythropoiesis has been divided into 3 stages: early erythropoiesis, terminal erythroid differentiation, and reticulocyte maturation.2 Early erythropoiesis involves commitment of multi-lineage progenitors into erythroid progenitor cells, with proliferation and differentiation into erythroid burst-forming unit cells, followed by erythroid colony-forming unit cells, then differentiation into proerythroblasts. Terminal erythroid differentiation begins with proerythroblasts differentiating into basophilic, then polychromatic, then orthochromatic erythroblasts that enucleate to become reticulocytes. Numerous changes occur during terminal erythroid differentiation. Erythroblasts decrease in size, synthesize increasing amounts of hemoglobin, undergo membrane reorganization and chromatin condensation, and then enucleate.7,8 In the final stage of erythropoiesis, reticulocytes mature into discoid erythrocytes, losing intracellular organelles, decreasing cell volume and surface area, and reorganizing the erythrocyte membrane.
Rapid advances in genomic technologies, particularly those coupled to high-throughput sequencing technologies, have revolutionized our understanding of gene expression, gene regulation, and mechanisms of human disease.9 RNA sequencing (RNA-seq) allows unbiased detection and quantification of transcriptomes using high-throughput sequencing.10,11 Beyond providing unbiased detection of transcripts, it provides information on transcript composition and abundance, including detection of novel transcripts, isoforms, alternative splice sites, allele-specific expression, and rare transcripts.11-13 RNA-seq has a low background signal and a large dynamic range, with high levels of reproducibility for both technical and biological replicates. The ability to determine detailed cellular transcriptomes has broad implications for interpreting the functional elements of the genome, revealing the molecular constituents of cells and tissues, and for understanding development and disease.
We have recently developed a fluorescence-activated cell sorting (FACS)-based method to obtain pure populations of human and murine erythroblasts at differing stages of terminal erythroid differentiation.14-16 RNA was prepared from these cells and subjected to RNA-seq analyses, creating unbiased differentiation stage–specific transcriptomes. Tight clustering of transcriptomes from differing stages validated the utility of the FACS-based isolation of erythroblasts at distinct stages of terminal differentiation. Marked differences were present between differentiation stages. Although there were many similarities, numerous differences were present between human and murine transcriptomes, with significant variation in the global patterns of gene expression. These data provide a significant resource for studies of normal and perturbed erythropoiesis, allowing a deeper understanding of mechanisms of erythroid development in various inherited and acquired erythroid disorders.
Materials and methods
Isolation of human and murine erythroblasts
CD34+ HSPCs were purified from cord blood by positive selection to a purity of 95% to 98% as described.15 A 3-phase CD34+ cell culture system was used to produce primary human erythroid cells at different stages of terminal differentiation. To obtain discrete populations of erythroid cells, a combination of cell surface markers for glycophorin A, band 3, and α4-integrin was used for FACS of cultured cells. This combination enables isolation of highly purified populations of erythroblasts at each distinct stage of terminal erythroid differentiation.15 Murine erythroblasts at distinct stages of terminal erythroid differentiation were sorted from bone marrow using TER119 antibody (anti-glycophorin A), CD44 antibody, and forward scatter (reflecting cell size) as markers.14,16
RNA isolation and preparation
RNA was prepared from primary human and murine erythroid cells for RNA-seq analyses as described.17 Quantitative real-time polymerase chain reaction (PCR) was performed to confirm expression levels of RNA transcripts (supplemental Table 1, available on the Blood Web site). Real-time PCR data were normalized as described.18
Library preparation and sequencing
RNA samples were treated with RNase-free DNase I (Takara, Otsu, Japan), quantified using a NanoDrop1000 (Thermo-Fisher, Waltham, MA), and assessed with an Agilent2100 Bioanalyzer (Agilent, Santa Clara, CA). The RNA integrity number of each sample was >9. DNA libraries were prepared according to the manufacturer’s instructions (Illumina, San Diego, CA). Each library was sequenced on the Illumina HiSeq2000 platform using a 50-bp single-end, nonstrand-specific sequencing strategy.
Data analyses
Detailed description of data analyses are provided in the supplemental Data.
Results
Cell isolation and RNA-seq
Highly purified human and murine erythroid cells corresponding to distinct stages of terminal erythroid differentiation were isolated using a recently developed FACS-based method as described. RNA was isolated from these cells and subjected to RNA-seq analyses to identify unbiased gene expression profiles across the entire transcriptome during terminal erythroid differentiation. Deep sequencing was performed on polyA+ messenger RNA (mRNA) from 3 biological replicates of each stage. Data were analyzed and subjected to quality control analyses. We analyzed transcriptomes using TopHat and Cufflinks packages generating high-confidence transcriptomes for each differentiation stage.13 Differences in levels of expression were analyzed by edgeR.19 Quantitative real-time PCR was performed to validate expression levels of representative mRNA transcripts detected by RNA-seq (supplemental Figure 1). Only annotated genes were analyzed.
Initially, we analyzed how many annotated genes were expressed at each differentiation stage. On average, we detected expression between 4804 human RefSeq genes in orthochromatic erythroblasts and 9606 RefSeq genes in proerythroblasts (supplemental Table 2) and between 6584 murine RefSeq genes in orthochromatic erythroblasts and 8838 RefSeq genes in proerythroblasts. Thus approximately 20% to 40% of known human genes and 28% to 38% of known murine genes were expressed during terminal erythroid differentiation.
Transcriptome profiles support FACS-based sorting methodology
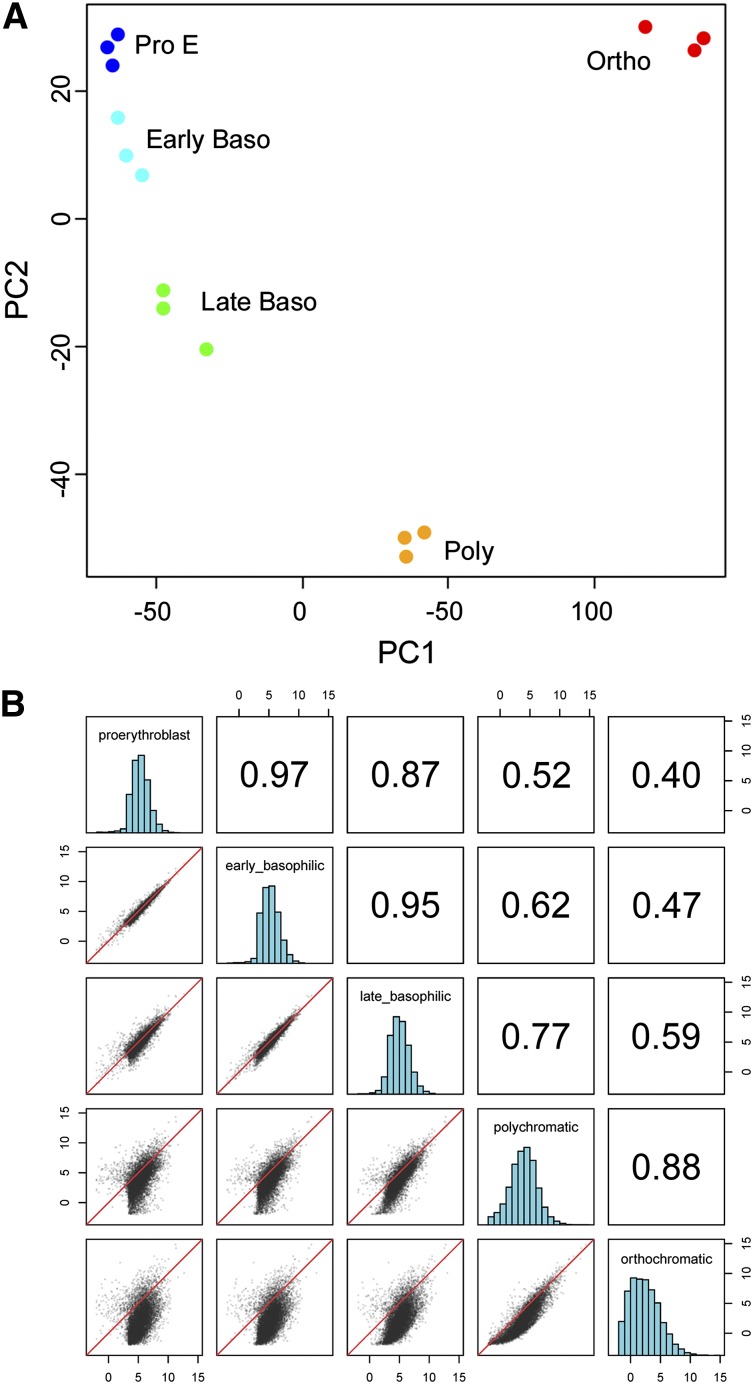
Principal component analysis) was performed on expressed genes in human erythroblasts (>10 count per million in 3 or more samples). Samples from individual stages of terminal erythroid differentiation clustered closely together (Figure 1A). Multidimensional scaling using edgeR, which places objects in dimensional space preserving the between-object distances, also revealed tight clustering of stage-specific biologic replicates (supplemental Figure 2). This tight clustering indicates that samples from each stage are very closely related. It also indicates that each stage of terminal erythroid differentiation is likely functionally distinct.
Figure 1.
Principal component analysis of expressed genes at differing stages of human terminal erythroid differentiation. (A) Samples representing 3 biologic replicates from individual stages of terminal erythroid differentiation clustered closely together. This tight clustering indicates that samples from each stage are very closely related and that the different stages of terminal erythroid differentiation are distinct. (B) Pairwise comparisons of replicates in scatterplot representation. Pearson correlations between pairwise comparisons using median expression of replicates from each differentiation stage. (Upper-right) The Pearson correlation values of genes with greater than 10 counts per million reads. (Lower left) Scatter plot data of log 2 counts per million reads for each gene of 1 sample compared with another. (Middle) Histograms of the log counts per million for each sample. The axis labels represent the log counts per million for the scatter and histogram plots. The y-axis for the histogram plots is scaled by the maximum y value. Baso, basophil; Ortho, orthochromatic; PC, principal component; poly, polychromatic; Pro E, proerythroblast.
Pairwise comparisons between adjacent stages of differentiation (Figure 1B; supplemental Figure 3) revealed very high Pearson correlation coefficients between replicates, range 0.94-0.99, indicating high reproducibility between stage-specific replicates (proerythroblast 0.95-0.97; early basophilic erythroblast 0.95-0.97; late basophilic erythroblast 0.95-0.97; polychromatic erythroblast 0.97-0.98; orthochromatic erythroblast 0.94-0.99). These comparisons also revealed dramatic changes in gene expression profiles across differentiation stages. The tight clustering of samples from individual stages of terminal erythroid differentiation indicate that transcriptome analyses support the FACS-based methodology used to define differentiation stages, even between biologically different replicates. It also indicates that differentiation stages are distinct, with both shared and dissimilar gene expression profiles defining each stage within transcriptional space. There are vast changes in gene expression across the differentiation stages, with each stage exhibiting unique transcriptomes.
Temporal patterns of gene expression during human terminal erythroid differentiation
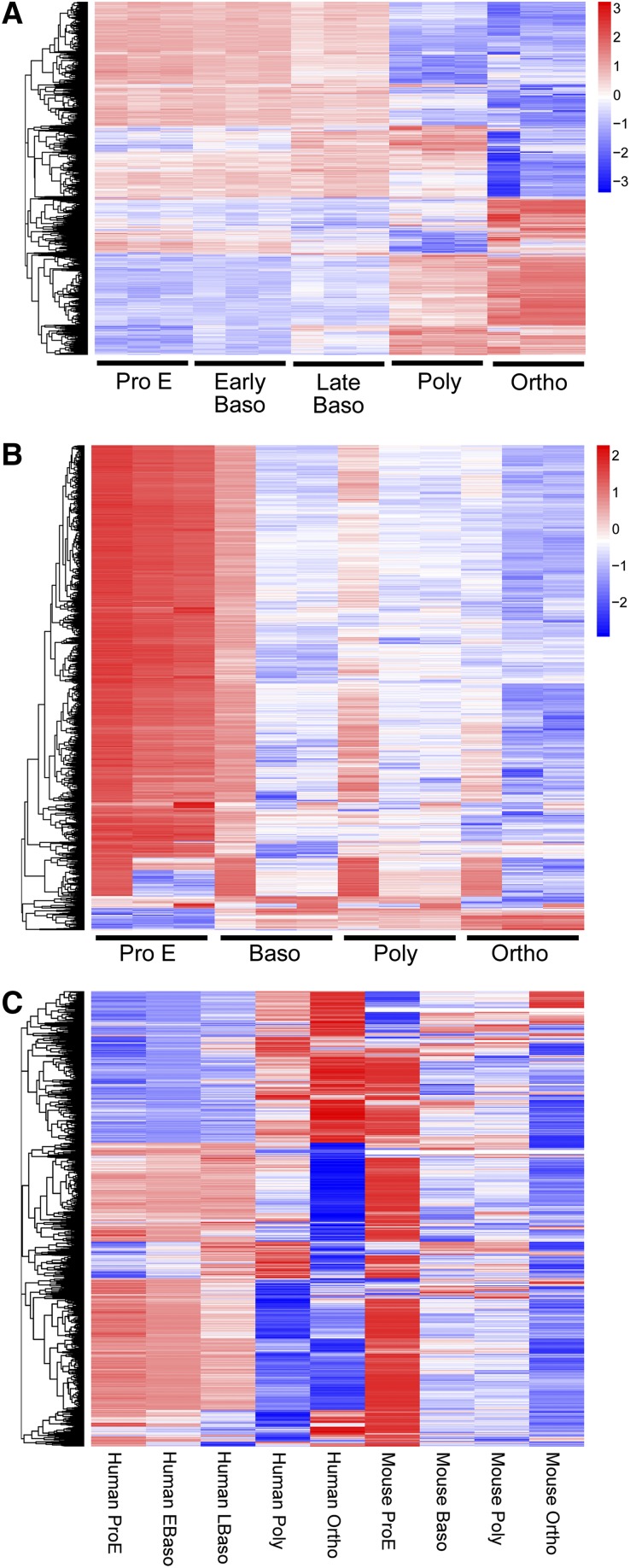
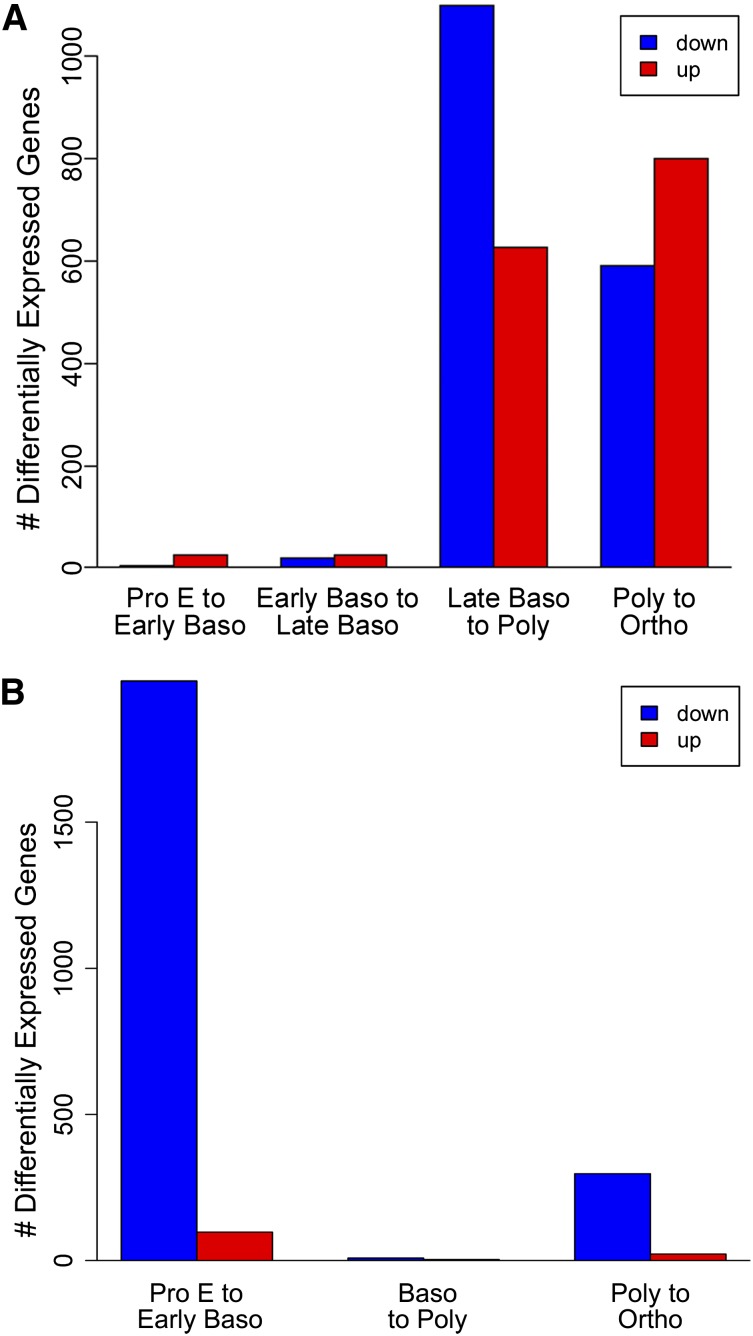
To further analyze transcriptome profiles across the stages of human terminal erythroid differentiation, we analyzed RNA-seq data using edgeR, a Bioconductor software package for examining differential expression of replicated count data.20-22 Pairwise comparisons of all adjacent stages of differentiation were performed. Large numbers of genes were differentially expressed at different stages of erythroid differentiation; of the 9931 genes expressed in 3 or more samples, more than a quarter (2702) were differentially expressed (false discovery rate [FDR] <0.01, fold change >4; supplemental Table 3). The greatest changes in differential gene expression were seen between the late basophilic to polychromatic and the polychromatic to orthochromatic stages (Figures 2A and 3A), demonstrating that the most dramatic changes occur at the late stages of terminal differentiation.
Figure 2.
Global differential gene expression between stages of human and murine terminal erythroid differentiation. Expression values of genes differentially expressed between differentiation stages are shown in heat map format. The red, white, and blue colors represent higher than average, close to average, and lower than average expression of a particular gene, respectively, as measured by row standardized Z-scores. The rows are organized by hierarchical clustering using agglomerative clustering with complete linkage and Euclidian distance metric. (A) Human. (B) Mouse. In human, there are varying patterns of coordinate gene expression. In contrast, in mouse, the majority of genes have a pattern of decreasing expression during terminal erythroid differentiation. The results from analyses in human (A) and mouse (B) were obtained in independent analyses. (C) Combined analyses. A combined data set of orthologous genes from each differentiation stage of terminal erythroid differentiation in human and mouse was created. Hierarchical clustering to identify groups of genes with similar and different patterns of gene expression between species was performed.
Figure 3.
Bar plot representation of differential gene expression at different stages of human terminal erythroid differentiation. (A) Human. The greatest changes in differential gene expression were seen between the late basophilic to polychromatic and the polychromatic to orthochromatic (Ortho) stages. (B) Mouse. The greatest changes in differential gene expression were seen between proerythroblast to basophilic and polychromatic to Ortho erythroblast stages, with genes being downregulated predominating between both stage transitions.
Venn diagram analyses revealed that differentially expressed genes found in 1 comparison were sometimes, but not usually, found in other comparisons (supplemental Figure 4), demonstrating there are stage-specific changes in differential gene expression.
Differentiation stages are transcriptionally enriched for genes of differing function
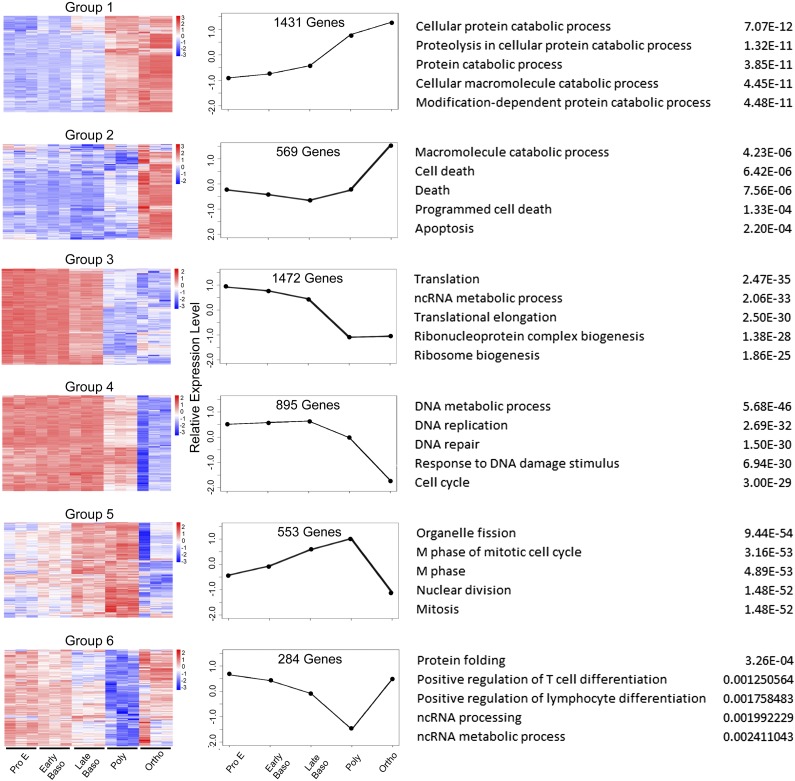
We analyzed the expression patterns of all differentially expressed, coregulated genes from the adjacent stage pairwise comparisons using k-means clustering. The differentially expressed genes clustered into 6 major groups, demonstrated in heat map and graphical format, based on patterns of expression at the differing time points (Figure 4). Two patterns revealed low- to mid-range levels of expression in proerythroblasts, increasing during differentiation (groups 1 and 2), whereas 2 patterns demonstrated high levels of expression in proerythroblasts, decreasing during differentiation (groups 3 and 4).The final 2 patterns were V-shaped mirror images, with either a peak or a nadir at the polychromatic erythroblast stage (groups 5 and 6).
Figure 4.
Clusters of gene expression across stages of human terminal erythroid differentiation. Differentially expressed, coregulated genes from adjacent stage pairwise comparisons were analyzed using k-means clustering. This identified 6 major groups, demonstrated in heat map and graphical format, based on patterns of expression at different stages. GO analysis of differentially expressed genes within clusters identified the top associated enriched GO terms with corresponding enrichment P values, shown on right.
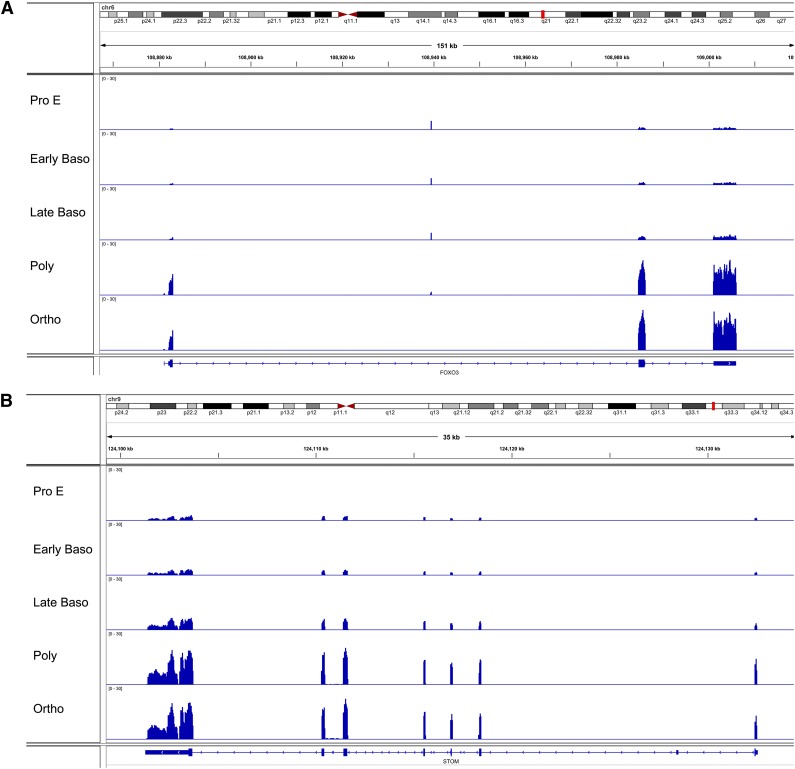
We performed gene ontology (GO) analysis of differentially expressed genes within these 6 clusters and identified associated enriched GO terms to gain insights into the biological processes regulated during terminal erythroid differentiation (Figure 4).23 In groups 1 and 2 with low- to mid-range levels of expression in proerythroblasts that increased during differentiation, GO terms significantly enriched (FDR <0.01) for differentially expressed genes related to cellular catabolism and cell death were identified. In groups 3 and 4 with high levels of expression in proerythroblasts that decreased during differentiation, GO terms significantly enriched for differentially expressed genes related to protein synthesis including translation and ribosome biogenesis, and DNA metabolism including replication, repair, and cell cycle were identified. Finally, for groups with V-shaped patterns, GO terms significantly enriched for differentially expressed genes were related to cell cycle and cell division (peak at polychromatic erythroblast stage) or protein folding and noncoding RNA metabolism (nadir at polychromatic erythroblast stage). Examples of genes with different patterns of expression across the stages of terminal erythroid differentiation—FOX03, STOM, PKLR, and STAT5A—are shown in Figure 5.
Figure 5.
Examples of patterns of gene expression in human terminal erythroid differentiation. Integrated genome viewer of RNA-seq tracks from representative genes. (A) FOX03. (B) STOM, stomatin. (C) PKLR, pyruvate kinase. (D) Signal transducer and activator of transcription 5A. (A-B) Genes from group 1, genes with increasing expression during differentiation. (C-D) Genes from group 3, genes with decreasing expression during differentiation. The tracks show read coverage values at each base normalized to the number of sample total reads/1 000 000. ncRNA, noncoding RNA.
Ingenuity Pathway Analysis of the differentially expressed genes within the 6 clusters was performed to gain additional insights into the biological processes regulated during terminal erythroid differentiation. The top functional networks, defined as scores >20, yielded results similar to the GO analyses (supplemental Table 4). For example, in groups 1 and 2, with low- to mid-range levels of expression in proerythroblasts increasing during differentiation, Ingenuity Pathway Analysis identified networks associated with RNA and DNA metabolism, cell cycle, and protein synthesis. Interestingly, for 2 of the top networks, the multifunctional serine/threonine protein kinase AKT was at the major organizing node (not shown). Ingenuity Pathway Analysis analyses of the term “molecular and cellular functions” yielded similar results (supplemental Table 5).
Marked variation in transcriptome composition during human terminal erythroid differentiation
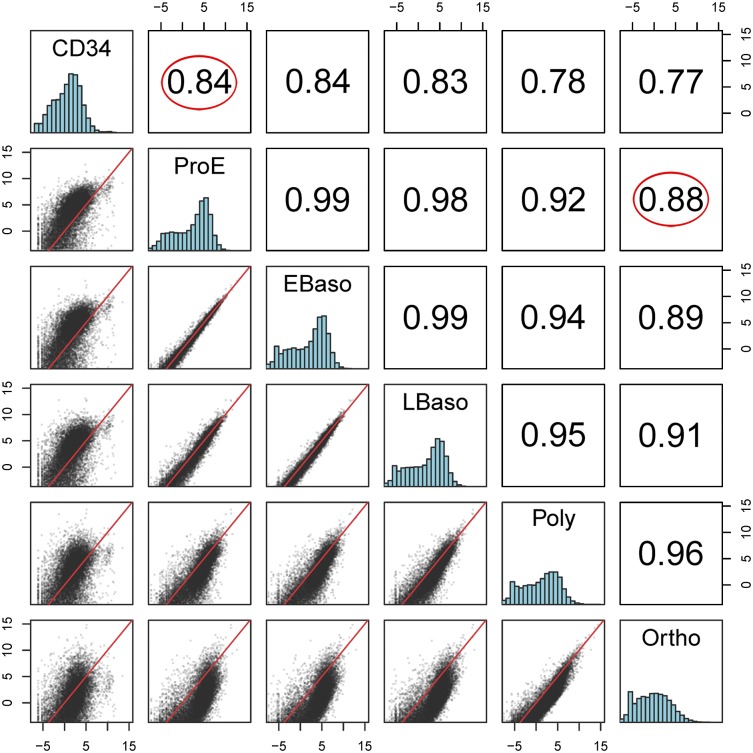
This analysis indicated that, at the transcriptome level, there are vast differences between stages of terminal erythroid differentiation. To better compare and contrast these differences, we compared cells from different stages of erythroid differentiation with HSPCs. HSPCs are at the apex of the hierarchy of hematopoietic cell development, commitment, and differentiation. They not only give rise to all hematopoietic cell types, they also have the ability to self-renew. We used Pearson correlation coefficient analysis and multidimensional scaling analysis with edgeR to compare HSPCs with proerythroblasts and orthochromatic erythroblasts. Multidimensional scaling allows visualization of the levels of similarity between datasets by placing each data point in dimensional space while preserving the space between object distances with each object assigned dimensional coordinates. As expected, there were significant differences in expression between HSPCs and proerythroblasts (Pearson correlation coefficient = 0.84; supplemental Figure 5). Remarkably, the degree of difference between HSPCs and proerythroblasts was nearly identical to the degree of difference between proerythroblasts and orthochromatic erythroblasts (Pearson correlation coefficient = 0.88; Figure 6).
Figure 6.
Pairwise comparisons of cell types and differentiation stages in scatterplot representation. Pearson correlations between pairwise comparisons using median expression of replicates from all genes for each cell type or differentiation stage (upper right) using median expression of replicates. (Lower left) The scatterplot data of log 2 counts per million. (Middle) Histograms of the log counts per million for each sample. The axes label represent the log counts per million for the scatter and histogram plots. The y-axis for the histogram plots is scaled by the maximum y value. Circles indicate the level of similarity/dissimilarity between HSPCs and proerythroblasts is nearly identical to the level of similarity/dissimilarity between proerythroblasts and orthochromatic (Ortho) erythroblasts.
Highly expressed transcripts encode hemoglobin-related proteins
We assembled lists of the highest expressed genes (by reads per kilobase of transcript per million) at each stage of erythroid differentiation (Table 1). These genes were primarily associated with hemoglobin synthesis, structure, and function. These included the globin genes, δ-aminolevulinate synthase 2 (ALAS2), α hemoglobin stabilizing protein (AHSP) (the hemoglobin chaperone), and proteins involved in iron metabolism including the transferrin receptor and light and heavy ferritin chains. In addition, many ribosomal genes were expressed at very high levels, particularly in proerythroblasts and early basophilic erythroblasts.
Table 1.
Top 25 expressed genes at stages of human terminal erythroid differentiation
| Rank | Proerythroblast | Early basophilic erythroblast | Late basophilic erythroblast | Polychromatic erythroblast | Orthochromatic erythroblast |
|---|---|---|---|---|---|
| 1 | HBG2 | HBB | HBB | HBB | HBB |
| 2 | HBB | HBG2 | HBG2 | HBG2 | HBG2 |
| 3 | RPS6 | TFRC | HBA1 | HBA2 | HBA2 |
| 4 | RPLP1 | RPS6 | HBA2 | HBA1 | HBA1 |
| 5 | TFRC | AHSP | HBG1 | HBG1 | HBG1 |
| 6 | TPT1 | HBA1 | TFRC | SLC4A1 | FTL |
| 7 | RPL8 | HBG1 | AHSP | FTL | ALAS2 |
| 8 | GNB2L1 | PRDX2 | SLC4A1 | ALAS2 | UBB |
| 9 | AHSP | TPT1 | PRDX2 | AHSP | SLC4A1 |
| 10 | EEF2 | RPLP1 | RPS6 | FTH1 | FTH1 |
| 11 | RPS4X | RPL8 | TPT1 | TFRC | BSG |
| 12 | RPL11 | FTL | BLVRB | HBM | SLC25A37 |
| 13 | RPL37A | HBA2 | FTL | BLVRB | TMCC2 |
| 14 | FTL | BLVRB | H1F0 | SLC25A37 | SLC25A39 |
| 15 | HBG1 | GNB2L1 | ALAS2 | TPT1 | SLC2A1 |
| 16 | PRDX2 | RPL11 | RPLP1 | UBB | RBM38 |
| 17 | RPL3 | RPL37A | RPL8 | PRDX2 | AHSP |
| 18 | RPL30 | RPS4X | HMGB2 | SLC25A39 | NCOA4 |
| 19 | RPS8 | EEF2 | RPL11 | UCP2 | BLVRB |
| 20 | BLVRB | S100A6 | RPL37A | SLC2A1 | OAZ1 |
| 21 | ACTB | RPS8 | GNB2L1 | BSG | BCL2L1 |
| 22 | RPS5 | ACTB | RPS4X | HMBS | FKBP8 |
| 23 | RPS11 | RPL30 | EEF2 | H1F0 | PIM1 |
| 24 | RPS18 | RPS12 | HBD | HEMGN | DCAF12 |
| 25 | RPL18 | RPL3 | ACTB | HBD | BPGM |
Transcription factor gene expression throughout human erythroid differentiation
Transcription factors are proteins that detect and bind to DNA regulatory sequences and participate in the assembly of multiprotein complexes that regulate gene expression. Recent studies have begun to dissect the transcriptional networks that regulate hematopoietic cell fate.24-26 We analyzed transcription factor gene expression across human terminal erythroid differentiation. Similar to global patterns of gene expression, transcription factors demonstrated varying patterns of differential expression throughout differentiation (supplemental Figure 6A; supplemental Table 6). Analysis of transcription factors by absolute expression revealed several critical erythroid transcription factors. The top 5 transcription factors were KLF1, NFE2, GFI1B, YBX1, and GATA1 (supplemental Figure 6B).
Transcriptomes in murine terminal erythroid differentiation
We used similar FACS-based strategies to obtain purified populations of murine proerythroblasts, basophilic, polychromatic, and orthochromatic erythroblasts and subjected them to RNA-seq. Analysis of murine transcriptome profiles across the stages of terminal erythroid differentiation using edgeR revealed that large numbers of genes were differentially expressed at different stages of erythroid differentiation; in 9096 of 23 283 genes expressed in 3 or more samples, 2288 were differentially expressed (FDR <0.01, fold change >4).
Overall, there was a gradual decrease in numbers of expressed genes from proerythroblasts to basophilic to polychromatic and on to orthochromatic erythroblast stages (Figures 2B and 3B).
Near global decrease in gene expression during murine terminal erythroid differentiation
We analyzed the expression patterns of all differentially expressed, coregulated genes from the adjacent stage pairwise comparisons using k-means clustering. The differentially expressed genes clustered into 3 major groups, demonstrated in heat map and graphical format, based on patterns of expression at the differing time points (supplemental Figure 7). There was 1 predominant pattern of gene expression with high levels of gene expression in proerythroblasts that decreased steadily during differentiation (group 1; supplemental Figure 7). Group 2 had an expression pattern similar to group 1. We performed GO analysis of differentially expressed genes within these 3 clusters and identified associated enriched GO terms to gain insight into the biological processes regulated during murine terminal erythroid differentiation (supplemental Figure 7).23 In groups 1 and 2, both with high levels of expression in proerythroblasts decreasing during differentiation, GO terms were significantly enriched (FDR = 0.01) for differentially expressed genes related to protein synthesis including translation and ribosome biogenesis, and DNA metabolism including replication, repair, and cell cycle. This is similar to the terms enriched in the clusters of human genes with decreasing expression during terminal erythroid differentiation.
We assembled lists of the highest expressed genes (by reads per kilobase of transcript per million) at each stage of erythroid differentiation (supplemental Table 7). These genes were primarily associated with hemoglobin synthesis and proteins involved in iron metabolism.
Comparison of human and murine transcriptomes
Additional analyses of the differences between human and murine terminal erythroid differentiation were performed to begin to understand the extent of similarities and differences as well as their potential functional consequences. Because initial human and mouse transcriptome analyses were done independently, an integrated dataset of expression values for orthologous genes between human and mouse for identical stages of terminal differentiation was created. Principal component analyses revealed general similarity in the global trend in patterns of gene expression between species during erythroid differentiation (supplemental Figure 8).
Hierarchical clustering of the combined species data set of orthologous genes from each stage revealed significant differences in gene expression between human and mouse (Figure 2C). These data demonstrating human/murine transcriptome differences were further analyzed to search for potential functional differences between species. Eleven clusters of orthologous genes (>100 genes/cluster) with different patterns of expression in human compared with mouse during terminal erythroid differentiation were identified and Database for Annotation, Visualization and Integrated Discovery (DAVID) analyses performed (supplemental Table 8). The goal of DAVID analyses is to identify enriched biological themes and discover functional-related gene groups. DAVID analyses identified numerous categories of varying cellular processes (supplemental Table 8). Although these analyses cannot precisely address how differing transcriptomes contribute to species-specific differences between human and mouse, the data indicate that numerous, complex, and differing patterns of gene expression likely regulate the complicated process of terminal erythroid differentiation in both species.
In addition to the global differences in patterns of gene expression during terminal erythroid differentiation between human and mouse noted previously, numerous other dissimilarities were noted. Whereas there were several patterns of transcription factor gene expression in human, patterns of transcription factor gene expression in mouse differed significantly, more similarly paralleling the gradual decrease in gene expression during murine terminal erythroid differentiation (supplemental Figure 9). Interestingly, growth differentiation factor 15 (GDF15), one of the top 25 expressed genes in human erythroid cells (Table 1), was not expressed in murine erythroid cells.
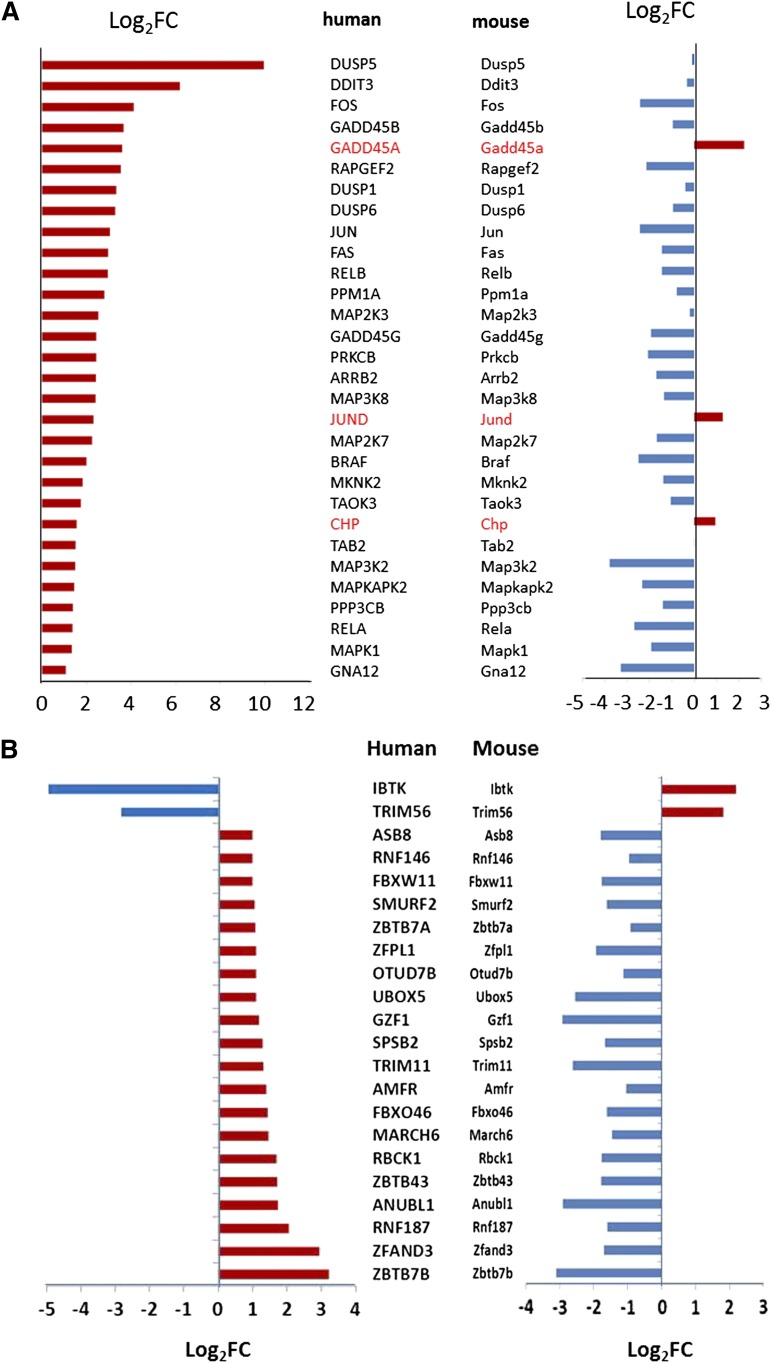
Another example of significant difference between human and murine terminal erythroid differentiation is in genes encoding proteins of the mitogen-activated protein kinase (MAPK) pathway. Of genes in this pathway, 113 of 268 genes were expressed in human and 95 of 269 genes were expressed in mouse. About half of the genes in human were downregulated during terminal erythroid differentiation (56 of 113), whereas three-quarters of genes in mouse were downregulated (72 of 95). In human, 30 of 113 expressed genes were upregulated during terminal erythroid differentiation, whereas only 3 genes were upregulated in mouse. Differentially expressed genes encoded proteins located at positions throughout the MAPK pathway (Figure 7A). Again, this pattern mirrors the global decrease in gene expression observed in murine terminal erythroid differentiation.
Figure 7.
Differences in human and murine patterns of gene expression. (A) Differentially expressed genes encoding proteins of the MAPK pathway. (B) Differentially expressed genes encoding proteins associated with E3 ubiquitin ligase and related proteins. Log2FC, Log 2 fold change.
In the ubiquitin-mediated proteolysis pathway, genes associated with E1 ubiquitin–activating enzyme and E2 ubiquitin–conjugating enzyme were, for the most part, similarly expressed between human and mouse (not shown). However, many genes associated with E3 ubiquitin ligase and related proteins were upregulated in human but downregulated in mouse (Figure 7B). This is another example of downregulation of a large segment of a biologically relevant pathway of genes in mouse.
Discussion
The stage-specific complexity of terminal erythroid differentiation has been studied for many years. Variation in many phenotypic features, such as metabolic properties, decrease in cell size, increases in hemoglobinization, alterations in membrane characteristics, epigenetic and nuclear changes with chromatin condensation, and, ultimately, enucleation, have led to the conclusion that terminal erythroid differentiation is a unique process, with each cell division simultaneously coupled with differentiation.2,7,8,14 In contrast to most cell types, in which each cell division generates 2 daughter cells that are nearly identical to the mother cell, during terminal erythroid differentiation, the 2 daughter cells are structurally and functionally different than the mother cell from which they are derived. This conclusion has been supported on a limited basis by gene expression analyses of varying populations of murine and human erythroid cells.8,27-35 Our transcriptome data strengthen these observations regarding the stage-specific complexity of erythroid differentiation.
The transcriptome data presented here also strongly support the recently described FACS-based strategy for identification of terminal differentiation stage-specific erythroid cells. Experiments were done in triplicate with different biologic replicates for each differentiation-specific stage to minimize individual-specific changes in expression of the ∼22 500 genes analyzed. This is a powerful technique that allows purification of pure populations of erythroid cells at specific stages of terminal erythroid differentiation for detailed studies of erythroid differentiation from primary human HSPCs.
Previous global gene expression studies of human and murine erythroid development and differentiation have used varying sources of erythroid cells representing primitive or definitive erythropoiesis.24,28,34,36-41 RNA was extracted from primary erythroid cells isolated without culture, or, after in vitro culture of nucleated cells from differing sources, eg, CD34+ HSPC or peripheral blood buffy coat. In most studies, cells were not purified by cell surface or other markers, but were selected at differing time points in vivo or in culture. In 1 study of global erythroid gene expression study, peripheral blood buffy coats were cultured in vitro then sorted by CD36, CD71, and CD235a expression and cell size to yield populations corresponding to erythroid colony-forming unit, proerythroblasts and intermediate and late erythroblasts. Transcriptomes constructed using stage-specific sorting strategies of primary, noncultured cells would be expected to yield the highest quality of transcriptome data. Because of limitations of human material, we used a 3-phase culture system starting with umbilical cord CD34+ cells to obtain erythroid cells. Depending on the source and age of the source of cells for analysis or culture, patterns of gene expression will represent fetal, neonatal, or adult programs of gene expression.40
These data sets can be used in a number of ways to interpret the transcriptional architecture of erythropoiesis. One is a better understanding of the basic biology of terminal erythroid differentiation at steady state, during stress erythropoiesis, and after various perturbations. For instance, these data can be interrogated to generate transcriptional circuits, as recently shown in hematopoiesis.24,25,42,43 In contrast to array-based studies, these RNA-seq–based studies allow creation and analysis of patterns of differentiation-stage, gene-specific isoform composition generated by RNA alternate splicing, cleavage, and polyadenylation.44-47 They also allow identification of long noncoding RNAs for use in functional analyses.48-50
Another important use for these data sets is to develop a better mechanistic understanding of disordered erythropoiesis, with the ability to better understand stage-specific defects. This includes disorders such as the thalassemia syndromes, bone marrow failure syndromes and aplastic anemia51-56 as well as acquired disorders such as the myelodysplasia syndromes,57-59 particularly subtypes with disordered terminal erythroid differentiation. Numerous abnormalities have been identified in these disorders, including perturbed apoptosis, cytokine signaling, and regulation of cellular growth.57,60-63 Comparative analyses between wild-type and variant cells may provide insight into disease pathobiology, allowing understanding of mechanisms of abnormal erythropoiesis over time in specific diseases, and may provide insights into identification of potential therapeutic targets.
These data also provide transcriptional context when interpreting data sets of genetic variants identified by genomewide sequencing in patients with hematologic disease from unknown causes (eg, when interpreting novel variants and analyzing gene expression in the process of variant analysis and interpretation).9
There is increasing understanding that there are fundamental differences between human and murine erythropoiesis. For instance, variation in glucose utilization, vitamin C metabolism, regulation of ion content and cell size, membrane protein composition and properties, and mechanisms of stress erythropoiesis are known differences between human and murine erythroid cells.30,64-70 Other differences include patterns of gene regulation (eg, the well-known dissimilarities in globin gene regulation) and differences in transcript isoform composition generated by alternate splicing, such as the differences in exon composition of the ALAS2 complementary DNA isoforms between mouse and man.71,72 One striking observation from our analyses was, in contrast to human, there was a near-global decrease in gene expression during murine terminal erythroid differentiation. This strongly suggests there are fundamental species-specific differences between human and murine erythropoiesis. The ability to compare and contrast gene expression profiles in human and murine terminal erythroid differentiation will provide novel insights into these differences between human and murine erythropoiesis while expanding our understanding of both.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (DK32094, DK26263, HL65448, and DK62039) and an American Society of Hematology Bridge Grant (X.A.).
Footnotes
There is an Inside Blood Commentary on this article in this issue.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession numbers GSE38110, GSE42551, GSE38169, and GSE42462).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: X.A. designed experiments, analyzed data, and wrote the manuscript; V.P.S. analyzed data and wrote the manuscript; J.L. performed experiments and analyzed data; J.L., J.P., and F.X. performed experiments; K.W. analyzed data; M.N. designed experiments and wrote the manuscript; and P.G.G. analyzed data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Patrick G. Gallagher, Departments of Pediatrics, Pathology, and Genetics, Yale University School of Medicine, 333 Cedar St, PO Box 208064, New Haven, CT 06520-8064; e-mail: patrick.gallagher@yale.edu.
References
- 1.Tsiftsoglou AS, Vizirianakis IS, Strouboulis J. Erythropoiesis: model systems, molecular regulators, and developmental programs. IUBMB Life. 2009;61(8):800–830. doi: 10.1002/iub.226. [DOI] [PubMed] [Google Scholar]
- 2.Dzierzak E, Philipsen S. Erythropoiesis: development and differentiation. Cold Spring Harb Perspect Med. 2013;3(4):a011601. doi: 10.1101/cshperspect.a011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.England SJ, McGrath KE, Frame JM, Palis J. Immature erythroblasts with extensive ex vivo self-renewal capacity emerge from the early mammalian fetus. Blood. 2011;117(9):2708–2717. doi: 10.1182/blood-2010-07-299743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGrath KE, Frame JM, Fromm GJ, et al. A transient definitive erythroid lineage with unique regulation of the β-globin locus in the mammalian embryo. Blood. 2011;117(17):4600–4608. doi: 10.1182/blood-2010-12-325357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palis J. Ontogeny of erythropoiesis. Curr Opin Hematol. 2008;15(3):155–161. doi: 10.1097/MOH.0b013e3282f97ae1. [DOI] [PubMed] [Google Scholar]
- 6.Palis J, Malik J, McGrath KE, Kingsley PD. Primitive erythropoiesis in the mammalian embryo. Int J Dev Biol. 2010;54(6-7):1011–1018. doi: 10.1387/ijdb.093056jp. [DOI] [PubMed] [Google Scholar]
- 7.Hattangadi SM, Wong P, Zhang L, Flygare J, Lodish HF. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood. 2011;118(24):6258–6268. doi: 10.1182/blood-2011-07-356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong P, Hattangadi SM, Cheng AW, Frampton GM, Young RA, Lodish HF. Gene induction and repression during terminal erythropoiesis are mediated by distinct epigenetic changes. Blood. 2011;118(16):e128–e138. doi: 10.1182/blood-2011-03-341404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sankaran VG, Gallagher PG. Applications of high-throughput DNA sequencing to benign hematology. Blood. 2013;122(22):3575–3582. doi: 10.1182/blood-2013-07-460337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mutz KO, Heilkenbrinker A, Lönne M, Walter JG, Stahl F. Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol. 2013;24(1):22–30. doi: 10.1016/j.copbio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 11.McGettigan PA. Transcriptomics in the RNA-seq era. Curr Opin Chem Biol. 2013;17(1):4–11. doi: 10.1016/j.cbpa.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011;8(6):469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 13.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K, Liu J, Heck S, Chasis JA, An X, Mohandas N. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci U S A. 2009;106(41):17413–17418. doi: 10.1073/pnas.0909296106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J, Liu J, Xue F, et al. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood. 2013;121(16):3246–3253. doi: 10.1182/blood-2013-01-476390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Zhang J, Ginzburg Y, et al. Quantitative analysis of murine terminal erythroid differentiation in vivo: novel method to study normal and disordered erythropoiesis. Blood. 2013;121(8):e43–e49. doi: 10.1182/blood-2012-09-456079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steiner LA, Maksimova Y, Schulz V, et al. Chromatin architecture and transcription factor binding regulate expression of erythrocyte membrane protein genes. Mol Cell Biol. 2009;29(20):5399–5412. doi: 10.1128/MCB.00777-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reimers M, Carey VJ. Bioconductor: an open source framework for bioinformatics and computational biology. Methods Enzymol. 2006;411:119–134. doi: 10.1016/S0076-6879(06)11008-3. [DOI] [PubMed] [Google Scholar]
- 21.Carey VJ, Davis AR, Lawrence MF, Gentleman R, Raby BA. Data structures and algorithms for analysis of genetics of gene expression with Bioconductor: GGtools 3.x. Bioinformatics. 2009;25(11):1447–1448. doi: 10.1093/bioinformatics/btp169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, et al. The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller MA, Addya S, Vadigepalli R, et al. Transcriptional regulatory network analysis of developing human erythroid progenitors reveals patterns of coregulation and potential transcriptional regulators. Physiol Genomics. 2006;28(1):114–128. doi: 10.1152/physiolgenomics.00055.2006. [DOI] [PubMed] [Google Scholar]
- 25.Novershtern N, Subramanian A, Lawton LN, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck D, Thoms JA, Perera D, et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122(14):e12–e22. doi: 10.1182/blood-2013-03-490425. [DOI] [PubMed] [Google Scholar]
- 27.Komor M, Güller S, Baldus CD, et al. Transcriptional profiling of human hematopoiesis during in vitro lineage-specific differentiation. Stem Cells. 2005;23(8):1154–1169. doi: 10.1634/stemcells.2004-0171. [DOI] [PubMed] [Google Scholar]
- 28.Merryweather-Clarke AT, Atzberger A, Soneji S, et al. Global gene expression analysis of human erythroid progenitors. Blood. 2011;117(13):e96–e108. doi: 10.1182/blood-2010-07-290825. [DOI] [PubMed] [Google Scholar]
- 29.Koury S, Yarlagadda S, Moskalik-Liermo K, et al. Differential gene expression during terminal erythroid differentiation. Genomics. 2007;90(5):574–582. doi: 10.1016/j.ygeno.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardison RC. Evolution of hemoglobin and its genes. Cold Spring Harb Perspect Med. 2012;2(12):a011627. doi: 10.1101/cshperspect.a011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scicchitano MS, McFarland DC, Tierney LA, Narayanan PK, Schwartz LW. In vitro expansion of human cord blood CD36+ erythroid progenitors: temporal changes in gene and protein expression. Exp Hematol. 2003;31(9):760–769. doi: 10.1016/s0301-472x(03)00185-1. [DOI] [PubMed] [Google Scholar]
- 32.Bonafoux B, Lejeune M, Piquemal D, et al. Analysis of remnant reticulocyte mRNA reveals new genes and antisense transcripts expressed in the human erythroid lineage. Haematologica. 2004;89(12):1434–1438. [PubMed] [Google Scholar]
- 33.Fujishima N, Hirokawa M, Aiba N, et al. Gene expression profiling of human erythroid progenitors by micro-serial analysis of gene expression. Int J Hematol. 2004;80(3):239–245. doi: 10.1532/ijh97.04053. [DOI] [PubMed] [Google Scholar]
- 34.Gubin AN, Njoroge JM, Bouffard GG, Miller JL. Gene expression in proliferating human erythroid cells. Genomics. 1999;59(2):168–177. doi: 10.1006/geno.1999.5855. [DOI] [PubMed] [Google Scholar]
- 35.Zhang D, Johnson MM, Miller CP, Pircher TJ, Geiger JN, Wojchowski DM. An optimized system for studies of EPO-dependent murine pro-erythroblast development. Exp Hematol. 2001;29(11):1278–1288. doi: 10.1016/s0301-472x(01)00725-1. [DOI] [PubMed] [Google Scholar]
- 36.Singleton BK, Burton NM, Green C, Brady RL, Anstee DJ. Mutations in EKLF/KLF1 form the molecular basis of the rare blood group In(Lu) phenotype. Blood. 2008;112(5):2081–2088. doi: 10.1182/blood-2008-03-145672. [DOI] [PubMed] [Google Scholar]
- 37.Pilon AM, Arcasoy MO, Dressman HK, et al. Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol Cell Biol. 2008;28(24):7394–7401. doi: 10.1128/MCB.01087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kingsley PD, Greenfest-Allen E, Frame JM, et al. Ontogeny of erythroid gene expression. Blood. 2013;121(6):e5–e13. doi: 10.1182/blood-2012-04-422394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isern J, He Z, Fraser ST, et al. Single-lineage transcriptome analysis reveals key regulatory pathways in primitive erythroid progenitors in the mouse embryo. Blood. 2011;117(18):4924–4934. doi: 10.1182/blood-2010-10-313676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sripichai O, Kiefer CM, Bhanu NV, et al. Cytokine-mediated increases in fetal hemoglobin are associated with globin gene histone modification and transcription factor reprogramming. Blood. 2009;114(11):2299–2306. doi: 10.1182/blood-2009-05-219386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peller S, Tabach Y, Rotschild M, et al. Identification of gene networks associated with erythroid differentiation. Blood Cells Mol Dis. 2009;43(1):74–80. doi: 10.1016/j.bcmd.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 42.Swiers G, Patient R, Loose M. Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Dev Biol. 2006;294(2):525–540. doi: 10.1016/j.ydbio.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 43.Loose M, Patient R. Global genetic regulatory networks controlling hematopoietic cell fates. Curr Opin Hematol. 2006;13(4):229–236. doi: 10.1097/01.moh.0000231419.15654.7f. [DOI] [PubMed] [Google Scholar]
- 44.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12(10):715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14(7):496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 46.Hou VC, Conboy JG. Regulation of alternative pre-mRNA splicing during erythroid differentiation. Curr Opin Hematol. 2001;8(2):74–79. doi: 10.1097/00062752-200103000-00003. [DOI] [PubMed] [Google Scholar]
- 47.Pimentel H, Parra M, Gee S, et al. A dynamic alternative splicing program regulates gene expression during terminal erythropoiesis [published online ahead of print January 17, 2014]. Nucleic Acids Res. 2014 doi: 10.1093/nar/gkt1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paralkar VR, Weiss MJ. Long noncoding RNAs in biology and hematopoiesis. Blood. 2013;121(24):4842–4846. doi: 10.1182/blood-2013-03-456111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aprea J, Prenninger S, Dori M, et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013;32(24):3145–3160. doi: 10.1038/emboj.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paralkar VR, Mishra T, Luan J, et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development [published online ahead of print January 17, 2014]. Blood. 2014 doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Libani IV, Guy EC, Melchiori L, et al. Decreased differentiation of erythroid cells exacerbates ineffective erythropoiesis in beta-thalassemia. Blood. 2008;112(3):875–885. doi: 10.1182/blood-2007-12-126938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rivella S. Ineffective erythropoiesis and thalassemias. Curr Opin Hematol. 2009;16(3):187–194. doi: 10.1097/MOH.0b013e32832990a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivella S. The role of ineffective erythropoiesis in non-transfusion-dependent thalassemia. Blood Rev. 2012;26(Suppl 1):S12–S15. doi: 10.1016/S0268-960X(12)70005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parikh S, Bessler M. Recent insights into inherited bone marrow failure syndromes. Curr Opin Pediatr. 2012;24(1):23–32. doi: 10.1097/MOP.0b013e32834eca77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chirnomas SD, Kupfer GM. The inherited bone marrow failure syndromes. Pediatr Clin North Am. 2013;60(6):1291–1310. doi: 10.1016/j.pcl.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamazaki H, Nakao S. Border between aplastic anemia and myelodysplastic syndrome. Int J Hematol. 2013;97(5):558–563. doi: 10.1007/s12185-013-1324-x. [DOI] [PubMed] [Google Scholar]
- 57.Houwerzijl EJ, Pol HW, Blom NR, van der Want JJ, de Wolf JT, Vellenga E. Erythroid precursors from patients with low-risk myelodysplasia demonstrate ultrastructural features of enhanced autophagy of mitochondria. Leukemia. 2009;23(5):886–891. doi: 10.1038/leu.2008.389. [DOI] [PubMed] [Google Scholar]
- 58.Stoll LM, Duffield AS, Johnson MW, Ali SZ. Acute myeloid leukemia with myelodysplasia-related changes with erythroid differentiation involving pleural fluid: a case report and brief cytopathologic review. Diagn Cytopathol. 2011;39(6):451–454. doi: 10.1002/dc.21470. [DOI] [PubMed] [Google Scholar]
- 59.Wang SA, Hasserjian RP. Erythroid proliferations in myeloid neoplasms. Hum Pathol. 2012;43(2):153–164. doi: 10.1016/j.humpath.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 60.van de Loosdrecht AA, Brada SJ, Blom NR, et al. Mitochondrial disruption and limited apoptosis of erythroblasts are associated with high risk myelodysplasia. An ultrastructural analysis. Leuk Res. 2001;25(5):385–393. doi: 10.1016/s0145-2126(00)00151-x. [DOI] [PubMed] [Google Scholar]
- 61.van de Loosdrecht AA, Vellenga E. Myelodysplasia and apoptosis: new insights into ineffective erythropoiesis. Med Oncol. 2000;17(1):16–21. doi: 10.1007/BF02826211. [DOI] [PubMed] [Google Scholar]
- 62.Fontenay-Roupie M, Bouscary D, Guesnu M, et al. Ineffective erythropoiesis in myelodysplastic syndromes: correlation with Fas expression but not with lack of erythropoietin receptor signal transduction. Br J Haematol. 1999;106(2):464–473. doi: 10.1046/j.1365-2141.1999.01539.x. [DOI] [PubMed] [Google Scholar]
- 63.Wang J, Fernald AA, Anastasi J, Le Beau MM, Qian Z. Haploinsufficiency of Apc leads to ineffective hematopoiesis. Blood. 2010;115(17):3481–3488. doi: 10.1182/blood-2009-11-251835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramanian S, Tsai R, Discher DE. The ‘metabolon,’ CD47, and the ‘phagocytic synapse’: molecular co-localization and species divergence. Transfus Clin Biol. 2006;13(1-2):31–38. doi: 10.1016/j.tracli.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 65.Huang ZY, Barreda DR, Worth RG, et al. Differential kinase requirements in human and mouse Fc-gamma receptor phagocytosis and endocytosis. J Leukoc Biol. 2006;80(6):1553–1562. doi: 10.1189/jlb.0106019. [DOI] [PubMed] [Google Scholar]
- 66.Wandersee NJ, Olson SC, Holzhauer SL, Hoffmann RG, Barker JE, Hillery CA. Increased erythrocyte adhesion in mice and humans with hereditary spherocytosis and hereditary elliptocytosis. Blood. 2004;103(2):710–716. doi: 10.1182/blood-2003-02-0492. [DOI] [PubMed] [Google Scholar]
- 67.Varki A, Kornfeld S. An autosomal dominant gene regulates the extent of 9-O-acetylation of murine erythrocyte sialic acids. A probable explanation for the variation in capacity to activate the human alternate complement pathway. J Exp Med. 1980;152(3):532–544. doi: 10.1084/jem.152.3.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen D, Kaul DK. Rheologic and hemodynamic characteristics of red cells of mouse, rat and human. Biorheology. 1994;31(1):103–113. doi: 10.3233/bir-1994-31109. [DOI] [PubMed] [Google Scholar]
- 69.Delpire E, Mount DB. Human and murine phenotypes associated with defects in cation-chloride cotransport. Annu Rev Physiol. 2002;64:803–843. doi: 10.1146/annurev.physiol.64.081501.155847. [DOI] [PubMed] [Google Scholar]
- 70.Montel-Hagen A, Kinet S, Manel N, et al. Erythrocyte Glut1 triggers dehydroascorbic acid uptake in mammals unable to synthesize vitamin C. Cell. 2008;132(6):1039–1048. doi: 10.1016/j.cell.2008.01.042. [DOI] [PubMed] [Google Scholar]
- 71.Conboy JG, Cox TC, Bottomley SS, Bawden MJ, May BK. Human erythroid 5-aminolevulinate synthase. Gene structure and species-specific differences in alternative RNA splicing. J Biol Chem. 1992;267(26):18753–18758. [PubMed] [Google Scholar]
- 72.Peterson KR. Hemoglobin switching: new insights. Curr Opin Hematol. 2003;10(2):123–129. doi: 10.1097/00062752-200303000-00004. [DOI] [PubMed] [Google Scholar]